1. Introduction
Peach (
Prunus persica L.) is one of the most economically important fruits, providing plentiful vitamins, minerals, fiber and antioxidant compounds for healthy diets. Peach is also acknowledged as an ideal model for genetics and genomics studies of tree fruit species. Peach originated about 2.5 millions of years ago (Mya) in the southwest range of the Tibetan Plateau in China, and its cultivation and domestication in China can be traced back to 4000 years ago [
1]. More than 1000 cultivars of
Prunus persica L. (
P. persica) were produced worldwide until now, with significant phenotypic changes in internal and external characteristics, such as fruit shape, fruit size, flavor, and flower type. However, due to selfing as well as important bottlenecks in its recent breeding history, peach has a lower level of genetic variability compared with other Prunus crops. The edible
Prunus ferganensis (P. ferganensis), which is native to arid regions of central Asia and featured with long unbranched leaf veins and longitudinal grooves on the pit, was a close relative of cultivated peach, and classified as a species currently [
2,
3,
4].
The flat peach was cultivated in China two thousand years ago and introduced to Western countries from China in the seventeenth century. Featuring a saucer fruit shape, flat peach was previously supposed to be a natural mutation variety of round peach [
5,
6]. In early era, the flat fruit shape trait was negatively selected in most breeding programs in 70 western countries due to its effects on fruit size and yield [
7]. However, compared with most round peaches, flat peaches demonstrated special germplasm characteristics for multiple unique or high quality traits [
8]. Most flat peach varieties have a sweet taste, low titratable acidity (less than 0.4%), very high sugar content and rich flavor (soluble solids content of 12–14% and soluble sugars content of 9.01 and 10.69%), these high quality traits attracted more and more interest from consumers and breeders [
8]. Compared with most native cultivars, the newly-bred varieties had a better quality, wider ripening period and improved fruit weight, especially richer fruit aroma. The mixture of terpene volatiles released by the peach fruits contained high levels of terpenes linalool [
9]. Among these improved cultivars, ‘Sahuahongpantao’, ‘Wanshudapantao’ and ‘124 Pan’ were proved to be good flat peach cultivars [
8]. Although
P. ferganensis has a flat shape, its fruit quality is not of commercial standard in terms of fruit firmness, quality, and skin color.
As a diploid species (2
n = 16) with a small genome (approximately 220 Mb, about twice that of Arabidopsis), peach was used as a model fruit species in comparative and functional genomics, especially Rosaceae family [
10]. The peach genome released by the International Peach Genome Initiative provides a foundation for population analyses of peach [
11]. In a previous study, we explored the reason for the flat fruit shape in peach and proposed that a 1.7 Mb chromosomal inversion downstream of a
P. persica OVATE family protein 1 (PpOFP1) was responsible for the flat fruit shape in peach [
12]. However, the reason why flat peach has a higher fruit quality remains unclear. To better exploit the resource of flat peach, the availability of whole-genome sequences is crucial.
To investigate the genome of flat peach, the cultivar ‘124 Pan’ was used for whole-genome sequencing and assembling. As a cross breeding variety produced by the Institute of Agricultural Science in Lixia-he Area of Jiangsu Province in 1957, ‘124 Pan’ was characterized by high fruit yield and quality, such as low acidity, high sugar content and rich flavor. Based on PacBio and Illumina reads with approximately 80-fold coverage of peach reference, we assembled a draft genome of ‘124 Pan’ by using a hybrid assembly algorithm. We compared the genome of ‘124 Pan’ with peach Lovell reference and discovered a large number of single nucleotide polymorphisms (SNPs), insertions or deletions of DNA segments (InDels), and structural variations (SVs). Through a series of comparative analysis, we confirmed that P. ferganensis was the ancestor of the domesticated peach. Gene family comparison revealed the expansion of terpene synthase genes (TPSs) in ‘124 Pan’, which might contribute to its good fruit flavor traits. This flat peach draft genome assembly provides an extra resource for peach improvement and comparative genomics research.
3. Discussion
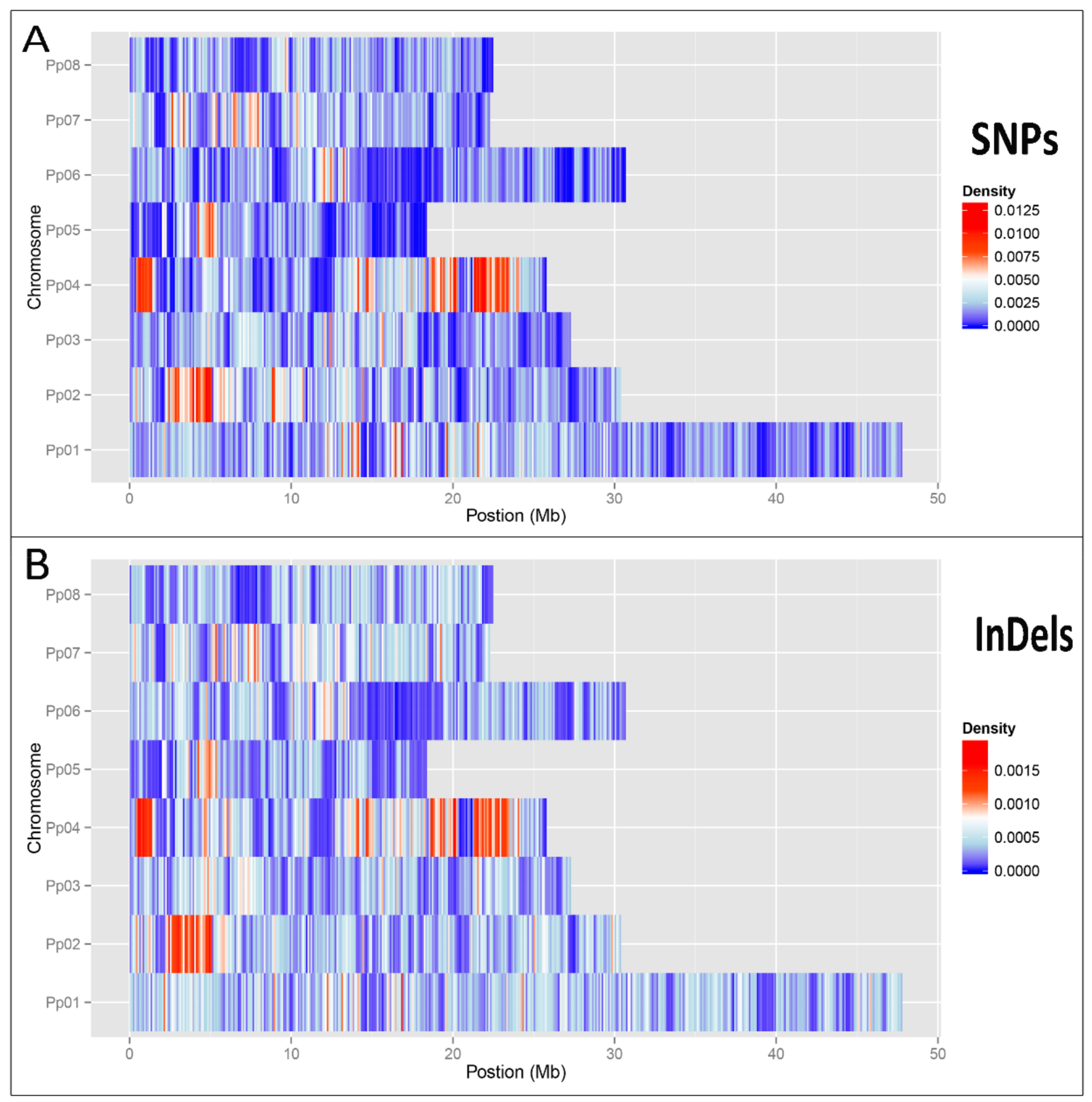
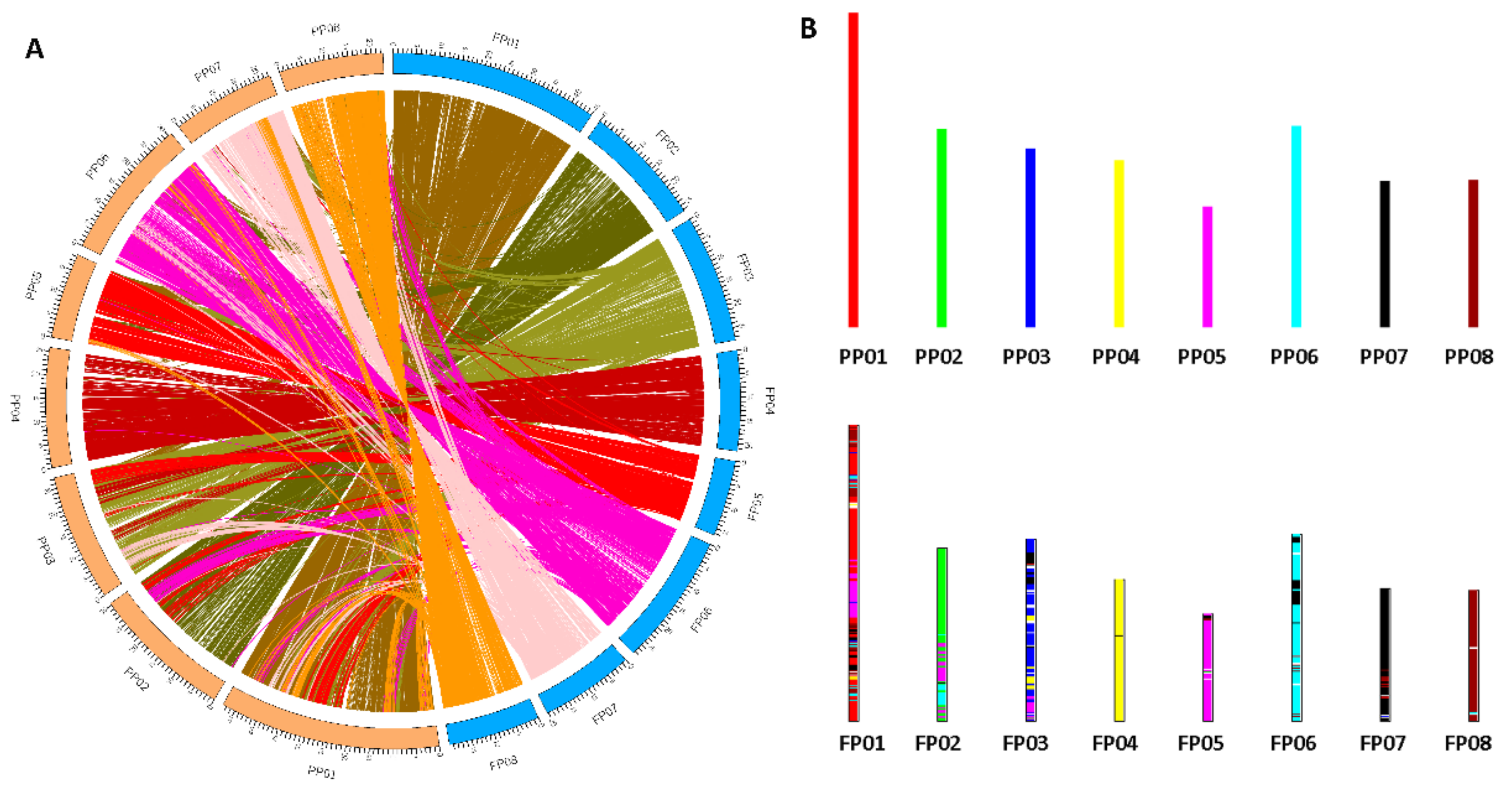
In this study, to explore the resource of flat peach, we obtained the draft genome of one representative flat peach ‘124 Pan’ based on combination of PacBio and Illumina reads. The flat peach genome offers an opportunity to comprehensively investigate the genome variations at the resolution of nucleotide. We characterized a comprehensive catalog of structure variations, including 95,124 InDels, 533,357 SNPs, and 18,422 SVs. These genome variations constitute a major resource of genomic variation and are known to have profound consequences on phenotypic variation [
17]. In plants, molecular genetic analyses have highlighted the functional importance of SVs on protein-coding and flanking noncoding regions of loci/genes linked to agriculturally important traits [
17,
18], the chromosomal inversion located in chromosome 6, with a size of 1.7 Mb, was proved to be responsible for the flat fruit shape in the peach [
12]. The genome comparison revealed that although ‘124 Pan’ and peach Lovell shared basically the same chromosome structures and organization, there were still frequent synteny blocks presented between different chromosomes, highlighting the chromosome rearrangements events in ‘124 Pan’. Analysis of Ks distribution within ‘124 Pan’ genome revealed that there was no recent WGD except a triplicated arrangement (ancestral γ event), which is consistent with previous studies [
2,
11], the self-collinearity within the genome further support this point. The phylogenetic tree based on single copy genes confirmed that
P. ferganensis was the ancestor of the domesticated peach, providing an additional evidence to support the assumption that peach domestication occurred in the region of Northwest China. The short divergence time of peach cultivars from ancestor revealed that although peach had gone through evolution by artificial and natural selection, the domesticated peach kept limited differentiation from
P. ferganensis. Considering the same fruit shape between the cultivated flat peach and
P. ferganensis, the flat fruit trait is more likely to have originally occurred in
P. ferganensis and then been introduced to peach cultivars. During artificial and natural selection, the round fruit trait occurred and was selected because the round peach attracted more attention owing to its larger fruit size and higher yield, the ancient flat fruit trait was later introduced to round cultivars to breed the modern flat peach with high flavor quality.
With KEGG enrichment analysis, we found several gene family expansions in the ‘124 Pan’ genome. For the top enriched term of terpene synthesis pathway, more copies of TPSs were detected in the ‘124 Pan’ genome than that of Lovell and
P. ferganensis. Peach TPSs were largely located in chromosome 4 that suffered drastic structural variations. For Lovell and
P. ferganensis, TPSs were mostly located in chromosomes 4 and 3, whereas for ‘124 Pan’, additional TPSs were detected in chromosomes 1 and 2. Considering the frequent structural variation in the peach genome, we speculate that higher numbers of TPSs in flat peach are shaped by a large segment duplication and rearrangements during domestication [
16]. Although Prunoideae has not experienced recent whole-genome duplication events, as observed in apple and pear [
19,
20], there were many translocations, large chromosomal inversions, and segment duplication regions in the genome, and the resulting chromosome rearrangements may play a key role in the speciation of Prunoideae and specialization of cultivars. For the very short evolutionary history of the modern peach, the diversity of proteins among the three peach genomes is actually small and the branch lengths in the phylogenetic tree (
Figure 6) are mostly very short. Although the copy number of TPSs in flat peach is increased, the amino acid variation remains still low.
The phenomenon of varied numbers of TPSs among different peach genomes is a typical gene copy number variant (CNV). CNVs are genomic rearrangements resulting from gains or losses of DNA segments, and usually produced by transposable elements mediated nonallelic homologous recombination [
21]. In plants, CNVs were mostly associated with tandem duplications and tend to occur in large families of functionally redundant genes [
21]. There are demonstrated cases in which CNVs contribute to domestication and diversification traits by affecting relevant gene dosage, function(s), and/or regulation, such as rice, maize, and potato [
21,
22]. For TPSs, previous expression analysis showed that most duplicated copies exhibited divergent expression patterns either in tissues or transcript intensity, indicating that expression divergence significantly contributed to TPSs survival after gene expansion by duplication [
16]. In addition, sequence variations within duplicated genes could generate protein diversity, hence, CNV may have enormous potential as a source of useful traits to improve cultivars [
21].
Fruit flavor is a complex trait that depends on the relative amount of sugars, non-volatiles, and volatiles (terpenes, phenylalanine- and fatty acid-derived compounds) [
23]. In plants, low-molecular-weight terpenes produced by TPSs are a large group of plant aromatic substances, in which monoterpenes and a few sesquiterpenes are aromatic constituents of many plants [
24]. As a member of terpenes, linalool was proved to be the highest content of terpenes aromatic constituents in peach fruits [
25,
26]. Previous studies demonstrated that TPSs played an important role in determining the quality of horticultural food products [
23,
27,
28]. A recent study also demonstrated the contribution of two TPSs located in chromosome 4 to carrot flavor [
29]. Till now, few studies have focused on the terpenoid volatiles and peach flavors traits. Our study provided a clue to the differences of the flavor quality among peach varieties, we speculate that the striking feature of the existence of an unusually large TPSs family related to terpene biosynthesis might contribute to the fruit flavor and aroma for ‘124 Pan’. However, the distribution of TPSs among various varieties and the genetic basis underlying this trait still need further investigation in future studies. What is more, terpenoids also play numerous roles in the interactions of plants with their environment, such as attracting pollinators and defending the plant against pathogens and herbivores [
24]. The plant–pathogen interaction pathway was also enriched in our study, hence, other probable relations of TPSs in peach cultivar with the improved defenses also need further investigation.