BcHTT4 Inhibits Branching of Non-Heading Chinese Cabbage at the Vegetative Stage
Abstract
:1. Introduction
2. Results
2.1. BcHTT4 May Be a Negative Regulator of Branching in NHCC
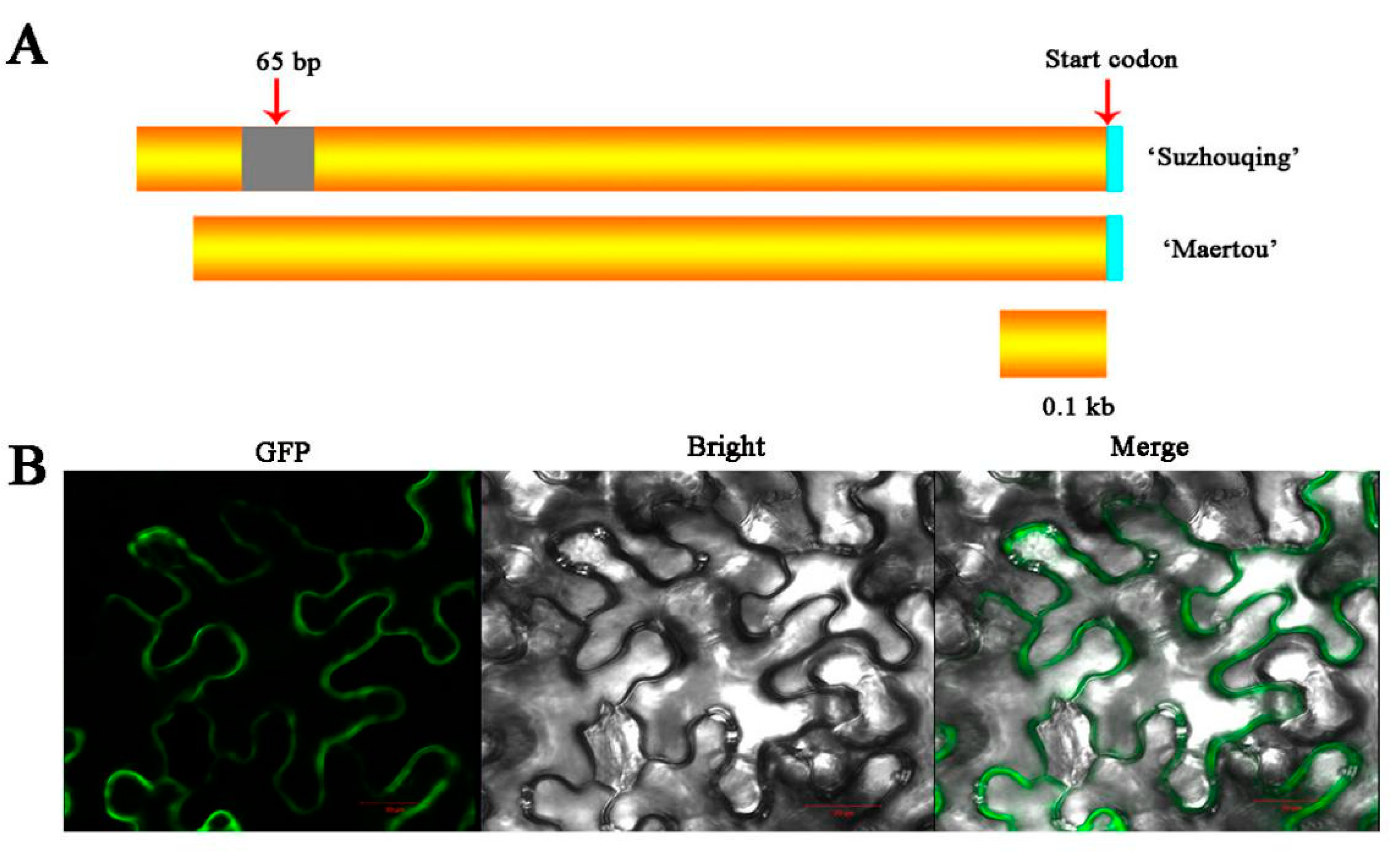
2.2. The Characteristics of BcHTT4
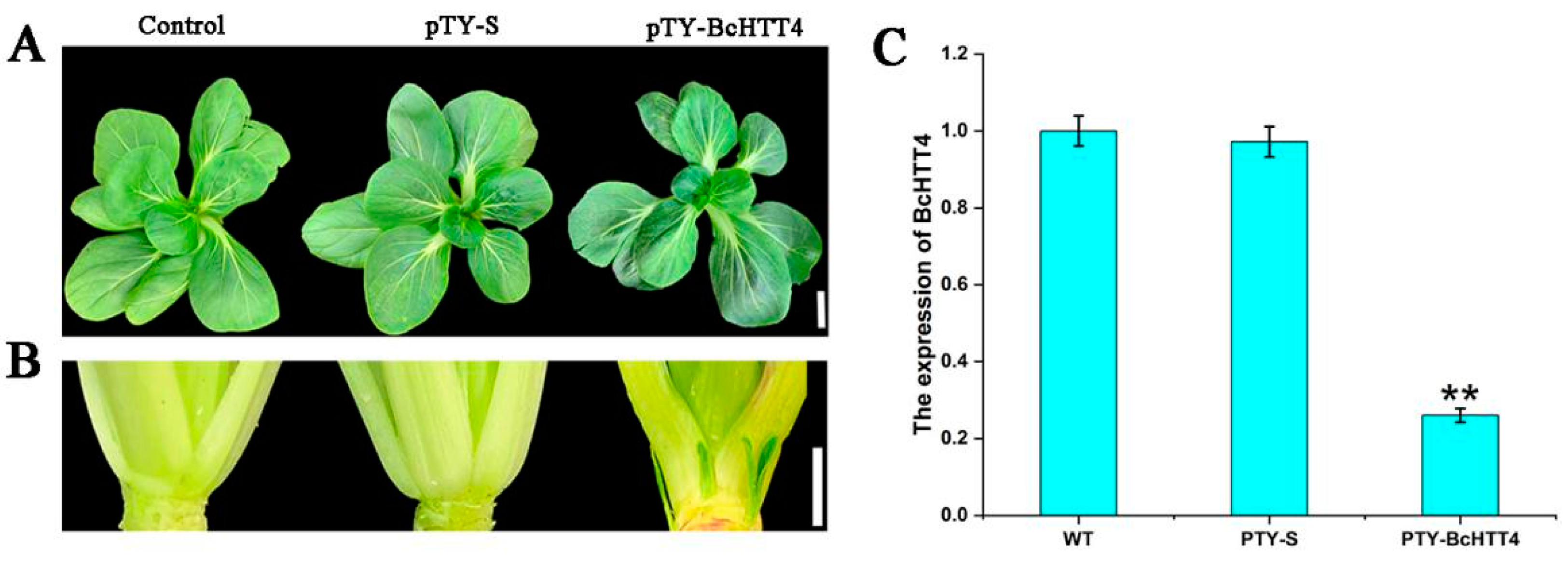
2.3. Silencing of the BcHTT4 Expression Promotes Branching of ‘Suzhouqing’
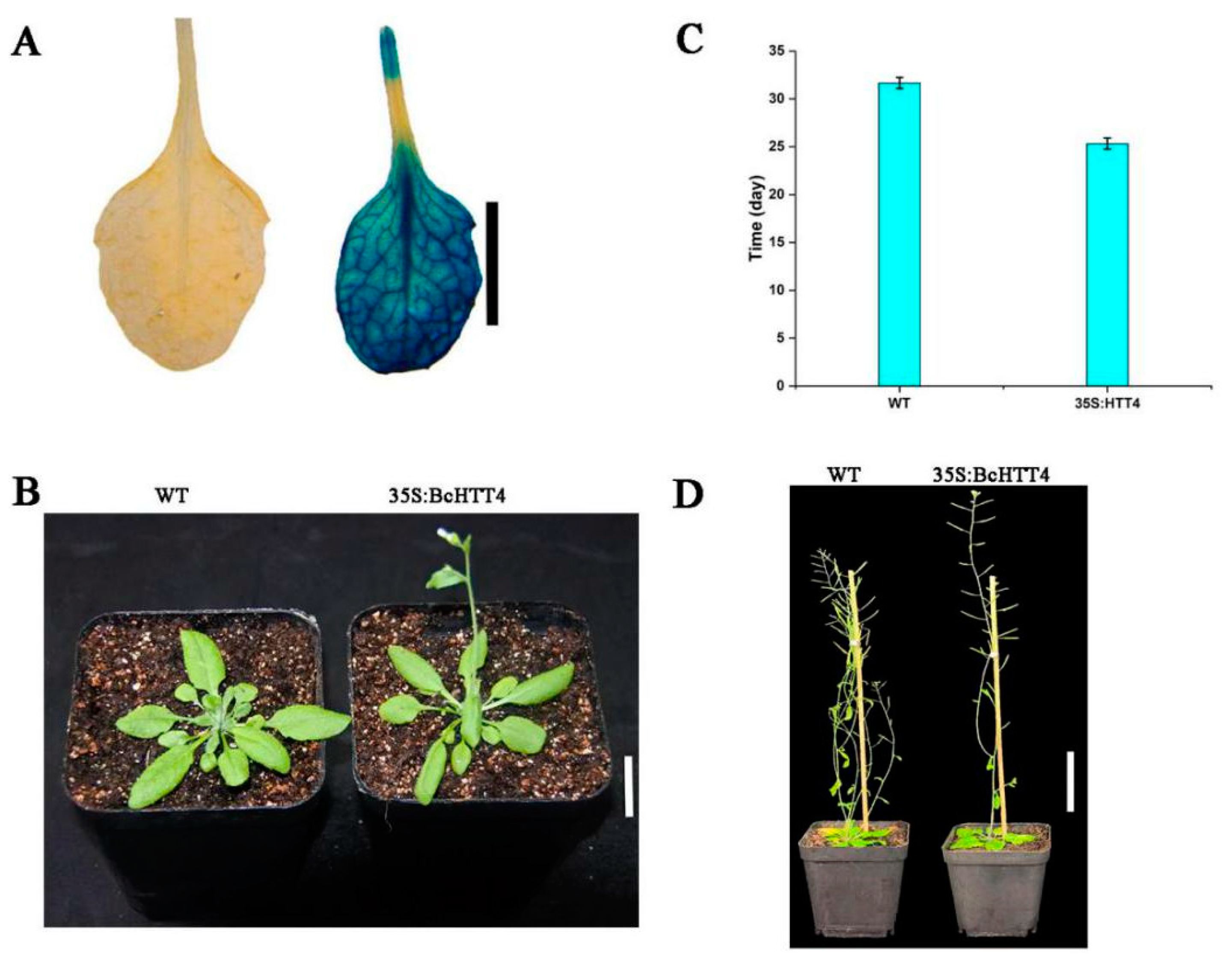
2.4. Overexpression of BcHTT4 in Arabidopsis Resulted in Early Flowering and Branching Inhibition
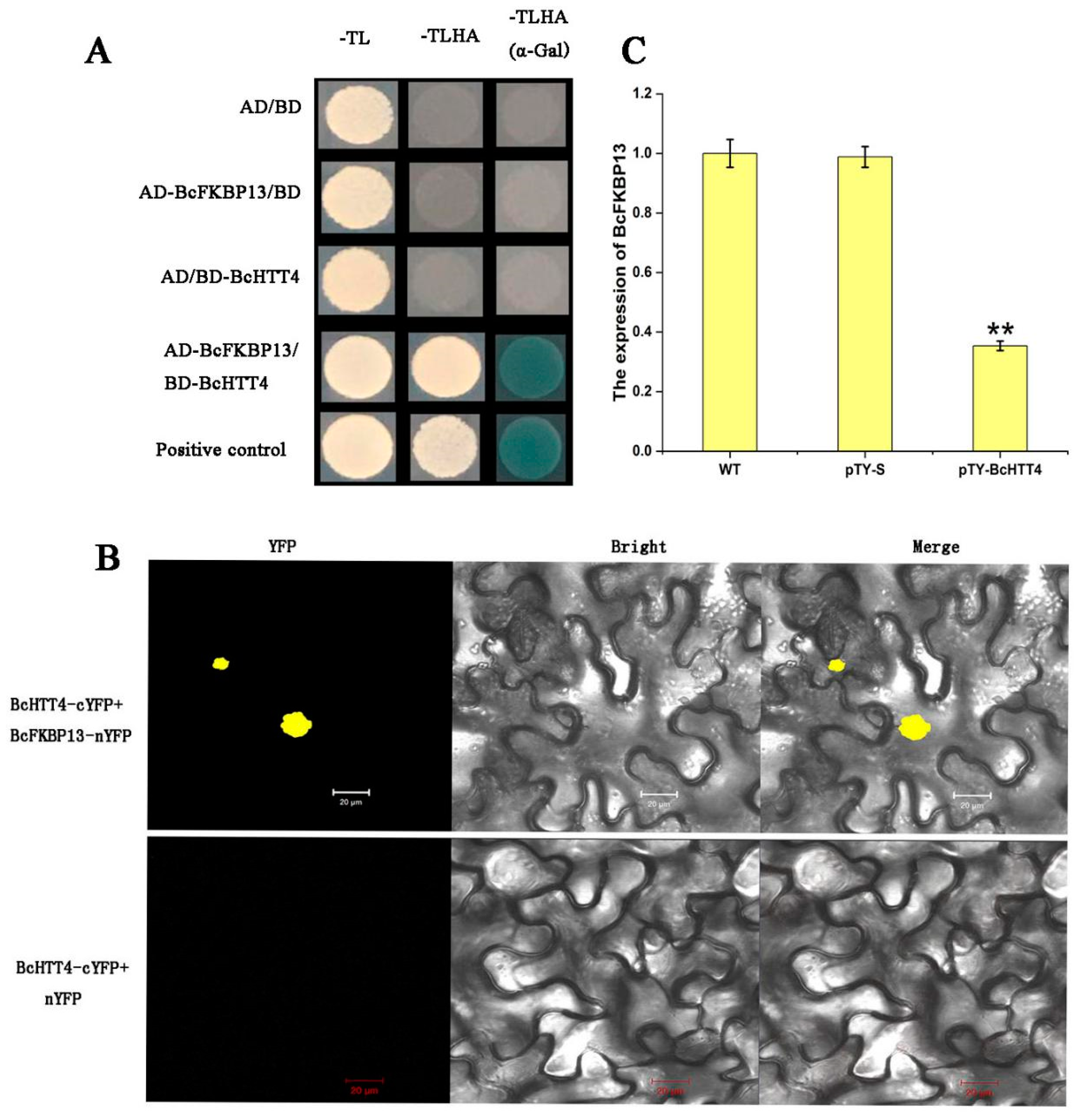
2.5. BcHTT4 Interacts with BcFKBP13
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Tissue Sampling of ‘Suzhouqing’ and ‘Maertou’
4.3. Quantitative Real-Time PCR
4.4. Acquisition of BcHTT4 Sequence
4.5. Subcellular Localization of BcHTT4-GFP Fusions
4.6. Silencing BcHTT4 Expression by VIGS Technology
4.7. Arabidopsis Transgenic Vector Construction and Transformation
4.8. The GUS Staining
4.9. Yeast Two-Hybrid Assay
4.10. BiFC Assays in Tobacco
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Doebley, J.F.; Gaut, B.S.; Smith, B.D. The molecular genetics of crop domestication. Cell 2006, 127, 1309–1321. [Google Scholar] [CrossRef] [Green Version]
- Doust, A.N.; Kellogg, E.A. Effect of genotype and environment on branching in weedy green millet (Setaria viridis) and domesticated foxtail millet (Setaria italica) (Poaceae). Mol. Ecol. 2006, 15, 1335–1349. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Zhang, Q. Genetic and molecular bases of rice yield. Ann. Rev. Plant Biol. 2010, 61, 421–442. [Google Scholar] [CrossRef]
- Li, X.; Qian, Q.; Fu, Z.; Wang, Y.; Xiong, G.; Zeng, D.; Wang, X.; Liu, X.; Teng, S.; Hiroshi, F.; et al. Control of tillering in rice. Nature 2003, 422, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Smith, S.M.; Li, J. Genetic regulation of shoot architecture. Annu. Rev. Plant Biol. 2018, 69, 437–468. [Google Scholar] [CrossRef]
- Stirnberg, P.; Furner, I.J.; Ottoline Leyser, H.M. MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 2007, 50, 80–94. [Google Scholar] [CrossRef]
- Booker, J.; Sieberer, T.; Wright, W.; Williamson, L.; Willett, B.; Stirnberg, P.; Turnbull, C.; Srinivasan, M.; Goddard, P.; Leyser, O. MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid derived branch-inhibiting hormone. Dev. Cell 2005, 8, 443–449. [Google Scholar] [CrossRef] [Green Version]
- Cai, T.; Meng, X.; Liu, X.; Liu, T.; Wang, H.; Jia, Z.; Yang, D.; Ren, X. Exogenous hormonal application regulates the occurrence of wheat tillers by changing endogenous hormones. Front. Plant Sci. 2018, 9, 1886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, S.E.; Cox, M.C.; Ross, J.J.; Krisantini, S.; Beveridge, C.A. Auxin dynamics after decapitation are not correlated with the initial growth of axillary buds. Plant Physiol. 2005, 138, 1665–1672. [Google Scholar] [CrossRef] [Green Version]
- Prusinkiewicz, P.; Crawford, S.; Smith, R.S.; Ljung, K.; Bennett, T.; Ongaro, V.; Leyser, O. Control of bud activation by an auxin transport switch. Proc. Nat. Acad. Sci. USA 2009, 106, 17431–17436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.K.; van Oosterom, E.; Dingkuhn, M.; Luquet, D.; Hammer, G. Regulation of tillering in sorghum: Environmental effects. Ann. Bot. 2010, 106, 57–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leduc, N.; Roman, H.; Barbier, F.; Péron, T.; Huché-Thélier, L.; Lothier, J.; Demotes-Mainard, S.; Sakr, S. Light signaling in bud outgrowth and branching in plants. Plants 2014, 3, 223–250. [Google Scholar] [CrossRef] [Green Version]
- Barbier, F.F.; Lunn, J.E.; Beveridge, C.A. Ready, steady, go! A sugar hit starts the race to shoot branching. Curr. Opin. Plant Biol. 2015, 25, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Evers, J.B. Sugar as a key component of the shoot branching regulation network. Plant Cell Environ. 2015, 38, 1455–1456. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.; Yu, H.; Duan, J.; Yuan, K.; Yu, C.; Meng, X.; Kou, L.; Chen, M.; Jing, Y.; Liu, G.; et al. SLR1 inhibits MOC1 degradation to coordinate tiller number and plant height in rice. Nat. Commun. 2019, 10, 2738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, G.; Lu, Z.; Xiong, J.; Wang, B.; Jing, Y.; Meng, X.; Liu, G.; Ma, H.; Liang, Y.; Chen, F.; et al. Tiller bud formation regulators MOC1 and MOC3 cooperatively promote tiller bud outgrowth by activating FON1 expression in rice. Mol. Plant 2019, 12, 1090–1102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.; Wang, R.; Qian, Q.; Yan, M.; Meng, X.; Fu, Z.; Yan, C.; Jiang, B.; Su, Z.; Li, J.; et al. DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 2009, 21, 1512–1525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greb, T.; Clarenz, O.; Schafer, E.; Muller, D.; Herrero, R.; Schmitz, G.; Theres, K. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 2003, 17, 1175–1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, F.; Wang, Q.; Schmitz, G.; Müller, D.; Theres, K. The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. Plant J. 2012, 71, 61–70. [Google Scholar] [CrossRef]
- Stirnberg, P.; Chatfield, S.P.; Leyser, H.M. AXR1 acts after lateral bud formation to inhibit lateral bud growth in Arabidopsis. Plant Physiol. 1999, 121, 839–847. [Google Scholar] [CrossRef] [Green Version]
- Stirnberg, P.; Zhao, S.; Williamson, L.; Ward, S.; Leyser, O. FHY3 promotes shoot branching and stress tolerance in Arabidopsis in an AXR1-dependent manner. Plant J. 2012, 71, 907–920. [Google Scholar] [CrossRef]
- Takeda, T.; Suwa, Y.; Suzuki, M.; Kitano, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Matsuoka, M.; Ueguchi, C. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 2003, 33, 513–520. [Google Scholar] [CrossRef]
- Cao, X.; Cui, H.; Li, J.; Xiong, A.; Hou, X.; Li, Y. Heritability and gene effects for tiller number and leaf number in non-heading Chinese cabbage estimated by joint segregation analysis. Scientia Horticulturae 2016, 203, 199–206. [Google Scholar] [CrossRef]
- Kumagai, M.H.; Donson, J.; della-Cioppa, G.; Harvey, D.; Hanley, K.; Grill, L.K. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc. Natl. Acad. Sci. USA 1995, 92, 1679–1683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baulcombe, D.C. RNA as a target and an initiator of posttranscriptional gene silencing in transgenic plants. Plant Mol. Biol. 1996, 32, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Baulcombe, D.C. Fast forward genetics based on virus induced gene silencing. Curr. Opin. Plant Biol. 1999, 2, 109–113. [Google Scholar] [CrossRef]
- Ramachandran, V.; Chen, X. Small RNA metabolism in Arabidopsis. Trends Plant Sci. 2008, 13, 368–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carthew, R.W.; Sontheimer, E.J. Origins and mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doench, J.G.; Petersen, C.P.; Sharp, P.A. siRNAs can function as miRNAs. Genes Dev. 2003, 17, 438–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef] [Green Version]
- Valencia-Sanchez, M.A.; Liu, J.; Hannon, G.J.; Parker, R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006, 20, 515–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kume, K.; Tsutsumi, K.; Saitoh, Y. TAS1 trans-acting siRNA targets are differentially regulated at low temperature, and TAS1 trans-acting siRNA mediates temperature-controlled At1g51670 expression. Biosci. Biotechnol. Biochem. 2010, 74, 1435–1440. [Google Scholar] [CrossRef] [Green Version]
- Vazquez, F.; Vaucheret, H.; Rajagopalan, R.; Lepers, C.; Gasciolli, V.; Mallory, A.C.; Hilbert, J.L.; Bartel, D.P.; Crété, P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 2004, 16, 69–79. [Google Scholar] [CrossRef]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mourelatos, Z. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002, 16, 720–728. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Rhoades, M.W.; Reinhart, B.J.; Lim, L.P.; Burge, C.B.; Bartel, B.; Bartel, D.P. Prediction of plant microRNA targets. Cell 2002, 110, 513–520. [Google Scholar] [CrossRef] [Green Version]
- Xie, K.B.; Wu, C.; Xiong, L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006, 14, 280–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, H.G.; Kirby, E.J.M. Effects of manipulation of number of tillers and water supply on grain yield in barley. J. Agric. Sci. 1977, 88, 391–397. [Google Scholar] [CrossRef]
- Schumacher, K.; Schmitt, T.; Rossberg, M.; Schmitz, G.; Theres, K. The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. USA 1999, 96, 290–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Liu, J.; Liu, Z.; Li, X.; Wu, F.; He, Y. Heat-induced TAS1 target1 mediates thermotolerance via heat stress transcription factor A1a-directed pathways in Arabidopsis. Plant Cell 2014, 26, 1764–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schreiber, S.L. Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science 1991, 251, 283–287. [Google Scholar] [CrossRef]
- Paić, A.T.; Fulgosi, H. Chloroplast immunophilins. Protoplasma 2016, 253, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Bailly, A.; Sovero, V.; Geisler, M. The twisted dwarf’s ABC: How immunophilins regulate auxin transport. Plant Signal. Behav. 2006, 1, 277–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aviezer-Hagai, K.; Skovorodnikova, J.; Galigniana, M.; Farchi-Pisanty, O.; Maayan, E.; Bocovza, S.; Efrat, Y.; von Koskull-Döring, P.; Ohad, N.; Breiman, A. Arabidopsis immunophilins ROF1 (AtFKBP62) and ROF2 (AtFKBP65) exhibit tissue specificity, are heat-stress induced, and bind HSP90. Plant Mol. Biol. 2007, 63, 237–255. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, C.; Yang, D.; Yu, H.; Liou, Y.C. Pin1At encoding a peptidyl-prolyl cis/trans isomerase regulates flowering time in Arabidopsis. Mol. Cell 2010, 37, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Edvardsson, A.; Shapiguzov, A.; Petersson, U.A.; Schröder, W.P.; Vener, A.V. Immunophilin AtFKBP13 sustains all peptidyl-prolyl isomerase activity in the thylakoid lumen from Arabidopsis thaliana deficient in AtCYP20-2. Biochemistry 2007, 46, 9432–9442. [Google Scholar] [CrossRef]
- Gupta, R.; Mould, R.M.; He, Z.; Luan, S. A chloroplast FKBP interacts with and affects the accumulation of Rieske subunit of cytochrome bf complex. Proc. Natl. Acad. Sci. USA 2002, 99, 15806–15811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Niwa, M.; Daimon, Y.; Kurotani, K.; Higo, A.; Pruneda-Paz, J.L.; Breton, G.; Mitsuda, N.; Kay, S.A.; Ohme-Takagi, M.; Endo, M.; et al. BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in Arabidopsis. Plant Cell 2013, 25, 1228–1242. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, M.; Xu, L.; Long, Y.; Huang, F.; Liu, T.; Li, Y.; Hou, X. BcHTT4 Inhibits Branching of Non-Heading Chinese Cabbage at the Vegetative Stage. Plants 2021, 10, 510. https://doi.org/10.3390/plants10030510
Guo M, Xu L, Long Y, Huang F, Liu T, Li Y, Hou X. BcHTT4 Inhibits Branching of Non-Heading Chinese Cabbage at the Vegetative Stage. Plants. 2021; 10(3):510. https://doi.org/10.3390/plants10030510
Chicago/Turabian StyleGuo, Mingliang, Lanlan Xu, Yan Long, Feiyi Huang, Tongkun Liu, Ying Li, and Xilin Hou. 2021. "BcHTT4 Inhibits Branching of Non-Heading Chinese Cabbage at the Vegetative Stage" Plants 10, no. 3: 510. https://doi.org/10.3390/plants10030510





