Mapping of a Major QTL, qBK1Z, for Bakanae Disease Resistance in Rice
Abstract
:1. Introduction
2. Results
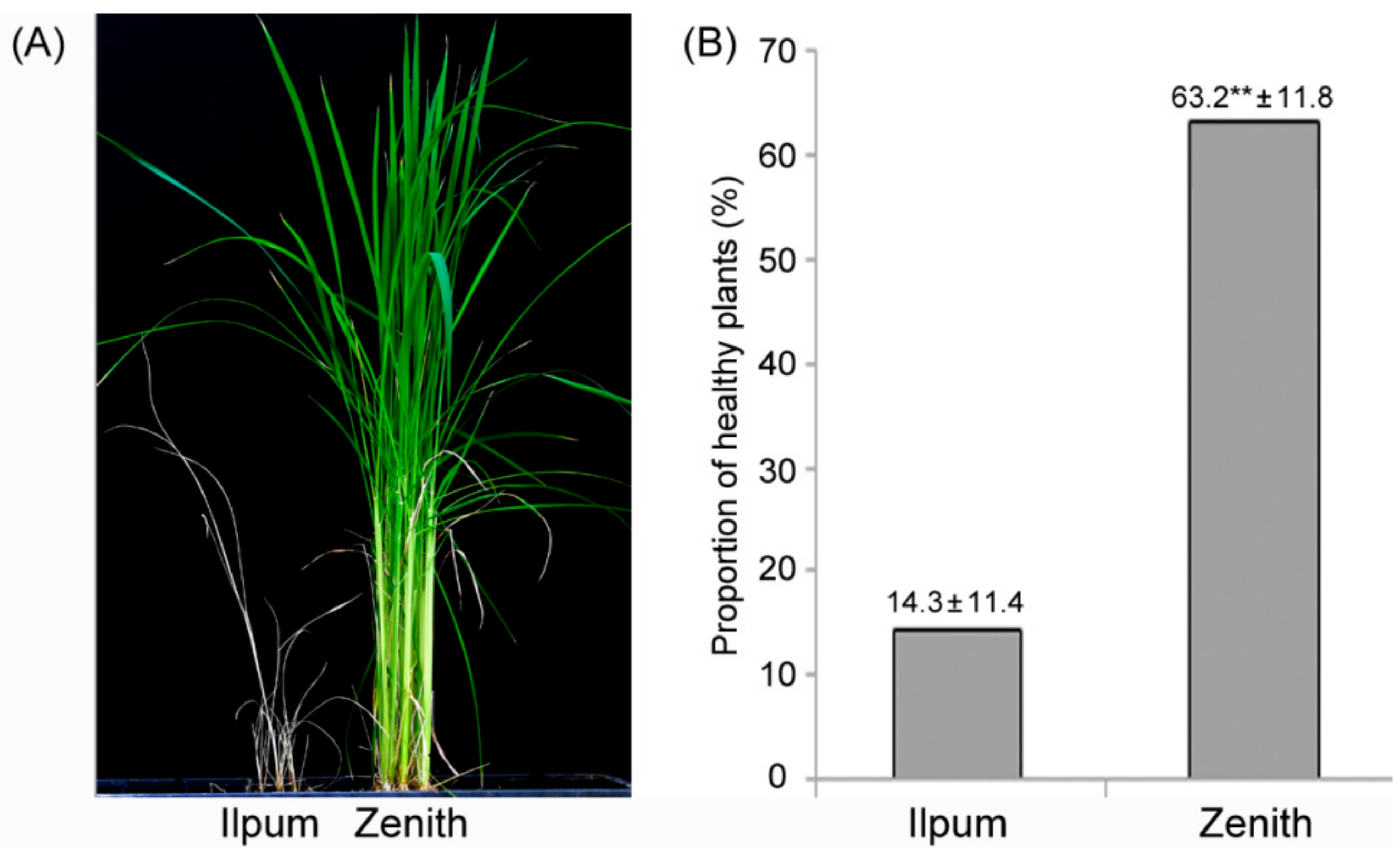
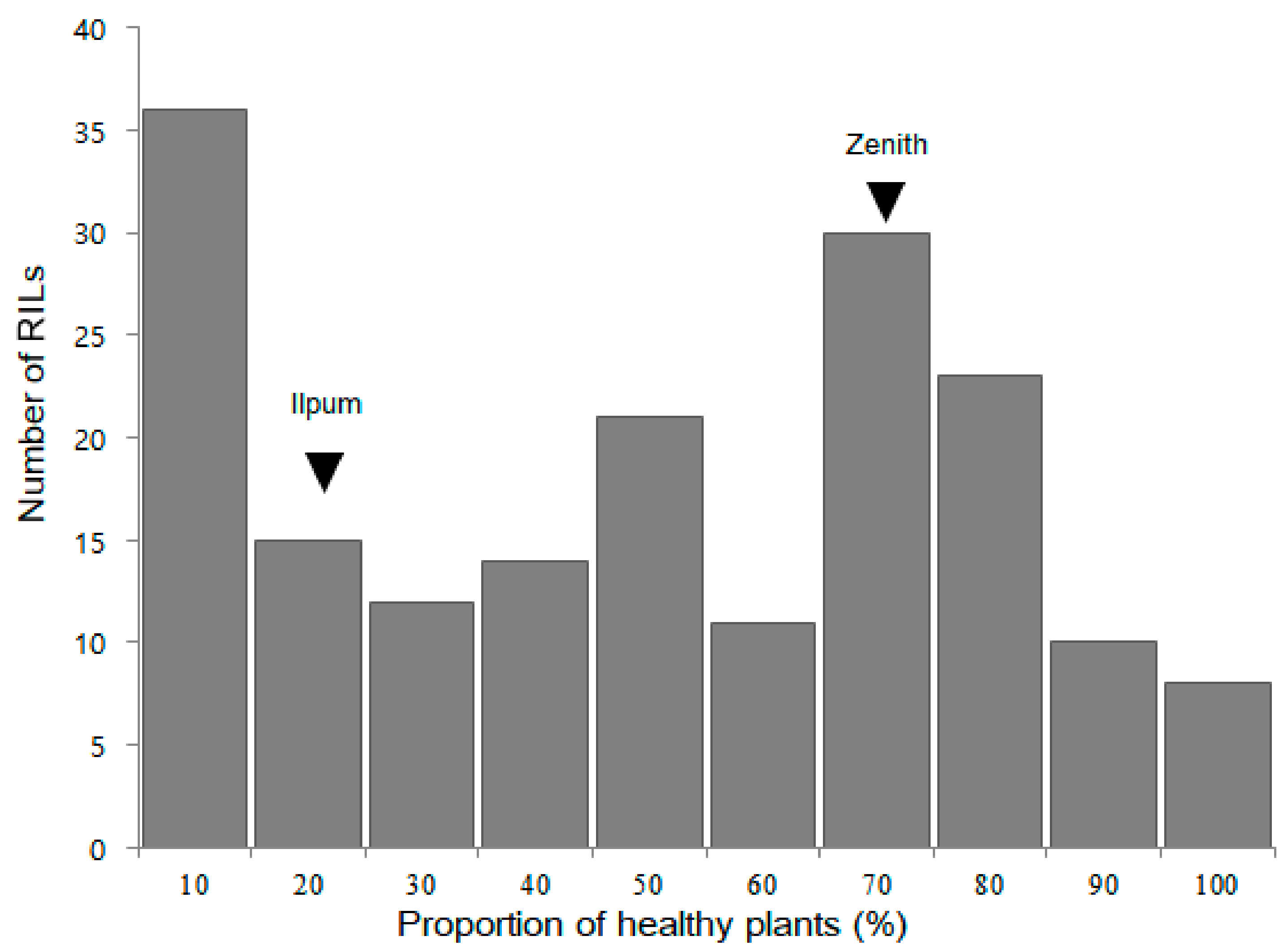
2.1. Bakanae Disease Bioassay in Parents and F2:9 RILs
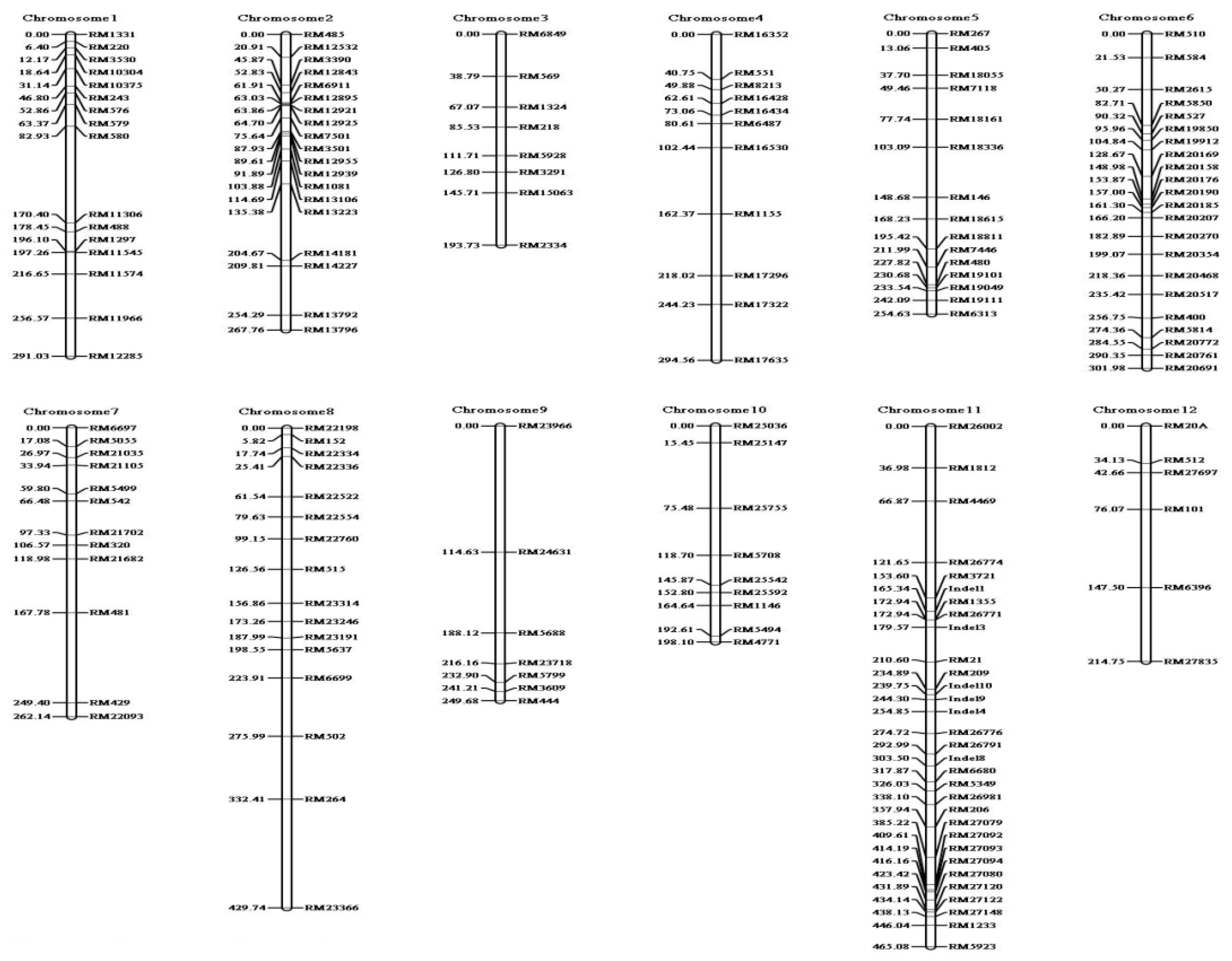
2.2. QTL Analysis and Mapping of qBK1z Using 180 F2:9 RILs
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Evaluation of Bakanae Resistance and Statistical Analysis
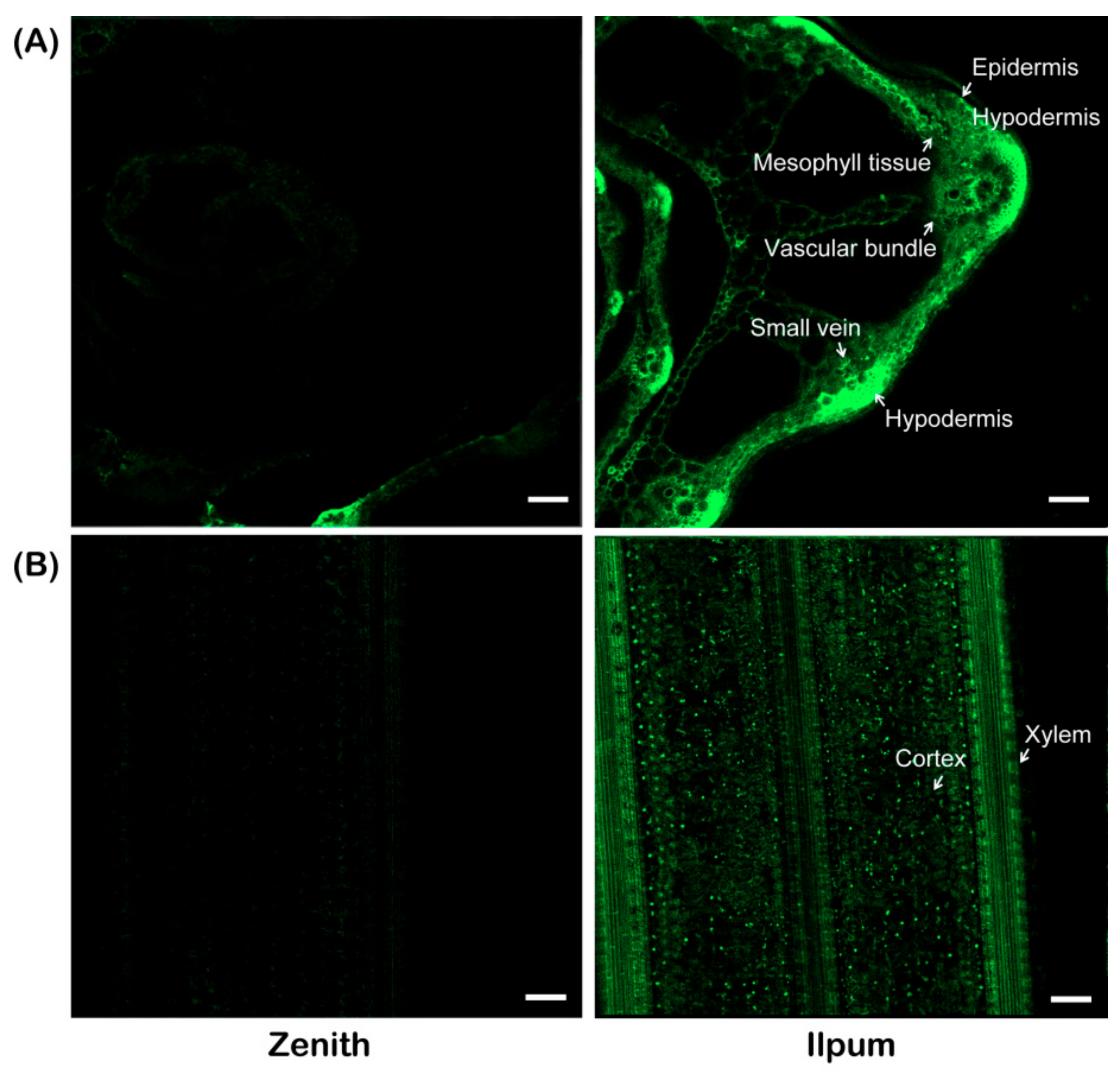
4.3. Localization of F. fujikuroi in Zenith and Ilpum Plants
4.4. DNA Extraction and Polymerase Chain Reaction
4.5. QTL Analysis of the F2:9 Population and Development of InDel Markers for Fine Mapping
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| Primer ID | Forward Primer (5′–3′) | Reverse Primer (5′–3′) | Location |
|---|---|---|---|
| Chr01_416669 | ACGCACCGTTTAGCAGTTTG | TGCATGCAACCTAATGACCC | 416,669 |
| Chr01_1435908 | GTTCTTCATGGTCGACAGCG | CCTTATAATTTGGGATGGAGGG | 1,435,908 |
| Chr01_2104309 | AAGCAAGTGGGAAACGAACC | GCATGATGTTTAAATGTGATGATTG | 2,104,309 |
| RM10116 | TCCCACTTGCAGACGTAGGTAGG | CACTGATCTGATGCAACTGTTTGG | 2,169,222 |
| Chr01_2319384 | GTTTGCGGGGCTAAGTTCTC | CTTCAGAAAAGAGGGTGGGC | 2,319,384 |
| Chr01_2898689 | CTCAAGCCTGTTGGTGTAGCA | AAAACCAACAACCTTAGGCCA | 2,898,689 |
| RM10208 | CGGAGCTCAATCGAATCTAGACC | TTCCTCATATCCTCCAGCTCTTAGC | 3,946,894 |
| RM10226 | GATCATCATCATCCATCCATCC | AAGGGAGAAATTGGTGGAGAGC | 4,005,199 |
| RM10287 | GTATTCCTTGCTGCTGCTGATGG | GACTGGAGATGTGATCGGAAACC | 4,966,447 |
| Chr01_5426210 | TGATGGCCTACACGATATGGA | TGTTACAAACAGGCGCAACA | 5,426,210 |
References
- Ito, S.; Kimura, J. Studies on the ‘bakanae’ disease of the rice plant. Rep. Hokkaido Natl. Agric. Exp. Stat. 1931, 27, 1–95. [Google Scholar]
- Gupta, A.K.; Solanki, I.S.; Bashyal, B.M.; Singh, Y.; Srivastava, K. Bakanae of rice—An emerging disease in Asia. J. Anim. Plant Sci. 2015, 25, 1499–1514. [Google Scholar]
- Wulff, E.G.; Sørensen, J.L.; Lübeck, M.; Nielsen, K.F.; Thrane, U.; Torp, J. Fusarium spp. associated with rice Bakanae: Ecology, genetic diversity, pathogenicity and toxigenicity. Environ. Microbiol. 2010, 12, 649–657. [Google Scholar] [CrossRef]
- Ora, N.; Faruq, A.N.; Islam, M.T.; Akhtar, N.; Rahman, M.M. Detection and identification of seed borne pathogens from some cultivated hybrid rice varieties in Bangladesh. Middle-East J. Sci. Res. 2011, 10, 482–488. [Google Scholar]
- Singh, R.; Sunder, S. Foot rot and bakanae disease of rice: An overview. Rev. Plant Pathol. 2012, 5, 565–604. [Google Scholar]
- Mew, T.W.; Gonzales, P.G. A Handbook of Rice Seedborne Fungi; International Rice Research Institute; Science Publishers, Inc.: Enfield, NH, USA; Los Baños, Philippines, 2002. [Google Scholar]
- Ou, S.H. Rice Diseases, 2nd ed.; Commonwealth Mycological Institute: Kew, UK, 1985. [Google Scholar]
- Sunder, S.; Virk, K.S. Studies on correlation between bakanae incidence and yield loss in paddy. Indian Phytopathol. 1997, 50, 99–101. [Google Scholar]
- Fiyaz, R.A.; Gopala Krishnan, S.; Rajashekara, H.; Yadav, A.K.; Bashyal, B.M.; Bhowmick, P.K.; Singh, N.K.; Prabhu, K.V.; Singh, A.K. Development of high throughput screening protocol and identification of novel sources of resistance against Bakanae disease in rice (Oryza sativa L.). Indian J. Genet. 2014, 74, 414–422. [Google Scholar]
- Singh, R.; Sunder, S. Foot rot and bakanae of rice: Retrospects and prospects. Intern. J. Trop. Plant Dis. 1997, 15, 153–176. [Google Scholar]
- Yasin, S.I.; Khan, T.Z.; Akhtar, K.M.; Muhammad, A.; Mushtaq, A. Economic evaluation of bakanae disease of rice. Mycopath 2003, 1, 115–117. [Google Scholar]
- Saremi, H.; Ammarellou, A.; Marefat, A.; Okhovat, S.M. Binam a rice cultivar, resistant for root rot disease on rice caused by Fusarium moniliforme in North-West, Iran. Intern. J. Bot. 2008, 4, 383–389. [Google Scholar] [CrossRef] [Green Version]
- Park, W.S.; Choi, H.W.; Han, S.S.; Shin, D.B.; Shim, H.K.; Jung, E.S.; Lee, S.W.; Lim, C.K.; Lee, Y.H. Control of bakanae disease of rice by seed soaking into the mixed solution of procholraz and fludioxnil. Res. Plant Dis. 2009, 15, 94–100. [Google Scholar] [CrossRef] [Green Version]
- Rosales, A.M.; Mew, T.W. Suppression of Fusarium moniliforme in rice by rice associated antagonistic bacteria. Plant Dis. 1997, 81, 49–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayasaka, T.; Ishiguro, K.; Shibutani, K.; Namai, T. Seed disinfection using hot water immersion to control several seed-borne diseases of rice plants. Jpn. J. Phytopathol. 2001, 67, 26–32. [Google Scholar] [CrossRef]
- Lee, S.B.; Hur, Y.J.; Cho, J.H.; Lee, J.H.; Kim, T.H.; Cho, S.M.; Song, Y.C.; Seo, Y.S.; Lee, J.K.; Kim, T.S.; et al. Molecular mapping of qBK1WD, a major QTL for bakanae disease resistance in rice. Rice 2018, 11, 3. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, K. Damage by “Bakanae” disease and its chemical control. Jpn. Pestic. Inf. 1988, 52, 13–15. [Google Scholar]
- Kim, J.M.; Hong, S.K.; Kim, W.G.; Lee, Y.K.; Yu, S.H.; Choi, H.W. Fungicide resistance of Gibberella fujikuroi isolates causing rice bakanae disease and their progeny isolates. Kor. J. Mycol. 2010, 38, 75–79. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.H.; Lee, M.J.; Choi, H.W.; Kim, S.T.; Park, J.W.; Myung, I.S.; Park, K.S.; Lee, S.W. Development of in vitro seedling screening method for selection of resistant rice against bakanae disease. Res. Plant Dis. 2011, 17, 288–294. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.D.; Guo, L.B.; Li, X.M.; Ji, Z.J.; Ma, L.Y.; Qian, Q. Analysis of QTLs for resistance to rice bakanae disease. Chin. J. Rice Sci. 2006, 6, 657–659. [Google Scholar]
- Hur, Y.J.; Lee, S.B.; Kim, T.H.; Kwon, T.; Lee, J.H.; Shin, D.J.; Park, S.K.; Hwang, U.H.; Cho, Y.H.; Yoon, Y.N.; et al. Mapping of qBK1, a major QTL for bakanae disease resistance in rice. Mol. Breed. 2015, 35. [Google Scholar] [CrossRef]
- Lee, S.B.; Kim, N.; Hur, Y.J.; Cho, S.M.; Kim, T.H.; Lee, J.Y.; Cho, J.H.; Lee, J.H.; Song, Y.C.; Seo, Y.S.; et al. Fine mapping of qBK1, a major QTL for bakanae disease resistance in rice. Rice 2019, 12, 36. [Google Scholar] [CrossRef] [Green Version]
- Fiyaz, R.A.; Yadav, A.K.; Krishnan, S.G.; Ellur, R.K.; Bashyal, B.M.; Grover, N.; Bhowmick, P.K.; Nagarajan, M.; Vinod, K.K.; Singh, N.K.; et al. Mapping quantitative trait loci responsible for resistance to Bakanae disease in rice. Rice 2016, 9, 3–10. [Google Scholar] [CrossRef] [Green Version]
- Ji, H.; Kim, T.H.; Lee, G.S.; Kang, H.J.; Lee, S.B.; Suh, S.C.; Kim, S.L.; Choi, I.; Baek, J.; Kim, K.H. Mapping of a major quantitative trait locus for bakanae disease resistance in rice by genome resequencing. Mol. Gen. Genom. 2017. [Google Scholar] [CrossRef]
- Volante, A.; Tondelli, A.; Aragona, M.; Valente, M.T.; Biselli, C.; Desiderio, F.; Bagnaresi, P.; Matic, S.; Gullino, M.L.; Infantino, A.; et al. Identification of bakanae disease resistance loci in japonica rice through genome wide association study. Rice 2017, 10. [Google Scholar] [CrossRef] [Green Version]
- Kang, D.Y.; Cheon, K.S.; Oh, J.; Oh, H.; Kim, S.L.; Kim, N.; Lee, E.; Choi, I.; Baek, J.; Kim, K.H.; et al. Rice genome resequencing reveals a major quantitative trait locus for resistance to bakanae disease caused by Fusarium fujikuroi. Int. J. Mol. Sci. 2019, 20, 2598. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.H.; Hur, Y.J.; Lee, S.B.; Kwon, T.M.; Hwang, U.H.; Park, S.K.; Youn, Y.N.; Lee, J.H.; Cho, J.H.; Shin, D.J.; et al. Large-scale screening of rice accessions to evaluate resistance to bakanae disease. J. Gen. Plant Pathol. 2014, 80, 408–414. [Google Scholar] [CrossRef]
- Lee, S.B.; Hur, Y.J.; Lee, J.H.; Kwon, T.M.; Shin, D.J.; Kim, T.H.; Han, S.I.; Choi, J.H.; Yoon, Y.N.; Kiswara, K.; et al. Molecular mapping of a quantitative trait locus qSTV11Z harboring rice stripe virus resistance gene, Stv-b. Plant Breed. 2017, 136, 61–66. [Google Scholar] [CrossRef] [Green Version]
- Cumagun, C.J.R.; Arcillas, E.; Gergon, E. UP-PCR analysis of the seedborne pathogen Fusarium fujikuroi causing bakanae disease in rice. Int. J. Agric. Biol. 2011, 13, 1029–1032. [Google Scholar]
- Bashyal, B.M.; Aggarwal, R.; Banerjee, S.; Gupta, S.; Sharma, S. Pathogenicity, ecology and genetic diversity of the Fusarium spp. associated with an emerging bakanae disease of rice (Oryza sativa L.) in India. In Microbial Diversity and Biotechnology in Food Security; Kharwar, R.N., Upadhyay, R., Dubey, N., Raghuwanshi, R., Eds.; Springer: New Delhi, India, 2014; pp. 307–314. [Google Scholar]
- Hur, Y.J.; Lee, S.B.; Shin, D.J.; Kim, T.H.; Cho, J.H.; Han, S.I.; Oh, S.H.; Lee, J.Y.; Son, Y.B.; Lee, J.H.; et al. Screening of rice germplasm for Bakanae disease resistance in rice. Korean J. Breed. Sci. 2016, 48, 22–28. [Google Scholar] [CrossRef] [Green Version]
- Rana, A.; Sahgal, M.; Johri, B.N. Fusarium oxysporum: Genomics, Diversity and Plant–Host Interaction. In Developments in Fungal Biology and Applied Mycology; Springer: Singapore, 2017; pp. 159–199. [Google Scholar]
- Jansen, C.; VonWettstein, D.; Schäfer, W.; Kogel, K.H.; Felk, A.; Maier, F.J. Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proc. Natl. Acad. Sci. USA 2005, 102, 16892–16897. [Google Scholar] [CrossRef] [Green Version]
- Elshafey, R.A.S.; Tahoon, A.M.; El-Emary, F.A. Analysis of varietal response to bakanae infection Fusarium fujikuroi and gibberellic acid through morphological, anatomical and hormonal changes in three rice varieties. J. Phytopathol. Pest Manag. 2018, 5, 63–87. [Google Scholar]
- Sundaram, R.M.; Vishnupriya, M.R.; Laha, G.S.; Rani, N.S.; Rao, P.S.; Balachandran, S.M.; Reddy, G.A.; Sharma, N.P.; Sonti, R.V. Introduction of bacterial blight resistance into Triguna, a high yielding, mid-early duration rice variety. Biotechnology 2009, 4, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Sidhu, J.S.; Huang, N.; Vikal, Y.; Li, Z.; Brar, D.S.; Dhaliwal, H.S.; Khush, G.S. Pyramiding three bacterial blight resistance genes (xa5, xa13 and Xa21) using marker-assisted selection into indica rice cultivar PR106. Theor. Appl. Genet. 2001, 102, 1011–1015. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Nayak, D.K.; Mohanty, S.; Behera, L.; Barik, S.R.; Pandit, E.; Lenka, S.; Anandan, A. Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety, Jalmagna. Rice 2015, 8, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, N.; Mitsunaga, T.; Hayashi, K.; Koizumi, S.; Fujita, Y. Effects of pyramiding quantitative resistance genes pi21, Pi34, and Pi35 on rice leaf blast disease. Plant Dis. 2015, 99, 904–909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lander, E.S.; Green, P.; Abrahamson, J.; Barlow, A.; Daly, M.J.; Lincoln, S.E.; Newburg, L. MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1987, 1, 174–181. [Google Scholar] [CrossRef]
- Wang, S.; Basten, C.J.; Zeng, Z.B. Windows QTL Cartographer 2.5. 2007. Available online: http://statgen.ncsu.edu/qtlcart/WQTLCart.htm (accessed on 12 August 2018).



| QTL | Chromosome | Position (cM) | Marker Interval | LOD a | PVE b (%) | Additive Effect | Dominant Effect |
|---|---|---|---|---|---|---|---|
| qBK1Z | 1 | 4.0 | RM1331–RM3530 | 13.43 | 30.93 | −22.10 | −1.09 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.-B.; Kim, N.; Jo, S.; Hur, Y.-J.; Lee, J.-Y.; Cho, J.-H.; Lee, J.-H.; Kang, J.-W.; Song, Y.-C.; Bombay, M.; et al. Mapping of a Major QTL, qBK1Z, for Bakanae Disease Resistance in Rice. Plants 2021, 10, 434. https://doi.org/10.3390/plants10030434
Lee S-B, Kim N, Jo S, Hur Y-J, Lee J-Y, Cho J-H, Lee J-H, Kang J-W, Song Y-C, Bombay M, et al. Mapping of a Major QTL, qBK1Z, for Bakanae Disease Resistance in Rice. Plants. 2021; 10(3):434. https://doi.org/10.3390/plants10030434
Chicago/Turabian StyleLee, Sais-Beul, Namgyu Kim, Sumin Jo, Yeon-Jae Hur, Ji-Youn Lee, Jun-Hyeon Cho, Jong-Hee Lee, Ju-Won Kang, You-Chun Song, Maurene Bombay, and et al. 2021. "Mapping of a Major QTL, qBK1Z, for Bakanae Disease Resistance in Rice" Plants 10, no. 3: 434. https://doi.org/10.3390/plants10030434
APA StyleLee, S. -B., Kim, N., Jo, S., Hur, Y. -J., Lee, J. -Y., Cho, J. -H., Lee, J. -H., Kang, J. -W., Song, Y. -C., Bombay, M., Kim, S. -R., Lee, J., Seo, Y. -S., Ko, J. -M., & Park, D. -S. (2021). Mapping of a Major QTL, qBK1Z, for Bakanae Disease Resistance in Rice. Plants, 10(3), 434. https://doi.org/10.3390/plants10030434







