Root-Derived Proteases as a Plant Tool to Access Soil Organic Nitrogen; Current Stage of Knowledge and Controversies
Abstract
:1. Introduction
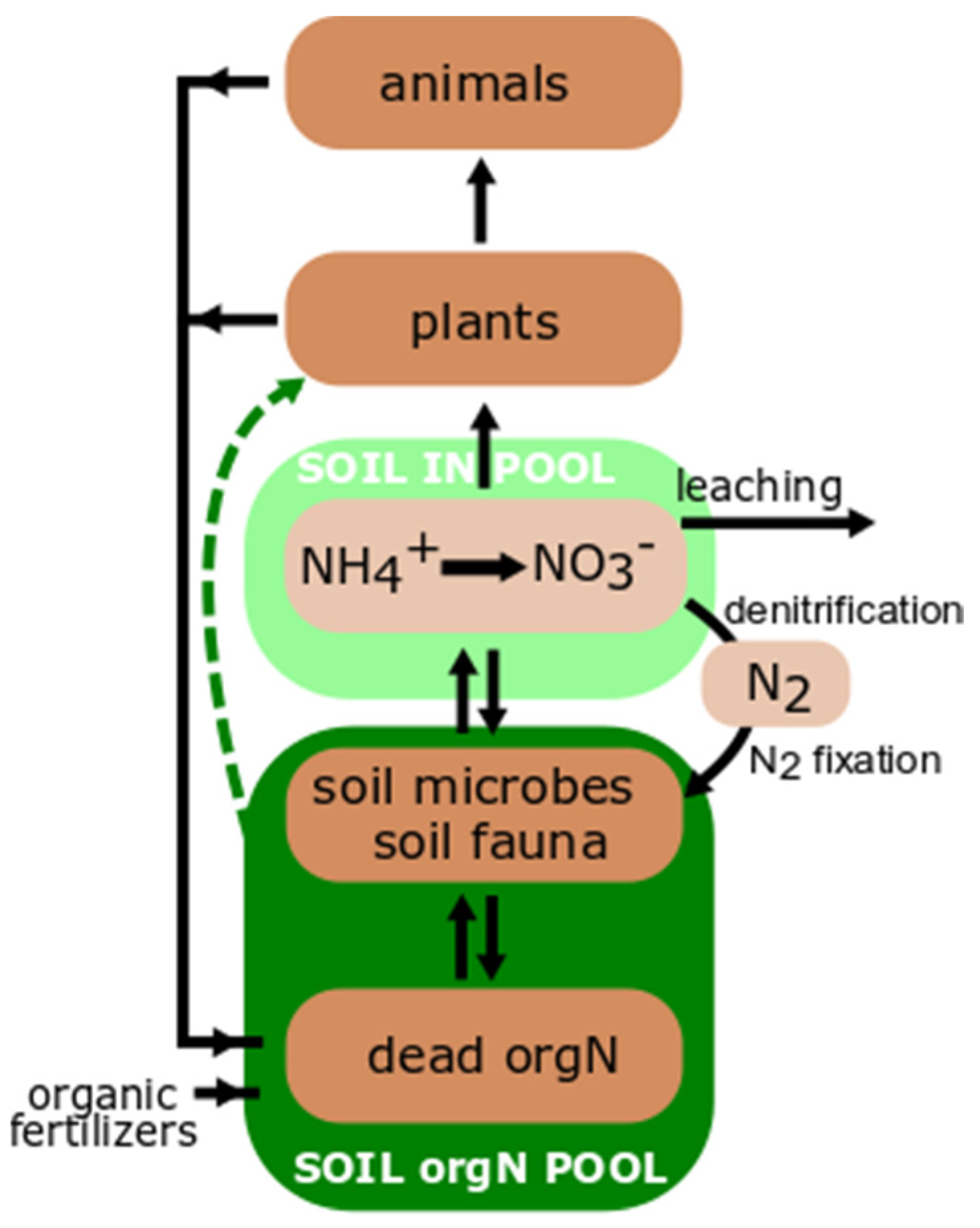
2. Nitrogen Cycle and Soil Proteolysis
3. Soil Proteases and Root-Derived Enzymes
4. How Nitrogen Forms Affect Root Proteases—May Acid Phosphatases and P-Deficiency Give Some Hints?
5. Controversies about Root-Derived Proteases
5.1. Experimental Design Aspect
5.2. Plant Physiology-Specific
5.3. Methodological Aspects
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Suddick, E.; Whitney, P.; Townsend, A.; Davidson, E. The Role of Nitrogen in Climate Change and the Impacts of Nitrogen-Climate Interactions in the United States: Foreword to Thematic Issue. Biogeochemistry 2012, 114, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Mulvaney, R.; Khan, S.; Ellsworth, T. Synthetic Nitrogen Fertilizers Deplete Soil Nitrogen: A Global Dilemma for Sustainable Cereal Production. J. Environ. Qual. 2009, 38, 2295–2314. [Google Scholar] [CrossRef] [Green Version]
- Lassaletta, L.; Billen, G.; Grizzetti, B.; Anglade, J.; Garnier, J. 50 Year Trends in Nitrogen Use Efficiency of World Cropping Systems: The Relationship between Yield and Nitrogen Input to Cropland. Environ. Res. Lett. 2014, 9, 105011. [Google Scholar] [CrossRef]
- Schlesinger, W.H. On the Fate of Anthropogenic Nitrogen. Proc. Natl. Acad. Sci. USA 2009, 106, 203–208. [Google Scholar] [CrossRef] [Green Version]
- Näsholm, T.; Kielland, K.; Ganeteg, U. Uptake of Organic Nitrogen by Plants. New Phytol. 2009, 182, 31–48. [Google Scholar] [CrossRef]
- Soper, F.; Paungfoo-Lonhienne, C.; Brackin, R.; Rentsch, D.; Schmidt, S.; Robinson, N. Arabidopsis and Lobelia anceps Access Small Peptides as a Nitrogen Source for Growth. Funct. Plant Biol. 2011, 38, 788–796. [Google Scholar] [CrossRef]
- Tegeder, M.; Rentsch, D. Uptake and Partitioning of Amino Acids and Peptides. Mol. Plant 2010, 3, 997–1011. [Google Scholar] [CrossRef] [PubMed]
- Paungfoo-Lonhienne, C.; Rentsch, D.; Robatzek, S.; Webb, R.I.; Sagulenko, E.; Näsholm, T.; Schmidt, S.; Lonhienne, T.G.A. Turning the Table: Plants Consume Microbes as a Source of Nutrients. PLoS ONE 2010, 5, e11915. [Google Scholar] [CrossRef] [Green Version]
- Paungfoo-Lonhienne, C.; Lonhienne, T.G.A.; Rentsch, D.; Robinson, N.; Christie, M.; Webb, R.I.; Gamage, H.K.; Carroll, B.J.; Schenk, P.M.; Schmidt, S. Plants Can Use Protein as a Nitrogen Source without Assistance from Other Organisms. Proc. Natl. Acad. Sci.USA 2008, 105, 4524–4529. [Google Scholar] [CrossRef] [Green Version]
- White, J.F.; Chen, Q.; Torres, M.S.; Mattera, R.; Irizarry, I.; Tadych, M.; Bergen, M. Collaboration between Grass Seedlings and Rhizobacteria to Scavenge Organic Nitrogen in Soils. AoB Plants 2015, 7, 6. [Google Scholar] [CrossRef]
- White, J.; Kingsley, K.; Verma, S.; Kowalski, K. Rhizophagy Cycle: An Oxidative Process in Plants for Nutrient Extraction from Symbiotic Microbes. Microorganisms 2018, 6, 95. [Google Scholar] [CrossRef] [Green Version]
- Godlewski, M.; Adamczyk, B. The Ability of Plants to Secrete Proteases by Roots. Plant Physiol. Biochem. 2007, 45, 657–664. [Google Scholar] [CrossRef]
- Greenfield, L.M.; Hill, P.W.; Paterson, E.; Baggs, E.M.; Jones, D.L. Do Plants Use Root-Derived Proteases to Promote the Uptake of Soil Organic Nitrogen? Plant Soil 2020, 456, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, J.; Gilroyed, B.; Reuter, T.; Badea, A.; Eudes, F.; Graf, R.; Laroche, A.; Kav, N.; Mcallister, T. Efficiency of Protein as a Nitrogen Source for Wheat and Morphological Changes in Roots Exposed to High Protein Concentrations. Can. J. Plant Sci. 2014, 94, 603–613. [Google Scholar] [CrossRef]
- Adamczyk, B.; Smolander, A.; Kitunen, V.; Godlewski, M. Proteins as Nitrogen Source for Plants. Plant Signal. Behav. 2010, 5, 817–819. [Google Scholar] [CrossRef] [Green Version]
- Van Der Hoorn, R.A.L. Plant Proteases: From Phenotypes to Molecular Mechanisms. Annu. Rev. Plant Biol. 2008, 59, 191–223. [Google Scholar] [CrossRef] [Green Version]
- Rawlings, N.D.; Waller, M.; Barrett, A.J.; Bateman, A. MEROPS: The Database of Proteolytic Enzymes, Their Substrates and Inhibitors. Nucleic Acids Res. 2014, 42, 503–509. [Google Scholar] [CrossRef] [Green Version]
- Galloway, J.; Leach, A.; Bleeker, A.; Erisman, J.W. A Chronology of Human Understanding of the Nitrogen Cycle. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20130120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porporato, A.; D’Odorico, P.; Laio, F.; Rodriguez-Iturbe, I. Hydrologic Controls on Soil Carbon and Nitrogen Cycles. I. Modeling Scheme. Adv. Water Resour. 2003, 26, 45–58. [Google Scholar] [CrossRef]
- Fowler, D.; Coyle, M.; Skiba, U.; Sutton, M.; Cape, J.; Reis, S.; Sheppard, L.; Jenkins, A.; Grizzetti, B.; Galloway, J.; et al. The Global Nitrogen Cycle in the Twenty-First Century. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20130164. [Google Scholar] [CrossRef]
- Knicker, H. Soil Organic N—An under-Rated Player for C Sequestration in Soils? Soil Biol. Biochem. 2011, 43, 1118–1129. [Google Scholar] [CrossRef]
- Schulten, H.-R.; Schnitzer, M. The Chemistry of Soil Organic Nitrogen: A Review. Biol. Fertil. Soils 1997, 26, 1–15. [Google Scholar] [CrossRef]
- Schimel, J.P.; Bennett, J. Nitrogen Mineralization: Challenges of a Changing Paradigm. Ecology 2004, 85, 591–602. [Google Scholar] [CrossRef]
- Dion, P.P.; Jämtgård, S.; Bertrand, A.; Pepin, S.; Dorais, M. Organic Nitrogen Uptake and Assimilation in Cucumis sativus Using Position-Specific Labeling and Compound-Specific Isotope Analysis. Front. Plant Sci. 2018, 9, 1596. [Google Scholar] [CrossRef] [PubMed]
- Bar-On, Y.M.; Phillips, R.; Milo, R. The Biomass Distribution on Earth. Proc. Natl. Acad. Sci. USA 2018, 115, 6506–6511. [Google Scholar] [CrossRef] [Green Version]
- Kuzyakov, Y.; Friedel, J.K.; Stahr, K. Review of Mechanisms and Quantification of Priming Effects. Soil Biol. Biochem. 2000, 32, 1485–1498. [Google Scholar] [CrossRef]
- Dijkstra, F.A.; Carrillo, Y.; Pendall, E.; Morgan, J.A. Rhizosphere Priming: A Nutrient Perspective. Front. Microbiol. 2013, 4, 216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, W.; Parton, W.J.; Gonzalez-Meler, M.A.; Phillips, R.; Asao, S.; McNickle, G.G.; Brzostek, E.; Jastrow, J.D. Synthesis and Modeling Perspectives of Rhizosphere Priming. New Phytol. 2013, 201, 31–44. [Google Scholar] [CrossRef] [Green Version]
- Whiteside, M.D.; Garcia, M.O.; Treseder, K.K. Amino Acid Uptake in Arbuscular Mycorrhizal Plants. PLoS ONE 2012, 7, e47643. [Google Scholar]
- Lambers, H.; Albornoz, F.; Arruda, A.; Barker, T.; Finnegan, P.; Gille, C.; Gooding, H.; Png, G.K.; Ranathunge, K.; Zhong, H.-T. Nutrient-Acquisition Strategies. In A Jewel in the Crown of a Global Biodiversity Hotspot; Lambers, H., Ed.; Kwongan Foundation and the Western Australian Naturalists’ Club Inc.: Perth, Australia, 2019; pp. 227–248. [Google Scholar]
- Smith, S.E.; Anderson, I.C.; Smith, F.A. Mycorrhizal Associations and Phosphorus Acquisition: From Cells to Ecosystems. Annu. Plant Rev. 2015, 48, 409–439. [Google Scholar] [CrossRef]
- Brundrett, M.C.; Tedersoo, L. Evolutionary History of Mycorrhizal Symbioses and Global Host Plant Diversity. New Phytol. 2018, 220, 1108–1115. [Google Scholar] [CrossRef] [PubMed]
- De-la-Peña, C.; Badri, D.V.; Lei, Z.; Watson, B.S.; Brandão, M.M.; Silva-Filho, M.C.; Sumner, L.W.; Vivanco, J.M. Root Secretion of Defense-Related Proteins Is Development-Dependent and Correlated with Flowering Time. J. Biol. Chem. 2010, 285, 30654–30665. [Google Scholar] [CrossRef] [Green Version]
- Wen, F.; Vanetten, H.D.; Tsaprailis, G.; Hawes, M.C. Extracellular Proteins in Pea Root Tip and Border Cell Exudates. Plant Physiol. 2007, 143, 773–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.W.; Bandurski, R.S. Exocellular Enzymes of Corn Roots. Plant Physiol. 1964, 39, 60–64. [Google Scholar] [CrossRef] [Green Version]
- Hawes, M.C.; Gunawardena, U.; Miyasaka, S.; Zhao, X. The Role of Root Border Cells in Plant Defense. Trends Plant Sci. 2000, 5, 128–133. [Google Scholar] [CrossRef]
- Miyasaka, S.C.; Hawes, M.C. Possible Role of Root Border Cells in Detection and Avoidance of Aluminum Toxicity. Plant Physiol. 2001, 125, 1978–1987. [Google Scholar] [CrossRef] [Green Version]
- Pan, J.W.; Ye, D.; Wang, L.L.; Hua, J.; Zhao, G.F.; Pan, W.H.; Han, N.; Zhu, M.Y. Root Border Cell Development Is a Temperature-Insensitive and Al-Sensitive Process in Barley. Plant Cell Physiol. 2004, 45, 751–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hawes, M.; Allen, C.; Turgeon, B.G.; Curlango-Rivera, G.; Tran, T.M.; Huskey, D.A.; Xiong, Z. Root Border Cells and Their Role in Plant Defense. Annu. Rev. Phytopathol. 2016, 54, 143–161. [Google Scholar] [CrossRef] [PubMed]
- Brigham, L.A.; Woo, H.H.; Nicoll, S.M.; Hawes, M.C. Differential Expression of Proteins and MRNAs from Border Cells and Root Tips of Pea. Plant Physiol. 1995, 109, 457–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tadano, T.; Ozawa, K.; Sakai, H.; Osaki, M.; Matsui, H. Secretion of Acid Phosphatase by the Roots of Crop Plants under Phosphorus-Deficient Conditions and Some Properties of the Enzyme Secreted by Lupin Roots. Plant Soil 1993, 155, 95–98. [Google Scholar] [CrossRef]
- Miller, S.S.; Liu, J.; Allan, D.L.; Menzhuber, C.J.; Fedorova, M.; Vance, C.P. Molecular Control of Acid Phosphatase Secretion into the Rhizosphere of Proteoid Roots from Phosphorus-Stressed White Lupin. Plant Physiol. 2001, 127, 594–606. [Google Scholar] [CrossRef] [PubMed]
- Tarafdar, J.C.; Claassen, N. Organic Phosphorus Utilization by Wheat Plants under Sterile Conditions. Biol. Fertil. Soils 2003, 39, 25–29. [Google Scholar] [CrossRef]
- Png, G.K.; Turner, B.L.; Albornoz, F.E.; Hayes, P.E.; Lambers, H.; Laliberté, E. Greater Root Phosphatase Activity in Nitrogen-Fixing Rhizobial but Not Actinorhizal Plants with Declining Phosphorus Availability. J. Ecol. 2017, 105, 1246–1255. [Google Scholar] [CrossRef] [Green Version]
- Yadav, R.; Tarafdar, J. Influence of Organic and Inorganic Phosphorus Supply on the Maximum Secretion of Acid Phosphatase by Plants. Biol. Fertil. Soils 2001, 34, 140–143. [Google Scholar] [CrossRef]
- Li, M.; Osaki, M.; Madhusudana Rao, I.; Tadano, T. Secretion of Phytase from the Roots of Several Plant Species under Phosphorus-Deficient Conditions. Plant Soil 1997, 195, 161–169. [Google Scholar] [CrossRef]
- Asmar, F. Variation in Activity of Root Extracellular Phytase between Genotypes of Barley. Plant Soil 1997, 195, 61–64. [Google Scholar] [CrossRef]
- Dubrovskaya, E.; Pozdnyakova, N.; Golubev, S.; Muratova, A.; Grinev, V.; Bondarenkova, A.; Turkovskaya, O. Peroxidases from Root Exudates of Medicago Sativa and Sorghum Bicolor: Catalytic Properties and Involvement in PAH Degradation. Chemosphere 2017, 169, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Badri, D.V.; Vivanco, J.M. Regulation and Function of Root Exudates. Plant Cell Environ. 2009, 32, 666–681. [Google Scholar] [CrossRef]
- Gramss, G.; Rudeschko, O. Activities of Oxidoreductase Enzymes in Tissue Extracts and Sterile Root Exudates of Three Crop Plants, and Some Properties of the Peroxidase Component. New Phytol. 1998, 138, 401–409. [Google Scholar] [CrossRef]
- Bieleski, R.L. Phosphate Pools, Phosphate Transport, and Phosphate Availability. Annu. Rev. Plant Physiol. 1973, 24, 225–252. [Google Scholar] [CrossRef]
- Margalef, O.; Sardans, J.; Fernández-Martínez, M.; Molowny-Horas, R.; Janssens, I.A.; Ciais, P.; Goll, D.; Richter, A.; Obersteiner, M.; Asensio, D.; et al. Global Patterns of Phosphatase Activity in Natural Soils. Sci. Rep. 2017, 7, 1337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamczyk, B.; Godlewski, M.; Zimny, J.; Zimny, A. Wheat (Triticum aestivum) Seedlings Secrete Proteases from the Roots and, after Protein Addition, Grow Well on Medium without Inorganic Nitrogen. Plant Biol. 2008, 10, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Adamczyk, B.; Godlewski, M.; Zimny, J.; Zimny, A. Growth and Protease Secretion by Roots of Wheat Seedlings Cultivated on Different Nitrogen Sources. Indian J. Plant Physiol. 2010, 15, 150–153. [Google Scholar]
- Lambers, H.; Juniper, D.; Cawthray, G.R.; Veneklaas, E.J.; Martínez-Ferri, E. The Pattern of Carboxylate Exudation in Banksia grandis (Proteaceae) Is Affected by the Form of Phosphate Added to the Soil. Plant Soil 2002, 238, 111–122. [Google Scholar] [CrossRef]
- Adamczyk, B.; Godlewski, M. Inter-Specific Variability in Protein Use by Two Vegetable Crop Species. Braz. J. Plant Physiol. 2010, 22, 171–176. [Google Scholar] [CrossRef] [Green Version]
- Chapin, F.S., III. New Cog in the Nitrogen Cycle. Nature 1995, 377, 199–200. [Google Scholar] [CrossRef]
- Lonhienne, T.; Trusov, Y.; Young, A.; Rentsch, D.; Näsholm, T.; Schmidt, S.; Paungfoo-Lonhienne, C. Effects of Externally Supplied Protein on Root Morphology and Biomass Allocation in Arabidopsis. Sci. Rep. 2014, 4, 5055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paungfoo-Lonhienne, C.; Schenk, P.; Lonhienne, T.; Brackin, R.; Meier, S.; Rentsch, D.; Schmidt, S. Nitrogen Affects Cluster Root Formation and Expression of Putative Peptide Transporters. J. Exp. Bot. 2009, 60, 2665–2676. [Google Scholar] [CrossRef]
- Beauzamy, L.; Derr, J.; Boudaoud, A. Quantifying Hydrostatic Pressure in Plant Cells by Using Indentation with an Atomic Force Microscope. Biophys. J. 2015, 108, 2448–2456. [Google Scholar] [CrossRef] [Green Version]
- Eatough, D.J.; Jensen, T.E.; Hansen, L.D.; Loken, H.F.; Rehfeld, S.J. The Binding of Ca2+ and Mg2+ to Human Serium Albumin: A Calorimetric Study. Thermochim. Acta 1978, 25, 289–297. [Google Scholar] [CrossRef]
- Playsted, C.W.S.; Johnston, M.E.; Ramage, C.M.; Edwards, D.G.; Cawthray, G.R.; Lambers, H. Functional Significance of Dauciform Roots: Exudation of Carboxylates and Acid Phosphatase under Phosphorus Deficiency in Caustis Blakei (Cyperaceae). New Phytol. 2006, 170, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Wasaki, J.; Ando, M.; Ozawa, K.; Omura, M.; Osaki, M.; Ito, H.; Matsui, H.; Tadano, T. Properties of Secretory Acid Phosphatase from Lupin Roots under Phosphorus-Deficient Conditions. Soil Sci. Plant Nutr. 1997, 43, 981–986. [Google Scholar] [CrossRef]
- Haran, S.; Logendra, S.; Seskar, M.; Bratanova, M.; Raskin, I. Characterization of Arabidopsis Acid Phosphatase Promoter and Regulation of Acid Phosphatase Expression. Plant Physiol. 2000, 124, 615–626. [Google Scholar] [CrossRef] [Green Version]
- Dietrich, R.; Ploß, K.; Heil, M. Constitutive and Induced Resistance to Pathogens in Arabidopsis thaliana Depends on Nitrogen Supply. Plant Cell Environ. 2004, 27, 896–906. [Google Scholar] [CrossRef]
- Nobile, C.; Houben, D.; Michel, E.; Firmin, S.; Lambers, H.; Kandeler, E.; Faucon, M.P. Phosphorus-Acquisition Strategies of Canola, Wheat and Barley in Soil Amended with Sewage Sludges. Sci. Rep. 2019, 9, 14878. [Google Scholar] [CrossRef]
- Sun, T.; Hobbie, S.E.; Berg, B.; Zhang, H.; Wang, Q.; Wang, Z.; Hättenschwiler, S. Contrasting Dynamics and Trait Controls in First-Order Root Compared with Leaf Litter Decomposition. Proc. Natl. Acad. Sci. USA 2018, 115, 10392–10397. [Google Scholar] [CrossRef] [Green Version]
- Honvault, N.; Houben, D.; Nobile, C.; Firmin, S.; Lambers, H.; Faucon, M.P. Tradeoffs among Phosphorus-Acquisition Root Traits of Crop Species for Agroecological Intensification. Plant Soil 2020, 1–14. [Google Scholar] [CrossRef]
- Adamczyk, B. Characterization of Proteases Secreted by Leek Roots. Russ. J. Plant Physiol. 2014, 61, 714–717. [Google Scholar] [CrossRef]
- Adamczyk, B.; Godlewski, M.; Smolander, A.; Kitunen, V. Degradation of Proteins by Enzymes Exuded by Allium porrum Roots—A Potentially Important Strategy for Acquiring Organic Nitrogen by Plants. Plant Physiol. Biochem. 2009, 47, 919–925. [Google Scholar] [CrossRef] [PubMed]
- Enggrob, K.L.; Jakobsen, C.M.; Pedersen, I.F.; Rasmussen, J. Newly Depolymerized Large Organic N Contributes Directly to Amino Acid Uptake in Young Maize Plants. New Phytol. 2019, 224, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Adamczyk, B.; Sietiö, O.-M.M.; Biasi, C.; Heinonsalo, J. Interaction between Tannins and Fungal Necromass Stabilizes Fungal Residues in Boreal Forest Soils. New Phytol. 2019, 223, 16–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamczyk, B.; Sietiö, O.-M.M.; Straková, P.; Prommer, J.; Wild, B.; Hagner, M.; Pihlatie, M.; Fritze, H.; Richter, A.; Heinonsalo, J. Plant Roots Increase Both Decomposition and Stable Organic Matter Formation in Boreal Forest Soil. Nat. Commun. 2019, 10, 3982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Enzyme | Plant Species | Potential Role | References |
|---|---|---|---|
| acid phosphatase | numerous species, for example: Arachis hypogaea, Brasicca oleracea, Glycine max, Lupinus sp., Oryza sativa, Pennisetum glaucum, Raphanus sativus, Sesamum indicum, Sinapis alba, Solanum lycopersicum, Sorghum, Triticum aestivum, Vigna aconitifolia, Vigna radiate | increase of plant-available P pool (digestion of organic P) | [30,41,42,43,44,45] |
| phytase | numerous species, for example: Agrostis gigantea, Dactylis glomerata, Lupinus albus, Medicago sativa, Oryza sativa, Phleum pretense, Solanum lycopersicum, Trifolium hybridum, Trifolium pratense, Trifolium repens | increase of plant-available P pool (digestion of inositol hexaphosphate) | [46,47] |
| chitinase glucanase myrosinase | Arabidopsis thaliana | defense | [33] |
| proteases | numerous species, for example: Allium porrum, Allium cepa, Zea mays, Cucurbita pepo, Cucumis sativus, Hippopohae rhamnoi-des, Geranium pusillum, Lactuca sativa, Ruta graveolens, Raphanus sativus | increase of plant- available N pool, defense | [33] |
| root-surface associated protease | Arabidopsis thaliana, Medicago sativa, Sinapis alba | unknown | [34] |
| Triticum eastivum, Zea mays | unknown | [13] | |
| peroxidase, laccase, monophenol mono-oxygenase, superoxide dismutase | Arabidopsis thaliana, Medicago sativa, Lepidium sativum, Sinapis alba | oxidative degradation of certain soil components, defense, regulation of allelopathic chemicals | [33,48,49,50] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adamczyk, B. Root-Derived Proteases as a Plant Tool to Access Soil Organic Nitrogen; Current Stage of Knowledge and Controversies. Plants 2021, 10, 731. https://doi.org/10.3390/plants10040731
Adamczyk B. Root-Derived Proteases as a Plant Tool to Access Soil Organic Nitrogen; Current Stage of Knowledge and Controversies. Plants. 2021; 10(4):731. https://doi.org/10.3390/plants10040731
Chicago/Turabian StyleAdamczyk, Bartosz. 2021. "Root-Derived Proteases as a Plant Tool to Access Soil Organic Nitrogen; Current Stage of Knowledge and Controversies" Plants 10, no. 4: 731. https://doi.org/10.3390/plants10040731





