Overexpression of Type 1 and 2 Diacylglycerol Acyltransferase Genes (JcDGAT1 and JcDGAT2) Enhances Oil Production in the Woody Perennial Biofuel Plant Jatropha curcas
Abstract
1. Introduction
2. Results
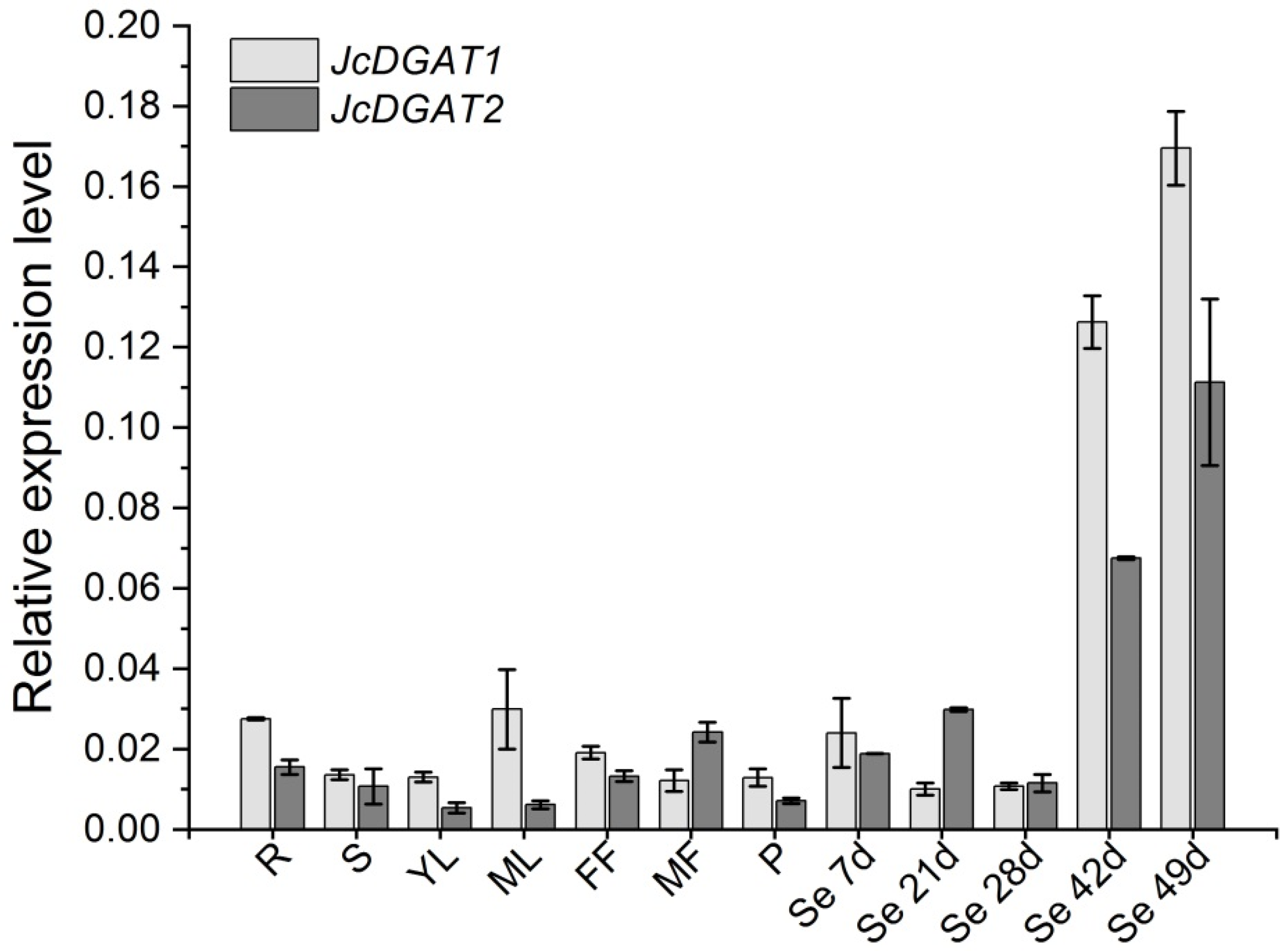
2.1. JcDGAT1 and JcDGAT2 are Highly Expressed at the Late Stages of Seed Development
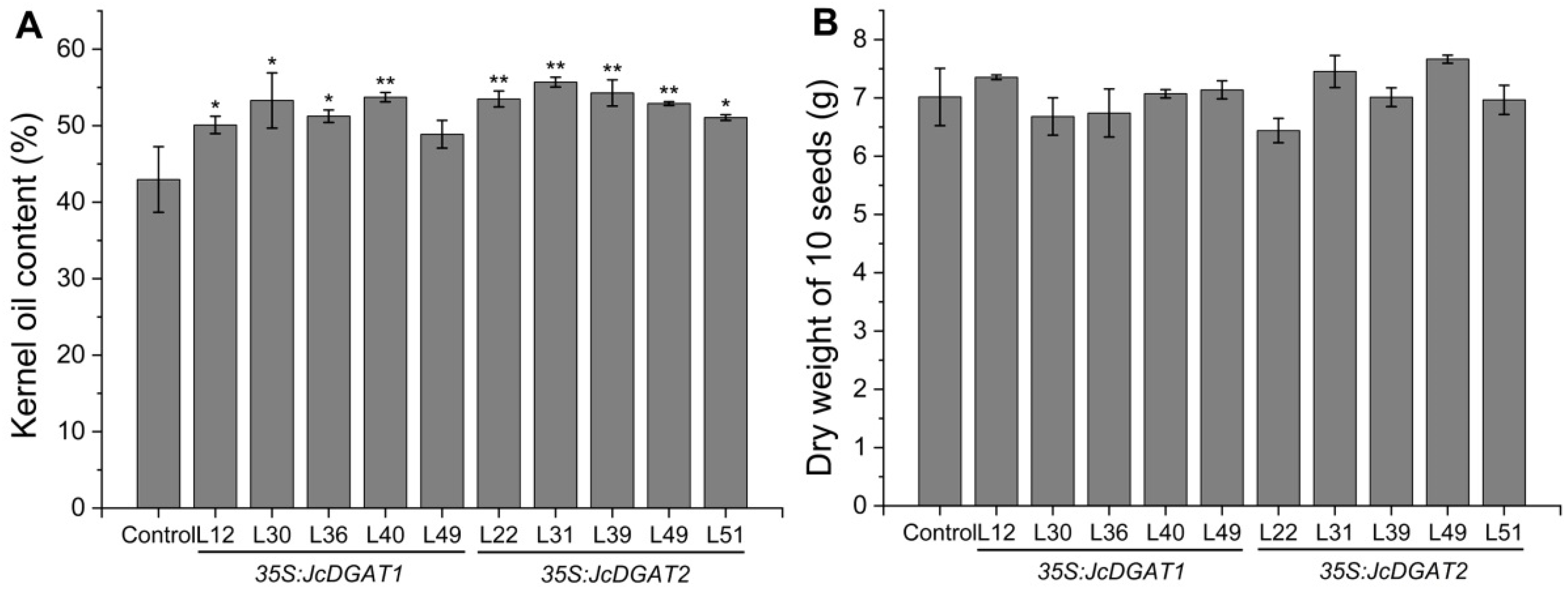
2.2. Overexpression of JcDGAT1 and JcDGAT2 Enhanced Seed Oil Production in Transgenic J. curcas
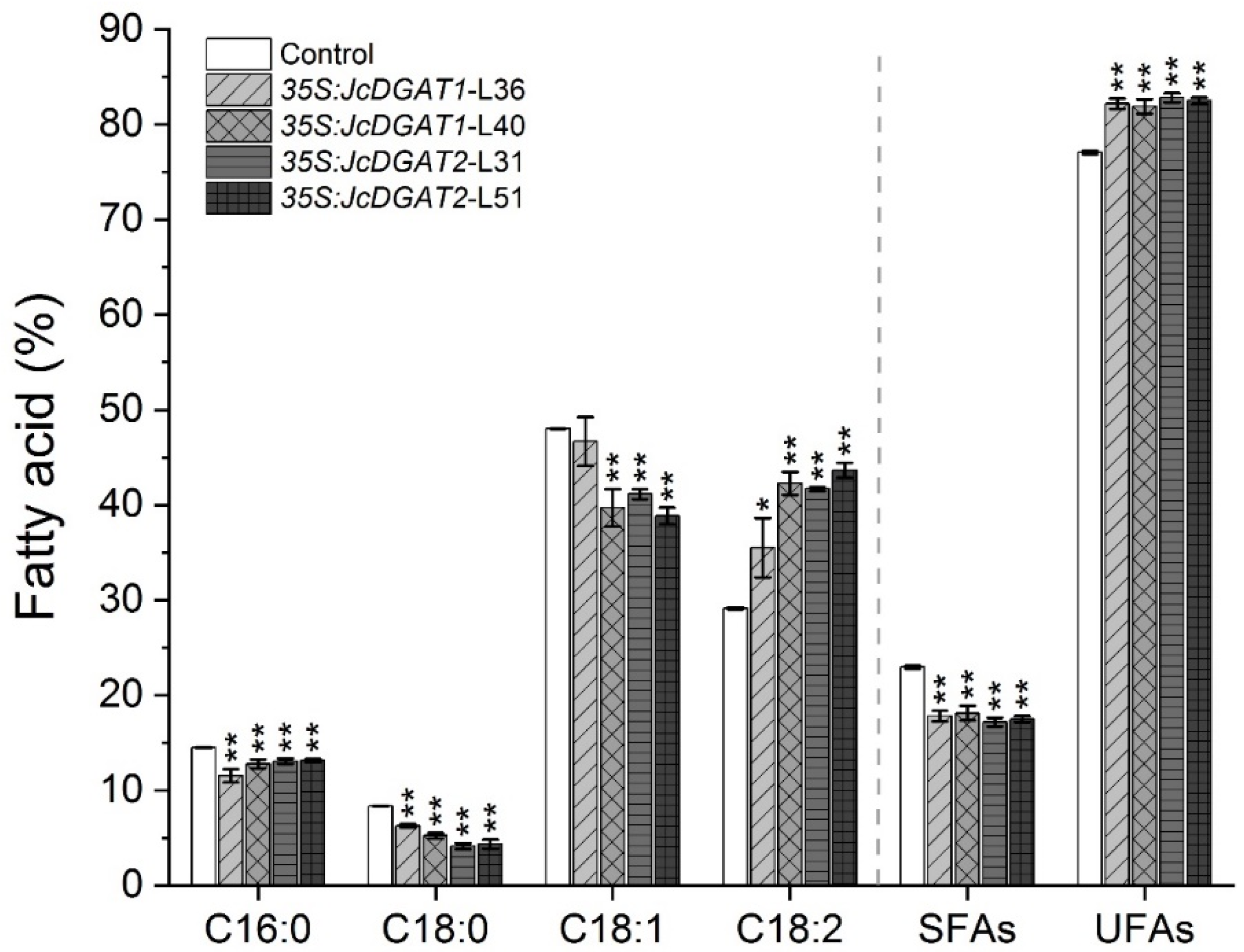
2.3. FA Compositions Significantly Changed in Seed Oil of Transgenic J. curcas
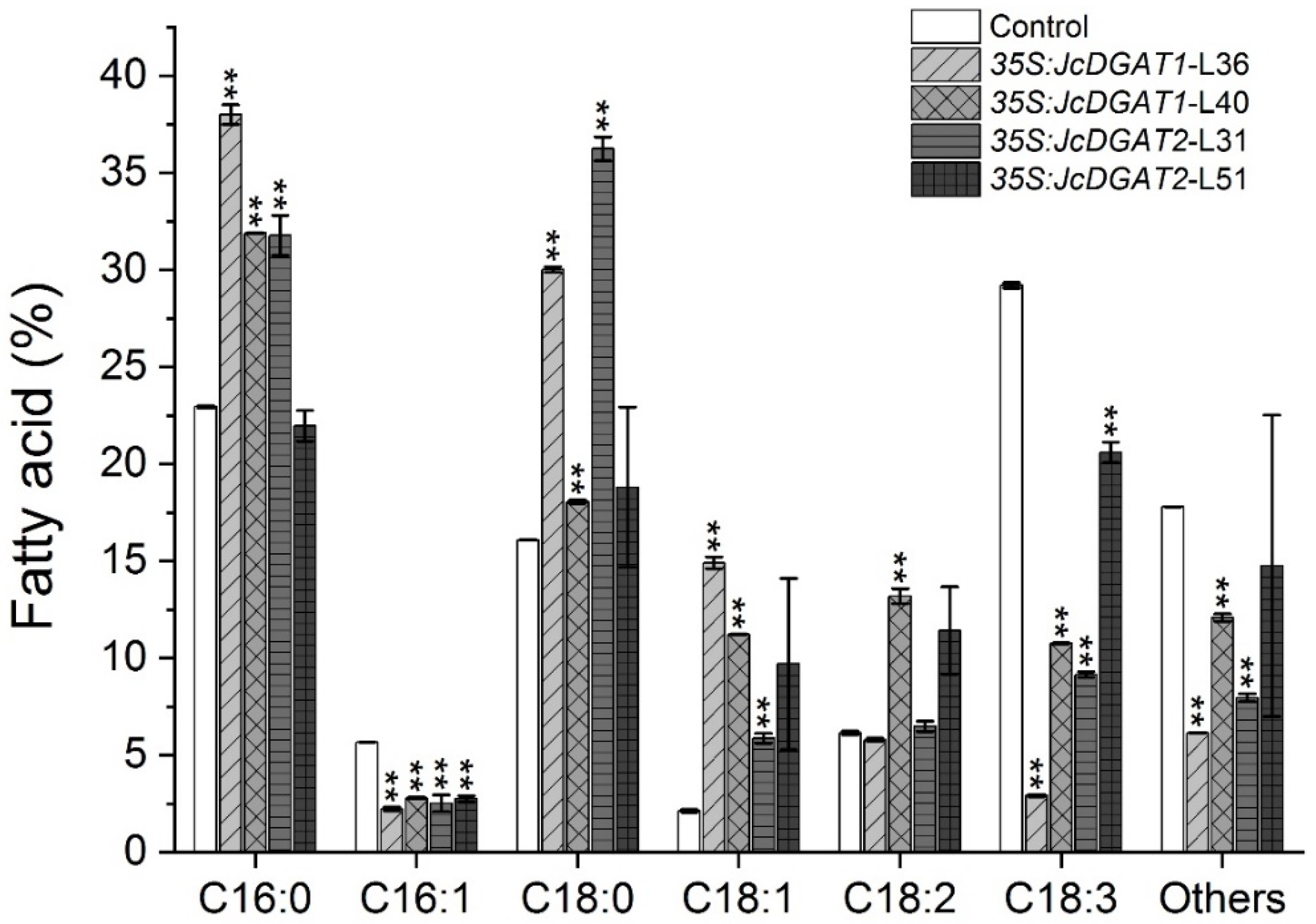
2.4. Overexpression of JcDGAT1 and JcDGAT2 Altered TAG Accumulation and FA Compositions in Transgenic J. curcas Leaves
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Plant Transformation
4.3. qRT-PCR Analysis
4.4. Lipid Analysis
4.5. TAG Analysis in Leaves
4.6. Quantifications of Protein, Starch, and Soluble Sugar
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, V.; Tripathi, A.M.; Tak, P.K.; Chouhan, S. Tree or shrub Jatropha curcas L.: Biofuel and potential herb. J. Biofuels 2016, 7, 89–101. [Google Scholar] [CrossRef]
- Abdulla, R.; Chan, E.S.; Ravindra, P. Biodiesel production from Jatropha curcas: A critical review. Crit. Rev. Biotechnol. 2011, 31, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Wu, P.; Zhang, S.; Song, C.; Chen, Y.; Li, M.; Jia, Y.; Fang, X.; Chen, F.; Wu, G. Global analysis of gene expression profiles in developing physic nut (Jatropha curcas L.) seeds. PLoS ONE 2012, 7, e36522. [Google Scholar] [CrossRef] [PubMed]
- Vaknin, Y.; Ghanim, M.; Samra, S.; Dvash, L.; Hendelsman, E.; Eisikowitch, D.; Samocha, Y. Predicting Jatropha curcas seed-oil content, oil composition and protein content using near-infrared spectroscopy—A quick and non-destructive method. Ind. Crop. Prod. 2011, 34, 1029–1034. [Google Scholar] [CrossRef]
- Jonas, M.; Ketlogetswe, C.; Gandure, J. Variation of Jatropha curcas seed oil content and fatty acid composition with fruit maturity stage. Heliyon 2020, 6, e03285. [Google Scholar] [CrossRef]
- Augustus, G.D.P.S.; Jayabalana, M.; Seilerb, G.J. Evaluation and bioinduction of energy components of Jatropha curcas. Biomass Bioenergy 2002, 23, 161–164. [Google Scholar] [CrossRef]
- Axelsson, L.; Franzén, M.; Ostwald, M.; Berndes, G.; Lakshmi, G.; Ravindranath, N.H. Jatropha cultivation in southern India: Assessing farmers’ experiences. Biofuels Bioprod. Biorefin. 2012, 6, 246–256. [Google Scholar] [CrossRef]
- Openshaw, K. A review of Jatropha curcas: An oil plant of unfulfilled promise. Biomass Bioenergy 2000, 19, 1–15. [Google Scholar] [CrossRef]
- Pan, B.-Z.; Xu, Z.-F. Benzyladenine treatment significantly increases the seed yield of the biofuel plant Jatropha curcas. J. Plant Growth Regul. 2011, 30, 166–174. [Google Scholar] [CrossRef]
- Barnwal, B.K.; Sharma, M.P. Prospects of biodiesel production from vegetable oils in India. Renew. Sustain. Energy Rev. 2005, 9, 363–378. [Google Scholar] [CrossRef]
- Bates, P.D. Understanding the control of acyl flux through the lipid metabolic network of plant oil biosynthesis. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2016, 1861, 1214–1225. [Google Scholar] [CrossRef] [PubMed]
- Lung, S.C.; Weselake, R.J. Diacylglycerol acyltransferase: A key mediator of plant triacylglycerol synthesis. Lipids 2006, 41, 1073–1088. [Google Scholar] [CrossRef] [PubMed]
- Turchetto-Zolet, A.C.; Maraschin, F.S.; de Morais, G.L.; Cagliari, A.; Andrade, C.M.B.; Marcia, M.-P.; Margis, R. Evolutionary view of acyl-CoA diacylglycerol acyltransferase (DGAT), a key enzyme in neutral lipid biosynthesis. BMC Evol. Biol. 2011, 11, 263. [Google Scholar] [CrossRef] [PubMed]
- Cao, H. Structure-function analysis of diacylglycerol acyltransferase sequences from 70 organisms. BMC Res. Notes 2011, 4, 249. [Google Scholar] [CrossRef] [PubMed]
- Shockey, J.M.; Gidda, S.K.; Chapital, D.C.; Kuan, J.-C.; Dhanoa, P.K.; Bland, J.M.; Rothstein, S.J.; Mullen, R.T.; Dyer, J.M. Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 2006, 18, 2294–2313. [Google Scholar] [CrossRef]
- Bhatt-Wessel, B.; Jordan, T.W.; Miller, J.H.; Peng, L. Role of DGAT enzymes in triacylglycerol metabolism. Arch. Biochem. Biophys. 2018, 655, 1–11. [Google Scholar] [CrossRef]
- Liu, Q.; Siloto, R.M.; Lehner, R.; Stone, S.J.; Weselake, R.J. Acyl-CoA:diacylglycerol acyltransferase: Molecular biology, biochemistry and biotechnology. Prog. Lipid Res. 2012, 51, 350–377. [Google Scholar] [CrossRef]
- Jako, C.; Kumar, A.; Wei, Y.; Zou, J.; Barton, D.L.; Giblin, E.M.; Covello, P.S.; Taylor, D.C. Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol. 2001, 126, 861–874. [Google Scholar] [CrossRef]
- Zhang, M.; Fan, J.; Taylor, D.C.; Ohlrogge, J.B. DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 2009, 21, 3885–3901. [Google Scholar] [CrossRef]
- Zhou, X.R.; Shrestha, P.; Yin, F.; Petrie, J.R.; Singh, S.P. AtDGAT2 is a functional acyl-CoA:diacylglycerol acyltransferase and displays different acyl-CoA substrate preferences than AtDGAT1. FEBS Lett. 2013, 587, 2371–2376. [Google Scholar] [CrossRef]
- Weselake, R.J.; Taylor, D.C.; Rahman, M.H.; Shah, S.; Laroche, A.; McVetty, P.B.E.; Harwood, J.L. Increasing the flow of carbon into seed oil. Biotechnol. Adv. 2009, 27, 866–878. [Google Scholar] [CrossRef]
- Wang, Z.; Huang, W.; Chang, J.; Sebastian, A.; Li, Y.; Li, H.; Wu, X.; Zhang, B.; Meng, F.; Li, W. Overexpression of SiDGAT1, a gene encoding acyl-CoA:diacylglycerol acyltransferase from Sesamum indicum L. increases oil content in transgenic Arabidopsis and soybean. Plant Cell Tissue Organ Cult. (PCTOC) 2014, 119, 399–410. [Google Scholar] [CrossRef]
- Xu, J.; Francis, T.; Mietkiewska, E.; Giblin, E.M.; Barton, D.L.; Zhang, Y.; Zhang, M.; Taylor, D.C. Cloning and characterization of an acyl-CoA-dependent diacylglycerol acyltransferase 1 (DGAT1) gene from Tropaeolum majus, and a study of the functional motifs of the DGAT protein using site-directed mutagenesis to modify enzyme activity and oil content. Plant Biotechnol. J. 2008, 6, 799–818. [Google Scholar] [CrossRef] [PubMed]
- Lock, Y.-Y.; Snyder, C.L.; Zhu, W.; Siloto, R.M.; Weselake, R.J.; Shah, S. Antisense suppression of type 1 diacylglycerol acyltransferase adversely affects plant development in Brassica napus. Physiol. Plant 2009, 137, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.-Y.; Yang, M.-F.; Xu, Y.-N. Silencing of DGAT1 in tobacco causes a reduction in seed oil content. Plant Sci. 2005, 169, 689–694. [Google Scholar] [CrossRef]
- Guiheneuf, F.; Leu, S.; Zarka, A.; Khozin-Goldberg, I.; Khalilov, I.; Boussiba, S. Cloning and molecular characterization of a novel acyl-CoA:diacylglycerol acyltransferase 1-like gene (PtDGAT1) from the diatom Phaeodactylum tricornutum. FEBS J. 2011, 278, 3651–3666. [Google Scholar] [CrossRef]
- Li, R.; Hatanaka, T.; Yu, K.; Wu, Y.; Fukushige, H.; Hildebrand, D. Soybean oil biosynthesis: Role of diacylglycerol acyltransferases. Funct. Integr. Genom. 2013, 13, 99–113. [Google Scholar] [CrossRef]
- Chen, B.; Wang, J.; Zhang, G.; Liu, J.; Manan, S.; Hu, H.; Zhao, J. Two types of soybean diacylglycerol acyltransferases are differentially involved in triacylglycerol biosynthesis and response to environmental stresses and hormones. Sci. Rep. 2016, 6, 28541. [Google Scholar] [CrossRef]
- Haslam, R.P.; Hamilton, M.L.; Economou, C.K.; Smith, R.; Hassall, K.L.; Napier, J.A.; Sayanova, O. Overexpression of an endogenous type 2 diacylglycerol acyltransferase in the marine diatom Phaeodactylum tricornutum enhances lipid production and omega-3 long-chain polyunsaturated fatty acid content. Biotechnol. Biofuels 2020, 13, 87. [Google Scholar] [CrossRef]
- Liu, D.; Ji, H.; Yang, Z. Functional characterization of three novel genes encoding diacylglycerol acyltransferase (DGAT) from oil-rich tubers of Cyperus esculentus. Plant Cell Physiol. 2020, 61, 118–129. [Google Scholar] [CrossRef]
- Burgal, J.; Shockey, J.; Lu, C.; Dyer, J.; Larson, T.; Graham, I.; Browse, J. Metabolic engineering of hydroxy fatty acid production in plants: RcDGAT2 drives dramatic increases in ricinoleate levels in seed oil. Plant Biotechnol. J. 2008, 6, 819–831. [Google Scholar] [CrossRef] [PubMed]
- Yurchenko, O.; Shockey, J.M.; Gidda, S.K.; Silver, M.I.; Chapman, K.D.; Mullen, R.T.; Dyer, J.M. Engineering the production of conjugated fatty acids in Arabidopsis thaliana leaves. Plant Biotechnol. J. 2017, 15, 1010–1023. [Google Scholar] [CrossRef]
- Li, R.; Yu, K.; Hatanaka, T.; Hildebrand, D.F. Vernonia DGATs increase accumulation of epoxy fatty acids in oil. Plant Biotechnol. J. 2010, 8, 184–195. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Yang, T.; Wang, R.; Liu, A. Characterisation of DGAT1 and DGAT2 from Jatropha curcas and their functions in storage lipid biosynthesis. Funct. Plant Biol. 2014, 41, 321–329. [Google Scholar] [CrossRef]
- Misra, A.; Khan, K.; Niranjan, A.; Nath, P.; Sane, V.A. Over-expression of JcDGAT1 from Jatropha curcas increases seed oil levels and alters oil quality in transgenic Arabidopsis thaliana. Phytochemistry 2013, 96, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.-B.; He, L.-L.; Niu, L.-J.; Xu, Z.-F. Isolation and characterization of an ubiquitin extension protein gene (JcUEP) promoter from Jatropha curcas. Planta 2015, 241, 823–836. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.-B.; Hu, X.-D.; Xu, Z.-F. Fatty acid biosynthesis and triacylglycerol accumulation in the biofuel plant Jatropha curcas. In Jatropha, Challenges for a New Energy Crop; Mulpuri, S., Carels, N., Bahadur, B., Eds.; Springer: Singapore, 2019; pp. 163–179. [Google Scholar] [CrossRef]
- Turchetto-Zolet, A.C.; Christoff, A.P.; Kulcheski, F.R.; Guilherm, L.-M.; Margis, R.; Marcia, M.-P. Diversity and evolution of plant diacylglycerol acyltransferase (DGATs) unveiled by phylogenetic, gene structure and expression analyses. Genet. Mol. Biol. 2016, 39, 524–538. [Google Scholar] [CrossRef]
- Rosli, R.; Chan, P.L.; Chan, K.L.; Amiruddin, N.; Low, E.L.; Singh, R.; Harwood, J.L.; Murphy, D.J. In silico characterization and expression profiling of the diacylglycerol acyltransferase gene family (DGAT1, DGAT2, DGAT3 and WS/DGAT) from oil palm, Elaeis guineensis. Plant Sci. 2018, 275, 84–96. [Google Scholar] [CrossRef]
- Yang, D.; Zhang, H.; Peng, K.; Chen, L.; He, H.; Huang, X.; Qin, J.; He, G.; Zhang, D. Differential gene regulation of lipid synthesis in the developing seeds of two biodiesel tree species, Jatropha and Vernicia. Int. J. Agric. Biol. 2016, 18, 1143–1152. [Google Scholar] [CrossRef]
- Khan, K.; Kumar, V.; Niranjan, A.; Shanware, A.; Sane, V.A. JcMYB1, a Jatropha R2R3MYB transcription factor gene, modulates lipid biosynthesis in transgenic plants. Plant Cell Physiol. 2019, 60, 462–475. [Google Scholar] [CrossRef]
- Chen, M.; Mooney, B.P.; Hajduch, M.; Joshi, T.; Zhou, M.; Xu, D.; Thelen, J.J. System analysis of an Arabidopsis mutant altered in de novo fatty acid synthesis reveals diverse changes in seed composition and metabolism. Plant Physiol. 2009, 150, 27–41. [Google Scholar] [CrossRef]
- Bourgis, F.; Kilaru, A.; Cao, X.; Ngando-Ebongue, G.F.; Drira, N.; Ohlrogge, J.B.; Arondel, V. Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proc. Natl. Acad. Sci. USA 2011, 108, 12527–12532. [Google Scholar] [CrossRef]
- Lu, S.; Sturtevant, D.; Aziz, M.; Jin, C.; Li, Q.; Chapman, K.D.; Guo, L. Spatial analysis of lipid metabolites and expressed genes reveals tissue-specific heterogeneity of lipid metabolism in high- and low-oil Brassica napus L. seeds. Plant J. 2018, 94, 915–932. [Google Scholar] [CrossRef]
- Dinamarca, J.; Levitan, O.; Kumaraswamy, K.G.; Lun, D.S.; Falkowski, P. Overexpression of a diacylglycerol acyltransferase gene in Phaeodactylum tricornutum directs carbon towards lipid biosynthesis. J. Phycol. 2017, 53, 405–414. [Google Scholar] [CrossRef]
- Hofvander, P.; Ischebeck, T.; Turesson, H.; Kushwaha, S.K.; Feussner, I.; Carlsson, A.S.; Andersson, M. Potato tuber expression of Arabidopsis WRINKLED1 increase triacylglycerol and membrane lipids while affecting central carbohydrate metabolism. Plant Biotechnol. J. 2016, 14, 1883–1898. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Vanhercke, T.; Shrestha, P.; Luo, J.; Akbar, S.; Konik-Rose, C.; Venugoban, L.; Hussain, D.; Tian, L.; Singh, S.; et al. Upregulated lipid biosynthesis at the expense of starch production in potato (Solanum tuberosum) vegetative tissues via simultaneous downregulation of ADP-Glucose Pyrophosphorylase and sugar dependent1 expressions. Front. Plant Sci. 2019, 10, 1444. [Google Scholar] [CrossRef] [PubMed]
- Knothe, G. A technical evaluation of biodiesel from vegetable oils vs. algae. Will algae-derived biodiesel perform? Green Chem. 2011, 13, 3048–3065. [Google Scholar] [CrossRef]
- Durrett, T.P.; Benning, C.; Ohlrogge, J. Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 2008, 54, 593–607. [Google Scholar] [CrossRef] [PubMed]
- Graef, G.; LaVallee, B.J.; Tenopir, P.; Tat, M.; Schweiger, B.; Kinney, A.J.; Van Gerpen, J.H.; Clemente, T.E. A high-oleic-acid and low-palmitic-acid soybean: Agronomic performance and evaluation as a feedstock for biodiesel. Plant Biotechnol. J. 2009, 7, 411–421. [Google Scholar] [CrossRef]
- Duffield, J.; Shapouri, H.; Graboski, M.; McCormick, R.; Wilson, R.U.S. biodiesel development: New markets for conventional and genetically modified agricultural products. Wash. DC USA Econ. Res. Serv. U. S. Dep. Agric. (USDA) 1998. [Google Scholar] [CrossRef]
- Qu, J.; Mao, H.-Z.; Chen, W.; Gao, S.-Q.; Bai, Y.-N.; Sun, Y.-W.; Geng, Y.-F.; Ye, J. Development of marker-free transgenic Jatropha plants with increased levels of seed oleic acid. Biotechnol. Biofuels 2012, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Maravi, D.K.; Kumar, S.; Sharma, P.K.; Kobayashi, Y.; Goud, V.V.; Sakurai, N.; Koyama, H.; Sahoo, L. Ectopic expression of AtDGAT1, encoding diacylglycerol O-acyltransferase exclusively committed to TAG biosynthesis, enhances oil accumulation in seeds and leaves of Jatropha. Biotechnol. Biofuels 2016, 9, 226. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.X.; Dörmann, P. Chloroplast membrane lipid biosynthesis and transport. Plant Cell Monogr. 2009, 13, 125–156. [Google Scholar]
- Kaup, M.T.; Froese, C.D.; Thompson, J.E. A role for diacylglycerol acyltransferase during leaf senescence. Plant Physiol. 2002, 129, 1616–1626. [Google Scholar] [CrossRef]
- Correa, S.M.; Alseekh, S.; Atehortua, L.; Brotman, Y.; Rios-Estepa, R.; Fernie, A.R.; Nikoloski, Z. Model-assisted identification of metabolic engineering strategies for Jatropha curcas lipid pathways. Plant J. 2020, 104, 76–95. [Google Scholar] [CrossRef] [PubMed]
- Tjellstrom, H.; Strawsine, M.; Ohlrogge, J.B. Tracking synthesis and turnover of triacylglycerol in leaves. J. Exp. Bot. 2015, 66, 1453–1461. [Google Scholar] [CrossRef]
- Katavic, V.; Reed, D.W.; Taylor, D.C.; Ciblin, E.M.; Barton, D.L.; Zou, J.; MacKenzie, S.L.; Covello, P.S.; Kunst, L. Alteration of seed fatty acid composition by an ethyl methanesulfonate-induced mutation in Arabidopsis thaliana affecting diacylglycerol acyltransferase activity. Plant Physiol. 1995, 108, 399–409. [Google Scholar] [CrossRef]
- Fu, Q.; Li, C.; Tang, M.; Tao, Y.B.; Pan, B.Z.; Zhang, L.; Niu, L.; He, H.; Wang, X.; Xu, Z.F. An efficient protocol for Agrobacterium-mediated transformation of the biofuel plant Jatropha curcas by optimizing kanamycin concentration and duration of delayed selection. Plant Biotechnol. Rep. 2015, 9, 405–416. [Google Scholar] [CrossRef]
- Ding, L.W.; Sun, Q.Y.; Wang, Z.Y.; Sun, Y.B.; Xu, Z.F. Using silica particles to isolate total RNA from plant tissues recalcitrant to extraction in guanidine thiocyanate. Anal. Biochem. 2008, 374, 426–428. [Google Scholar] [CrossRef]
- Zhang, L.; He, L.L.; Fu, Q.T.; Xu, Z.F. Selection of reliable reference genes for gene expression studies in the biofuel plant Jatropha curcas using real-time quantitative PCR. Int. J. Mol. Sci. 2013, 14, 24338–24354. [Google Scholar] [CrossRef]
- Ye, J.; Wang, C.; Sun, Y.; Qu, J.; Mao, H.; Chua, N.H. Overexpression of a transcription factor increases lipid content in a woody perennial Jatropha curcas. Front. Plant Sci. 2018, 9, 1479. [Google Scholar] [CrossRef] [PubMed]
- Annarao, S.; Sidhu, O.P.; Roy, R.; Tuli, R.; Khetrapal, C.L. Lipid profiling of developing Jatropha curcas L. seeds using 1H NMR spectroscopy. Bioresour. Technol. 2008, 99, 9032–9035. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Sun, M.; Jayawardana, K.; Wu, D.; Chen, G. Characterization of a PLDζ2 homology gene from developing castor bean endosperm. Lipids 2020, 55, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Guo, Q.; Akbar, S.; Zhi, Y.; El Tahchy, A.; Mitchell, M.; Li, Z.; Shrestha, P.; Vanhercke, T.; Ral, J.P.; et al. Genetic enhancement of oil content in potato tuber (Solanum tuberosum L.) through an integrated metabolic engineering strategy. Plant Biotechnol. J. 2017, 15, 56–67. [Google Scholar] [CrossRef]
- Wang, H.; Pan, J.; Li, Y.; Lou, D.; Hu, Y.; Yu, D. The DELLA-CONSTANS transcription factor cascade integrates gibberellic acid and photoperiod signaling to regulate flowering. Plant Physiol. 2016, 172, 479–488. [Google Scholar] [CrossRef]


| Genotype | Oil (%, w/w) | Protein (%, w/w) | Starch (%, w/w) | Soluble Sugar (%, w/w) |
|---|---|---|---|---|
| Control | 42.97 ± 4.29 | 19.15 ± 0.95 | 5.95 ± 0.93 | 3.15 ± 0.18 |
| 35S:JcDGAT1-L36 | 51.25 ± 0.82 * | 17.24 ± 2.63 | 6.06 ± 1.20 | 2.96 ± 0.01 |
| 35S:JcDGAT1-L40 | 53.73 ± 0.61 ** | 15.61 ± 0.26 ** | 4.65 ± 0.52 | 2.17 ± 0.14 ** |
| 35S:JcDGAT2-L31 | 55.70 ± 0.64 ** | 15.83 ± 0.60 ** | 5.32 ± 0.93 | 2.79 ± 0.42 |
| 35S:JcDGAT2-L51 | 51.07 ± 0.37 * | 17.66 ± 0.89 | 6.03 ± 1.01 | 2.88 ± 0.20 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.-T.; He, H.; Xu, C.-J.; Fu, Q.; Tao, Y.-B.; Xu, R.; Xu, Z.-F. Overexpression of Type 1 and 2 Diacylglycerol Acyltransferase Genes (JcDGAT1 and JcDGAT2) Enhances Oil Production in the Woody Perennial Biofuel Plant Jatropha curcas. Plants 2021, 10, 699. https://doi.org/10.3390/plants10040699
Zhang T-T, He H, Xu C-J, Fu Q, Tao Y-B, Xu R, Xu Z-F. Overexpression of Type 1 and 2 Diacylglycerol Acyltransferase Genes (JcDGAT1 and JcDGAT2) Enhances Oil Production in the Woody Perennial Biofuel Plant Jatropha curcas. Plants. 2021; 10(4):699. https://doi.org/10.3390/plants10040699
Chicago/Turabian StyleZhang, Tian-Tian, Huiying He, Chuan-Jia Xu, Qiantang Fu, Yan-Bin Tao, Ronghua Xu, and Zeng-Fu Xu. 2021. "Overexpression of Type 1 and 2 Diacylglycerol Acyltransferase Genes (JcDGAT1 and JcDGAT2) Enhances Oil Production in the Woody Perennial Biofuel Plant Jatropha curcas" Plants 10, no. 4: 699. https://doi.org/10.3390/plants10040699
APA StyleZhang, T.-T., He, H., Xu, C.-J., Fu, Q., Tao, Y.-B., Xu, R., & Xu, Z.-F. (2021). Overexpression of Type 1 and 2 Diacylglycerol Acyltransferase Genes (JcDGAT1 and JcDGAT2) Enhances Oil Production in the Woody Perennial Biofuel Plant Jatropha curcas. Plants, 10(4), 699. https://doi.org/10.3390/plants10040699







