Application of Plant Growth Regulators Modulates the Profile of Chlorogenic Acids in Cultured Bidens pilosa Cells
Abstract
:1. Introduction
2. Results and Discussion
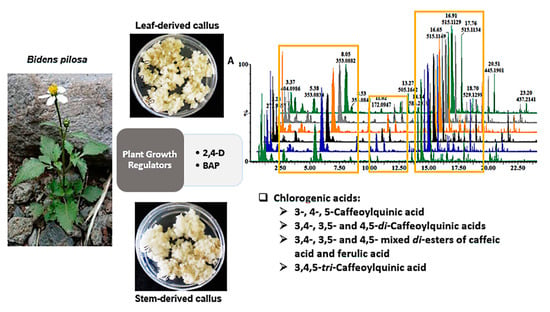
2.1. Manipulation of Undifferentiated Bidens pilosa Cells with Plant Growth Regulators
2.2. Total Phenolic Content in Response to Different Plant Growth Regulator Combinations
2.3. Analysis of Altered Callus Metabolomes in Response to Different Plant Growth Regulator Combinations
2.4. Multivariate Statistical Analysis of Phytochemical Profiles/Constituents of Callus Maintained on Different Plant Growth Regulator Combinations
2.5. Comparative Analysis of Metabolites Identified in Callus Maintained on Media with Different PGR Ratios
3. Materials and Methods
3.1. Callus Initiation and Cultivation on Different Ratios of Plant Growth Regulators
3.2. Metabolite Extraction
3.3. Total Phenolic Content (TPC) Assay
3.4. Ultra-High-Performance Liquid Chromatography—High-Definition Mass Spectrometry (UHPLC–HDMS)
3.5. Multivariate Data Analysis, Metabolite Annotation and Relative Quantification
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seca, A.M.L.; Pinto, D.C.G.A. Biological potential and medical use of secondary metabolites. Medicines 2019, 6, 66. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, R.; Rana, C.S. Plant secondary metabolites: A review. Int. J. Eng. Res. Gen. Sci. 2015, 3, 661–670. [Google Scholar] [CrossRef] [Green Version]
- Bartolome, A.P.; Villaseñor, I.M.; Yang, W.C. Bidens pilosa L. (Asteraceae): Botanical properties, traditional uses, phytochemistry, and pharmacology. Evid. Based Complement. Altern. Med. 2013, 2013, 340215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khoza, B.S.; Gbashi, S.; Steenkamp, P.A.; Njobeh, P.B.; Madala, N.E. Identification of hydroxylcinnamoyl tartaric acid esters in Bidens pilosa by UPLC-tandem mass spectrometry. S. Afr. J. Bot. 2016, 103, 95–100. [Google Scholar] [CrossRef]
- Ramabulana, A.-T.; Steenkamp, P.; Madala, N.; Dubery, I.A. Profiling of chlorogenic acids from Bidens pilosa and differentiation of closely related positional isomers with the aid of UHPLC-QTOF-MS / MS-based in-source collision-induced dissociation. Metabolites 2020, 10, 178. [Google Scholar] [CrossRef]
- Ong, K.W.; Hsu, A.; Tan, B.K.H. Anti-diabetic and anti-lipidemic effects of chlorogenic acid are mediated by AMPK activation. Biochem. Pharmacol. 2013, 85, 1341–1351. [Google Scholar] [CrossRef]
- Makola, M.M.; Dubery, I.A.; Koorsen, G.; Steenkamp, P.A.; Kabanda, M.M.; Du Preez, L.L.; Madala, N.E. The effect of geometrical isomerism of 3,5-dicaffeoylquinic acid on its binding affinity to HIV-integrase enzyme: A molecular docking study. Evid. Based Complement. Altern. Med. 2016, 2016, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Lodise, O.; Patil, K.; Karshenboym, I.; Prombo, S.; Chukwueke, C.; Pai, S.B. Inhibition of prostate cancer cells by 4,5-dicaffeoylquinic acid through cell cycle arrest. Prostate Cancer 2019, 2019, 4520645. [Google Scholar] [CrossRef] [Green Version]
- Ramirez-Estrada, K.; Vidal-Limon, H.; Hidalgo, D.; Moyano, E.; Golenioswki, M.; Cusidó, R.M.; Palazon, J. Elicitation, an effective strategy for the biotechnological production of bioactive high-added value compounds in plant cell factories. Molecules 2016, 21, 182. [Google Scholar] [CrossRef]
- Marchev, A.S.; Yordanova, Z.P.; Georgiev, M.I. Green (cell) factories for advanced production of plant secondary metabolites. Crit. Rev. Biotechnol. 2020, 40, 443–458. [Google Scholar] [CrossRef] [PubMed]
- Phillips, G.C.; Garda, M. Plant tissue culture media and practices: An overview. Plant 2019, 55, 242–257. [Google Scholar] [CrossRef]
- Gaspar, T.; Keveks, C.; Penel, C.; Greppin, H.; Reid, D.M.; Thorpe, T.A. Plant hormones and plant growth regulators in plant tissue culture. In Vitro Cell. Dev. Biol. Plant 1996, 32, 272–289. [Google Scholar] [CrossRef]
- Moubayidin, L.; Mambro, R.D.; Sabatini, S. Cytokinin-auxin crosstalk. Trends Plant Sci. 2009, 14, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.H.; Liu, Y.B.; Zhang, X.S. Auxin-cytokinin interaction regulates meristem development. Mol. Plant 2011, 4, 616–625. [Google Scholar] [CrossRef]
- Lee, Z.H.; Hirakawa, T.; Yamaguchi, N.; Ito, T. The roles of plant hormones and their interactions with regulatory genes in determining meristem activity. Int. J. Mol. Sci. 2019, 20, 4065. [Google Scholar] [CrossRef] [Green Version]
- Coenen, C.; Lomax, T.L. Auxin-cytokinin interactions in higher plants: Old problems and new tools. Trends Plant Sci. 1997, 2, 351–355. [Google Scholar] [CrossRef]
- Shin, J.; Bae, S.; Seo, P.J. De novo shoot organogenesis during plant regeneration. J. Exp. Bot. 2020, 71, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Pernisová, M.; Klíma, P.; Horák, J.; Válková, M.; Malbeck, J.; Souček, P.; Reichman, P.; Hoyerová, K.; Dubová, J.; Friml, J.; et al. Cytokinins modulate auxin-induced organogenesis in plants via regulation of the auxin efflux. Proc. Natl. Acad. Sci. USA 2009, 106, 3609–3614. [Google Scholar] [CrossRef] [Green Version]
- Schaller, G.E.; Bishopp, A.; Kieber, J.J. The yin-yang of hormones: Cytokinin and auxin interactions in plant development. Plant Cell 2015, 27, 44–63. [Google Scholar] [CrossRef] [Green Version]
- Hamany Djande, C.Y.; Steenkamp, P.A.; Steenkamp, A.; Piater, L.A.; Madala, N.E. Habituated Moringa oleifera callus retains metabolic responsiveness to external plant growth regulators. Plant Cell. Tissue Organ Cult. 2019. [Google Scholar] [CrossRef]
- Malik, S.; Zia, M.; Rehman, R.; Chaudhary, F. In vitro plant regeneration from direct and indirect organogenesis of Momordica charantia. Pakistan J. Biol. Sci. 2007, 10, 4118–4122. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Liu, W.; Liu, J.; Qin, P.; Xu, L. Auxin control of root organogenesis from callus in tissue culture. Front. Plant Sci. 2017, 8, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Cardoso, J.C.; de Oliveira, M.E.B.S.; de Cardoso, F.I. Advances and challenges on the in vitro production of secondary metabolites from medicinal plants. Hortic. Bras. 2019, 37, 124–132. [Google Scholar] [CrossRef] [Green Version]
- Alagarsamy, K.; Shamala, L.F.; Wei, S. Influence of media supplements on inhibition of oxidative browning and bacterial endophytes of Camellia sinensis var. sinensis. 3 Biotech 2018, 8, 1–7. [Google Scholar] [CrossRef]
- Murata, M.; Nishimura, M.; Murai, N.; Haruta, M.; Itoh, Y. A Transgenic apple callus showing reduced polyphenol oxidase activity and lower browning potential. Biosci. Biotechnol. Biochem. 2014, 65, 383–388. [Google Scholar] [CrossRef]
- Wu, J.; Lin, L. Ultrasound-induced stress responses of Panax ginseng Cells: Enzymatic browning and phenolics production. Biotechnol. Prog. 2002, 18, 862–866. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, G.; Dantu, P.K. Evaluation of callus browning and develop a strategically callus culturing of Boerhaavia diffusa L. J. Plant Dev. 2015, 22, 47–58. [Google Scholar]
- Pischke, M.S.; Huttlin, E.L.; Hegeman, A.D.; Sussman, M.R. A Transcriptome-based characterization of habituation in plant tissue culture. Plant Physiol. 2006, 140, 1255–1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaspar, T.; Kevers, C.; Bisbis, B.; Franck, T.; Crevecoeur, M.; Greppin, H.; Dommes, J. Special symposium: In vitro plant recalcitrance loss of plant organogenic totipotency in the course of in vitro neoplastic progression. In Vitro Cell Dev. Biol. Plant 2000, 36, 171–181. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.Q.; Lian, H.; Tang, H.; Dolezal, K.; Zhou, C.M.; Yu, S.; Chen, J.H.; Chen, Q.; Liu, H.; Ljung, K.; et al. An intrinsicmicroRNA timer regulates progressive decline in shoot regenerative capacity in plants. Plant Cell 2015, 27, 349–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeuchi, M.; Ogawa, Y.; Iwase, A.; Sugimoto, K. Plant regeneration: Cellular origins and molecular mechanisms. Development 2016, 143, 1442–1451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef] [Green Version]
- Stojakowska, A.; Malarz, J.; Kiss, A.K. Hydroxycinnamates from elecampane (Inula helenium L.) callus culture. Acta Physiol. Plant. 2016, 38, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Ramabulana, A.T.; Steenkamp, P.A.; Madala, N.E.; Dubery, I.A. Profiling of altered metabolomic states in Bidens pilosa leaves in response to treatment by methyl jasmonate and methyl salicylate. Plants 2020, 9, 1275. [Google Scholar] [CrossRef] [PubMed]
- Luczkiewicz, M.; Kokotkiewicz, A.; Glod, D. Plant growth regulators affect biosynthesis and accumulation profile of isoflavone phytoestrogens in high-productive in vitro cultures of Genista tinctoria. Plant Cell. Tissue Organ Cult. 2014, 118, 419–429. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Lee, D.E.; Lee, H.S.; Kim, S.K.; Lee, W.S.; Kim, S.H.; Kim, M.W. Influence of auxins, cytokinins, and nitrogen on production of rutin from callus and adventitious roots of the white mulberry tree (Morus alba L.). Plant Cell. Tissue Organ Cult. 2011, 105, 9–19. [Google Scholar] [CrossRef]
- Jamwal, K.; Bhattacharya, S.; Puri, S. Plant growth regulator mediated consequences of secondary metabolites in medicinal plants journal of applied research on medicinal and aromatic plants plant growth regulator mediated consequences of secondary metabolites in medicinal plants. J. Appl. Res. Med. Aromat. Plants 2018, 9, 26–38. [Google Scholar] [CrossRef]
- Wiklund, S.; Johansson, E.; Sjo, L.; Shockcor, J.P.; Gottfries, J.; Moritz, T.; Trygg, J. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal. Chem. 2008, 80, 115–122. [Google Scholar] [CrossRef]
- Bartel, J.; Krumsiek, J.; Theis, F.J. Statistical methods for the analysis of high-throughput metabolomics data. Comput. Struct. Biotechnol. J. 2013, 4, e201301009. [Google Scholar] [CrossRef] [Green Version]
- Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. A conversation on data mining strategies in LC-MS untargeted metabolomics: Pre-processing and pre-treatment steps. Metabolites 2016, 6, 40. [Google Scholar] [CrossRef] [Green Version]
- Godzien, J.; Ciborowski, M.; Angulo, S.; Barbas, C. From numbers to a biological sense: How the strategy chosen for metabolomics data treatment may affect final results. A Practical example based on urine fingerprints obtained by LC-MS. Electrophoresis 2013, 34, 2812–2826. [Google Scholar] [CrossRef]
- Granato, D.; Santos, J.S.; Escher, G.B.; Ferreira, B.L.; Maggio, R.M. Use of principal component analysis (PCA) and hierarchical cluster analysis (HCA) for multivariate association between bioactive compounds and functional properties in foods: A critical perspective. Trends Food Sci. Technol. 2018, 72, 83–90. [Google Scholar] [CrossRef]
- Rodriguez, M.Z.; Comin, C.H.; Casanova, D.; Bruno, O.M.; Amancio, D.R.; Costa, L.D.F.; Rodrigues, F.A. Clustering algorithms: A comparative approach. PLoS ONE 2019, 14, 1–34. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, 486–494. [Google Scholar] [CrossRef] [Green Version]
- Park, C.H.; Yeo, H.J.; Park, Y.J.; Morgan, A.M.A.; Arasu, M.V.; Al-Dhabi, N.A.; Park, S.U.; Cravotto, G. Influence of indole-3-acetic acid and gibberellic acid on phenylpropanoid accumulation in common buckwheat (Fagopyrum esculentum Moench) sprouts. Molecules 2017, 22, 374. [Google Scholar] [CrossRef] [Green Version]
- Teixeira, J.; Gaspar, A.; Garrido, E.M.; Garrido, J.; Borges, F. Hydroxycinnamic acid antioxidants: An electrochemical overview. Biomed. Res. Int. 2013, 2013, 251754. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.M.; Mohamed, H.E.; Mostafa, E.M. Nutrient starvation enhances the phenolic compounds and antioxidant activity in Azolla caroliniana plant. Egypt. J. Bot. 2020, 60, 239–247. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A Revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 474–497. [Google Scholar] [CrossRef]
- Van der Berg, R.A.; Hoofsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. Cancer Res. 2006, 7, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.; Lan, W.; Zheng, G.; Gao, X. Metabolomics: A high-throughput platform for metabolite profile exploration. Methods Mol. Biol. 2018, 1754, 265–292. [Google Scholar] [CrossRef]
- Sumner, L.W.; Samuel, T.; Noble, R.; Gmbh, S.D.; Barrett, D.; Beale, M.H.; Hardy, N. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dictionary of Natural Products. Available online: http://dnp.chemnetbase.com/faces/chemical/ChemicalSearch.xhtml (accessed on 20 July 2020).
- Kyoto Encyclopedia of Genes and Genomes. Available online: https://www.genome.jp/kegg/ (accessed on 20 July 2020).
- Chemspider Chemspider. Available online: http://www.chemspider.com (accessed on 20 July 2020).
- MetaboAnalyst MetaboAnalyst. Available online: https://www.metaboanalyst.ca/ (accessed on 15 August 2020).
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, 652–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Condition Number | 2,4-D (mg/L) | BAP (mg/L) | Ratio (2,4-D: BAP) |
|---|---|---|---|
| 1 | 0.20 | 2.00 | 1:10 |
| 2 | 2.00 | 0.20 | 10:1 |
| 3 | 0.00 | 0.00 | - |
| 4 | 0.45 | 1.00 | 1:2 |
| 5 | 0.30 | 4.00 | 1:20 |
| 6 | 0.20 | 8.00 | 1:40 |
| Condition Number | 2,4-D (mg/L) | BAP (mg/L) | TPC (Leaf Calli) | TPC (Stem Calli) |
|---|---|---|---|---|
| 1 | 0.20 | 2.00 | 18.02 ± 0.08 | 25.86 ± 0.07 |
| 2 | 2.00 | 0.20 | 22.25 ± 0.04 | 28.29 ± 0.02 |
| 3 | 0.00 | 0.00 | 29.20 ± 0.06 | 31.15 ± 0.02 |
| 4 * | 0.45 | 1.00 | 20.38 ± 0.02 | 31.15 ± 0.03 |
| 5 | 0.30 | 4.00 | 27.12 ± 0.07 | 33.30 ± 0.05 |
| 6 | 0.20 | 8.00 | 25.16 ± 0.08 | 25.30 ± 0.05 |
| No. | m/z | Rt (min) | Diagnostic Fragment Ions | Molecular Formulae | Metabolite * | Abbreviation |
|---|---|---|---|---|---|---|
| 1 | 195.590 | 0.90 | 191, 162, 108 | C6H12O7 | Gluconic acid | Gluc |
| 2 | 133.010 | 0.99 | 115 | C4H6O5 | Malic acid | Mal |
| 3 | 191.014 | 1.18 | 111 | C6H8O7 | Citric acid | CTA |
| 4 | 331.064 | 1.72 | 168, 125 | C13H16O10 | Galloyl-hexoside | Gall |
| 5 | 164.067 | 1.96 | 147 | C9H11NO2 | Phenylalanine | Phe |
| 6 | 315.069 | 2.07 | 153, 152, 109, 108 | C13H16O9 | 2,5-Dihydroxybenzoic acid | 2,5-DHBA |
| 7 | 353.0842 | 2.77 | 191, 179, 135 | C16H18O9 | trans-3-Caffeoylquinic acid | trans-3-CQA |
| 8 | 203.077 | 3.22 | 142, 116 | C11H12N2O2 | Tryptophan | Trp |
| 9 | 353.0881 | 5.48 | 191 | C16H18O9 | trans-5-Caffeoylquinic acid | trans-5-CQA |
| 10 | 353.0831 | 5.87 | 191, 179, 173, 135 | C16H18O9 | trans-4-Caffeoylquinic acid | trans-4-CQA |
| 11 | 367.0980 | 10.03 | 193, 173 | C17H20O9 | 4-Feruloylquinic acid | 4-FQA |
| 12 | 515.1166 | 14.01 | 353, 335, 191, 179, 135 | C25H24O12 | 3,4-di-Caffeoylquinic acid | 3,4-diCQA |
| 13 | 515.1195 | 14.34 | 353, 191, 179, 135 | C25H24O12 | 3,5-di-Caffeoylquinic acid | 3,5-diCQA |
| 14 | 515.1219 | 15.17 | 353, 335, 191, 179, 173, 135 | C25H24O12 | 4,5-di-Caffeoylquinic acid | 4,5-diCQA |
| 15 | 529.1398 | 15.54 | 367, 353, 335, 193, 179, 173, 134 | C26H26O12 | 3-Feruloyl-4-caffeoylquinic acid | 3F-4CQA |
| 16 | 529.1013 | 15.72 | 367, 335, 193, 173 | C26H26O12 | 3-Caffeoyl-4-feruloylquinic acid | 3C-4FQA |
| 17 | 529.0983 | 16.00 | 367, 193, 134 | C26H26O12 | 3-Feruloyl-5-caffeoylquinic acid | 3F-5CQA |
| 18 | 529.1345 | 16.11 | 367, 353, 191, 179 | C26H26O12 | 3-Caffeoyl-5-feruloylquinic acid | 3C-5FQA |
| 19 | 529.1345 | 16.53 | 367, 193, 173 | C26H26O12 | 4-Feruloyl-5-caffeoylquinic acid | 4F-5CQA |
| 20 | 529.117 | 16.68 | 367, 353, 191, 179, 173, 135 | C26H26O12 | 4-Caffeoyl-5-feruloylquinic acid | 4C-5FQA |
| 21 | 677.1561 | 17.51 | 515, 353,179, 173 | C34H30O15 | tri-Caffeoylquinic acid | tri-CQA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramabulana, A.-T.; Steenkamp, P.A.; Madala, N.E.; Dubery, I.A. Application of Plant Growth Regulators Modulates the Profile of Chlorogenic Acids in Cultured Bidens pilosa Cells. Plants 2021, 10, 437. https://doi.org/10.3390/plants10030437
Ramabulana A-T, Steenkamp PA, Madala NE, Dubery IA. Application of Plant Growth Regulators Modulates the Profile of Chlorogenic Acids in Cultured Bidens pilosa Cells. Plants. 2021; 10(3):437. https://doi.org/10.3390/plants10030437
Chicago/Turabian StyleRamabulana, Anza-Tshilidzi, Paul A. Steenkamp, Ntakadzeni E. Madala, and Ian A. Dubery. 2021. "Application of Plant Growth Regulators Modulates the Profile of Chlorogenic Acids in Cultured Bidens pilosa Cells" Plants 10, no. 3: 437. https://doi.org/10.3390/plants10030437
APA StyleRamabulana, A.-T., Steenkamp, P. A., Madala, N. E., & Dubery, I. A. (2021). Application of Plant Growth Regulators Modulates the Profile of Chlorogenic Acids in Cultured Bidens pilosa Cells. Plants, 10(3), 437. https://doi.org/10.3390/plants10030437