Cellular Phosphorylation Signaling and Gene Expression in Drought Stress Responses: ABA-Dependent and ABA-Independent Regulatory Systems
Abstract
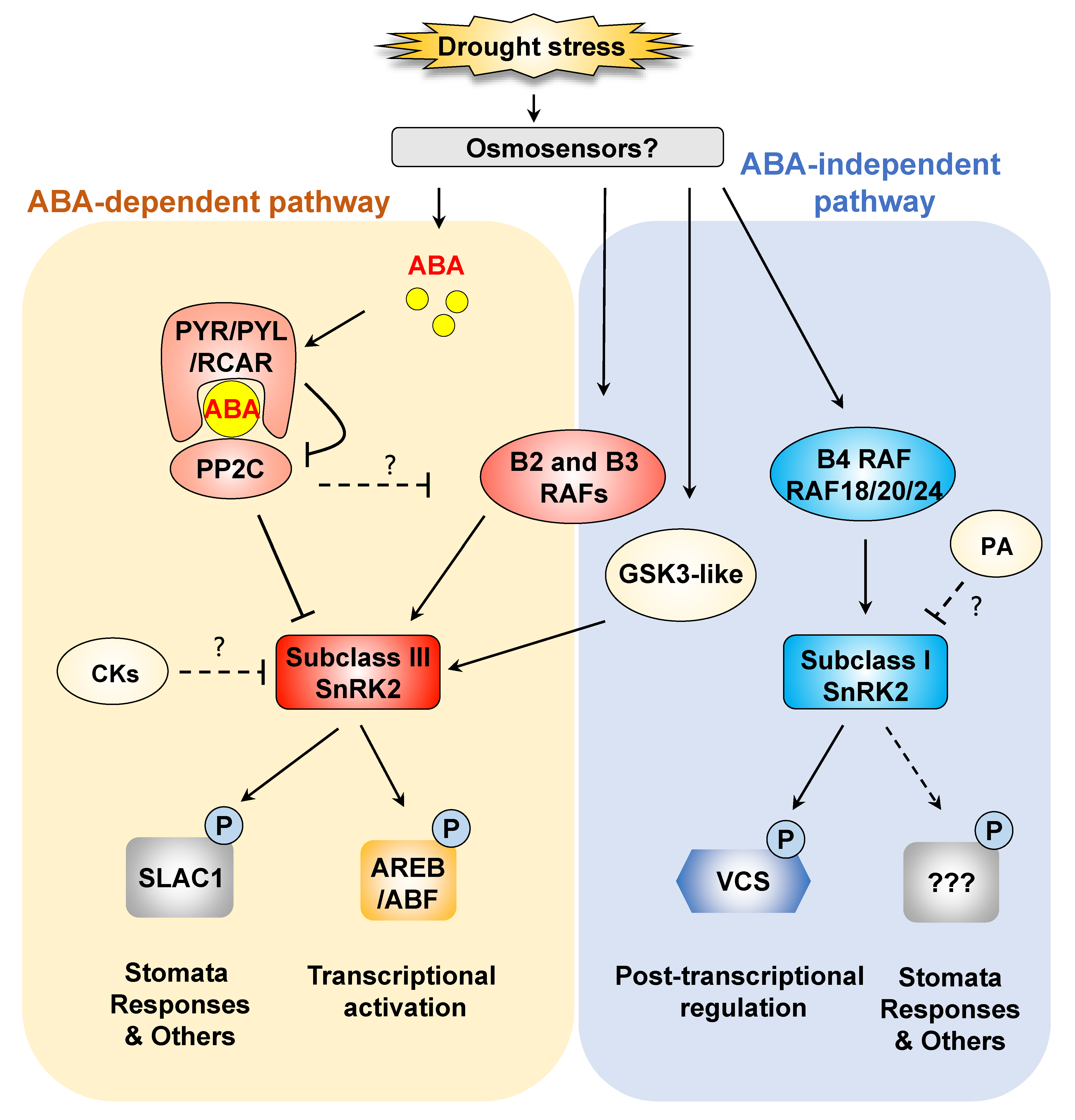
:1. Introduction
2. Importance of ABA-Dependent Responses under Drought Stress
2.1. ABA-Dependent Phosphorylation Signaling in the Dehydration Stress Response: Important Roles of Subclass III SnRK2 Protein Kinases
2.2. Regulatory Mechanisms of Subclass III SnRK2s
2.3. Other Regulatory Mechanisms of ABA Receptors and Coreceptors
3. ABA-Independent Signaling in Response to Dehydration Stress: Important Roles of Subclass I SnRK2s
3.1. Subclass I SnRK2-Mediated Dehydration Stress Signaling
3.2. Regulatory Components of Subclass I SnRK2s
4. Transcriptional Regulation of Cellular Signaling in Drought Stress Responses
4.1. Transcription Factors That Mediate ABA-Responsive Gene Expression
4.2. Transcription Factors for Drought- and Heat-Responsive Gene Expression in ABA-Independent Signaling
4.3. Crosstalk in Transcriptional Regulation between Drought and Other Stress Responses
5. ABA Crosstalk with Other Hormones under Drought Stress
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umezawa, T.; Fujita, M.; Fujita, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Engineering drought tolerance in plants: Discovering and tailoring genes to unlock the future. Curr. Opin. Biotechnol. 2006, 17, 113–122. [Google Scholar] [CrossRef]
- Hirayama, T.; Shinozaki, K. Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 2010, 61, 1041–1052. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 2014, 5, 170. [Google Scholar] [CrossRef] [Green Version]
- Umezawa, T.; Nakashima, K.; Miyakawa, T.; Kuromori, T.; Tanokura, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 2010, 51, 1821–1839. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, F.; Kuromori, T.; Sato, H.; Shinozaki, K. Regulatory Gene Networks in Drought Stress Responses and Resistance in Plants. Adv. Exp. Med. Biol. 2018, 1081, 189–214. [Google Scholar] [CrossRef]
- Soma, F.; Mogami, J.; Yoshida, T.; Abekura, M.; Takahashi, F.; Kidokoro, S.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-unresponsive SnRK2 protein kinases regulate mRNA decay under osmotic stress in plants. Nat. Plants 2017, 3, 16204. [Google Scholar] [CrossRef]
- Kawa, D.; Meyer, A.J.; Dekker, H.L.; Abd-El-Haliem, A.; Gevaert, K.; Van De Slijke, E.; Maszkowska, J.; Bucholc, M.; Dobrowolska, G.; De Jaeger, G.; et al. SnRK2 protein kinases and mRNA decapping machinery control root development and response to salt. Plant Physiol. 2019, 182, 361–377. [Google Scholar] [CrossRef] [Green Version]
- McLoughlin, F.; Galvan-Ampudia, C.S.; Julkowska, M.M.; Caarls, L.; van der Does, D.; Laurière, C.; Munnik, T.; Haring, M.A.; Testerink, C. The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. Plant J. 2012, 72, 436–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabregas, N.; Yoshida, T.; Fernie, A.R. Role of Raf-like kinases in SnRK2 activation and osmotic stress response in plants. Nat. Commun. 2020, 11, 6184. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Dittrich, M.; Mueller, H.M.; Bauer, H.; Peirats-Llobet, M.; Rodriguez, P.L.; Geilfus, C.M.; Carpentier, S.C.; Al Rasheid, K.A.S.; Kollist, H.; Merilo, E.; et al. The role of Arabidopsis ABA receptors from the PYR/PYL/RCAR family in stomatal acclimation and closure signal integration. Nat. Plants 2019, 5, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Mustilli, A.C.; Merlot, S.; Vavasseur, A.; Fenzi, F.; Giraudat, J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 2002, 14, 3089–3099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, R.; Hobo, T.; Ichimura, K.; Mizoguchi, T.; Takahashi, F.; Aronso, J.; Ecker, J.R.; Shinozaki, K. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 2002, 43, 1473–1483. [Google Scholar] [CrossRef]
- Yoshida, R.; Umezawa, T.; Mizoguchi, T.; Takahashi, S.; Takahashi, F.; Shinozaki, K. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 2006, 281, 5310–5318. [Google Scholar] [CrossRef] [Green Version]
- Boudsocq, M.; Droillard, M.J.; Barbier-Brygoo, H.; Laurière, C. Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol. Biol. 2007, 63, 491–503. [Google Scholar] [CrossRef]
- Mizoguchi, M.; Umezawa, T.; Nakashima, K.; Kidokoro, S.; Takasaki, H.; Fujita, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two closely related subclass II SnRK2 protein kinases cooperatively regulate drought-inducible gene expression. Plant Cell Physiol. 2010, 51, 842–847. [Google Scholar] [CrossRef] [Green Version]
- Fujii, H.; Chinnusamy, V.; Rodrigues, A.; Rubio, S.; Antoni, R.; Park, S.Y.; Cutler, S.R.; Sheen, J.; Rodriguez, P.L.; Zhu, J.K. In vitro reconstitution of an abscisic acid signalling pathway. Nature 2009, 462, 660–664. [Google Scholar] [CrossRef] [Green Version]
- Fujita, Y.; Nakashima, K.; Yoshida, T.; Katagiri, T.; Kidokoro, S.; Kanamori, N.; Umezawa, T.; Fujita, M.; Maruyama, K.; Ishiyama, K.; et al. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009, 50, 2123–2132. [Google Scholar] [CrossRef] [Green Version]
- Nakashima, K.; Fujita, Y.; Kanamori, N.; Katagiri, T.; Umezawa, T.; Kidokoro, S.; Maruyama, K.; Yoshida, T.; Ishiyama, K.; Kobayashi, M.; et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009, 50, 1345–1363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F.; Yamaguchi-Shinozaki, K.; Ishihama, Y.; Hirayama, T.; Shinozaki, K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyazono, K.; Miyakawa, T.; Sawano, Y.; Kubota, K.; Kang, H.J.; Asano, A.; Miyauchi, Y.; Takahashi, M.; Zhi, Y.; Fujita, Y.; et al. Structural basis of abscisic acid signalling. Nature 2009, 462, 609–614. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.; et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, Y.; Yamamoto, S.; Minami, H.; Kagaya, Y.; Hattori, T. Differential activation of the rice sucrose nonfermenting1–related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 2004, 16, 1163–1177. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.; Hong, J.; Ha, J.; Kang, J.; Kim, S.Y. ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 2000, 275, 1723–1730. [Google Scholar] [CrossRef] [Green Version]
- Uno, Y.; Furihata, T.; Abe, H.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 2000, 97, 11632–11637. [Google Scholar] [CrossRef] [Green Version]
- Fujii, H.; Verslues, P.E.; Zhu, J.K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 2007, 19, 485–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furihata, T.; Maruyama, K.; Fujita, Y.; Umezawa, T.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 2006, 103, 1988–1993. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, T.; Fujita, Y.; Maruyama, K.; Mogami, J.; Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 2015, 38, 35–49. [Google Scholar] [CrossRef] [Green Version]
- Ruschhaupt, M.; Mergner, J.; Mucha, S.; Papacek, M.; Doch, I.; Tischer, S.V.; Hemmler, D.; Chiasson, D.; Edel, K.H.; Kudla, J.; et al. Rebuilding core abscisic acid signaling pathways of Arabidopsis in yeast. EMBO J. 2019, 38, e101859. [Google Scholar] [CrossRef]
- Umezawa, T.; Sugiyama, N.; Takahashi, F.; Anderson, J.C.; Ishihama, Y.; Peck, S.C.; Shinozaki, K. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 2013, 6, rs8. [Google Scholar] [CrossRef]
- Wang, P.; Xue, L.; Batelli, G.; Lee, S.; Hou, Y.J.; Van Oosten, M.J.; Zhang, H.; Tao, W.A.; Zhu, J.K. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. USA 2013, 110, 11205–11210. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tong, X.; Qiu, J.; Li, Z.; Zhao, J.; Hou, Y.; Tang, L.; Zhang, J. A phosphoproteomic landscape of rice (Oryza sativa) tissues. Physiol. Plant. 2017, 160, 458–475. [Google Scholar] [CrossRef] [Green Version]
- Lv, D.W.; Subburaj, S.; Cao, M.; Yan, X.; Li, X.; Appels, R.; Sun, D.F.; Ma, W.; Yan, Y.M. Proteome and phosphoproteome characterization reveals new response and defense mechanisms of Brachypodium distachyon leaves under salt stress. Mol. Cell. Proteom. 2014, 13, 632–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, C.C.; Zhu, Y.; Arrington, J.V.; Paez, J.S.; Wang, P.; Zhu, P.; Chen, I.H.; Zhu, J.K.; Tao, W.A. Universal Plant Phosphoproteomics Workflow and Its Application to Tomato Signaling in Response to Cold Stress. Mol. Cell. Proteom. 2018, 17, 2068–2080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Hsu, C.C.; Du, Y.; Zhu, P.; Zhao, C.; Fu, X.; Zhang, C.; Paez, J.S.; Macho, A.P.; Tao, W.A.; et al. Mapping proteome-wide targets of protein kinases in plant stress responses. Proc. Natl. Acad. Sci. USA 2020, 117, 3270–3280. [Google Scholar] [CrossRef] [PubMed]
- Mergner, J.; Frejno, M.; List, M.; Papacek, M.; Chen, X.; Chaudhary, A.; Samaras, P.; Richter, S.; Shikata, H.; Messerer, M.; et al. Mass-spectrometry-based draft of the Arabidopsis proteome. Nature 2020, 579, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Boudsocq, M.; Barbier-Brygoo, H.; Laurière, C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 2004, 279, 41758–41766. [Google Scholar] [CrossRef] [Green Version]
- Vlad, F.; Droillard, M.J.; Valot, B.; Khafif, M.; Rodrigues, A.; Brault, M.; Zivy, M.; Rodriguez, P.L.; Merlot, S.; Laurière, C. Phospho-site mapping, genetic and in planta activation studies reveal key aspects of the different phosphorylation mechanisms involved in activation of SnRK2s. Plant J. 2010, 63, 778–790. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Juste, J.; Alrefaei, A.F.; Rodriguez, P.L. Plant Osmotic Stress Signaling: MAPKKKs Meet SnRK2s. Trends Plant Sci. 2020, 25, 1179–1182. [Google Scholar] [CrossRef]
- Komatsu, K.; Takezawa, D.; Sakata, Y. Decoding ABA and osmostress signalling in plants from an evolutionary point of view. Plant Cell Environ. 2020, 43, 2894–2911. [Google Scholar] [CrossRef]
- Hsu, C.C.; Tsai, C.F.; Tao, W.A.; Wang, P. Phosphoproteomic Strategy for Profiling Osmotic Stress Signaling in Arabidopsis. J. Vis. Exp. 2020, 160, e61489. [Google Scholar] [CrossRef]
- Saruhashi, M.; Kumar Ghosh, T.; Arai, K.; Ishizaki, Y.; Hagiwara, K.; Komatsu, K.; Shiwa, Y.; Izumikawa, K.; Yoshikawa, H.; Umezawa, T.; et al. Plant Raf-like kinase integrates abscisic acid and hyperosmotic stress signaling upstream of SNF1-related protein kinase2. Proc. Natl. Acad. Sci. USA 2015, 112, E6388–E6396. [Google Scholar] [CrossRef] [Green Version]
- Stevenson, S.R.; Kamisugi, Y.; Trinh, C.H.; Schmutz, J.; Jenkins, J.W.; Grimwood, J.; Muchero, W.; Tuskan, G.A.; Rensing, S.A.; Lang, D.; et al. Genetic Analysis of Physcomitrella patens Identifies ABSCISIC ACID NON-RESPONSIVE, a Regulator of ABA Responses Unique to Basal Land Plants and Required for Desiccation Tolerance. Plant Cell 2016, 28, 1310–1327. [Google Scholar] [CrossRef] [PubMed]
- Yasumura, Y.; Pierik, R.; Kelly, S.; Sakuta, M.; Voesenek, L.A.; Harberd, N.P. An Ancestral Role for CONSTITUTIVE TRIPLE RESPONSE1 Proteins in Both Ethylene and Abscisic Acid Signaling. Plant Physiol. 2015, 169, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Shinozawa, A.; Otake, R.; Takezawa, D.; Umezawa, T.; Komatsu, K.; Tanaka, K.; Amagai, A.; Ishikawa, S.; Hara, Y.; Kamisugi, Y.; et al. SnRK2 protein kinases represent an ancient system in plants for adaptation to a terrestrial environment. Commun. Biol. 2019, 2, 30. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, Y.; Zhang, J.; Hsu, P.K.; Ceciliato, P.H.O.; Zhang, L.; Dubeaux, G.; Munemasa, S.; Ge, C.; Zhao, Y.; Hauser, F.; et al. MAP3Kinase-dependent SnRK2-kinase activation is required for abscisic acid signal transduction and rapid osmotic stress response. Nat. Commun. 2020, 11, 12. [Google Scholar] [CrossRef]
- Katsuta, S.; Masuda, G.; Bak, H.; Shinozawa, A.; Kamiyama, Y.; Umezawa, T.; Takezawa, D.; Yotsui, I.; Taji, T.; Sakata, Y. Arabidopsis Raf-like kinases act as positive regulators of subclass III SnRK2 in osmostress signaling. Plant J. 2020, 103, 634–644. [Google Scholar] [CrossRef]
- Nguyen, Q.T.C.; Lee, S.J.; Choi, S.W.; Na, Y.J.; Song, M.R.; Hoang, Q.T.N.; Sim, S.Y.; Kim, M.S.; Kim, J.I.; Soh, M.S.; et al. Arabidopsis Raf-Like Kinase Raf10 Is a Regulatory Component of Core ABA Signaling. Mol. Cells 2019, 42, 646–660. [Google Scholar] [CrossRef]
- Lee, S.J.; Lee, M.H.; Kim, J.I.; Kim, S.Y. Arabidopsis putative MAP kinase kinase kinases Raf10 and Raf11 are positive regulators of seed dormancy and ABA response. Plant Cell Physiol. 2015, 56, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Soma, F.; Takahashi, F.; Suzuki, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Plant Raf-like kinases regulate the mRNA population upstream of ABA-unresponsive SnRK2 kinases under drought stress. Nat. Commun. 2020, 11, 1373. [Google Scholar] [CrossRef] [Green Version]
- Lin, Z.; Li, Y.; Zhang, Z.; Liu, X.; Hsu, C.C.; Du, Y.; Sang, T.; Zhu, C.; Wang, Y.; Satheesh, V.; et al. A RAF-SnRK2 kinase cascade mediates early osmotic stress signaling in higher plants. Nat. Commun. 2020, 11, 613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vilela, B.; Nájar, E.; Lumbreras, V.; Leung, J.; Pagès, M. Casein Kinase 2 Negatively Regulates Abscisic Acid-Activated SnRK2s in the Core Abscisic Acid-Signaling Module. Mol. Plant 2015, 8, 709–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Chang, H.; Hu, S.; Lu, X.; Yuan, C.; Zhang, C.; Wang, P.; Xiao, W.; Xiao, L.; Xue, G.P.; et al. Plastid casein kinase 2 knockout reduces abscisic acid (ABA) sensitivity, thermotolerance, and expression of ABA- and heat-stress-responsive nuclear genes. J. Exp. Bot. 2014, 65, 4159–4175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagatoshi, Y.; Fujita, M.; Fujita, Y. Casein kinase 2 α and β subunits inversely modulate ABA signal output in Arabidopsis protoplasts. Planta 2018, 248, 571–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Z.; Liu, J.; Wang, H.; Yang, C.; Chen, Y.; Li, Y.; Pan, S.; Dong, R.; Tang, G.; Barajas-Lopez, J.E.D.; et al. GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 9651–9656. [Google Scholar] [CrossRef] [Green Version]
- Cheng, C.; Wang, Z.; Ren, Z.; Zhi, L.; Yao, B.; Su, C.; Liu, L.; Li, X. SCFAtPP2-B11 modulates ABA signaling by facilitating SnRK2.3 degradation in Arabidopsis thaliana. PLoS Genet. 2017, 13, e1006947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, A.; Kim, J.K.; Jan, M.; Khan, H.A.; Khan, I.U.; Shen, M.; Park, J.; Lim, C.J.; Hussain, S.; Baek, D.; et al. Rheostatic Control of ABA Signaling through HOS15-Mediated OST1 Degradation. Mol. Plant 2019, 12, 1447–1462. [Google Scholar] [CrossRef] [PubMed]
- Tarnowski, K.; Klimecka, M.; Ciesielski, A.; Goch, G.; Kulik, A.; Fedak, H.; Poznanski, J.; Lichocka, M.; Pierechod, M.; Engh, R.A.; et al. Two SnRK2-interacting Calcium Sensor Isoforms Negatively Regulate SnRK2 Activity by Different Mechanisms. Plant Physiol. 2019, 182, 1142–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bucholc, M.; Ciesielski, A.; Goch, G.; Anielska-Mazur, A.; Kulik, A.; Krzywińska, E.; Dobrowolska, G. SNF1-related protein kinases 2 are negatively regulated by a plant-specific calcium sensor. J. Biol. Chem. 2011, 286, 3429–3441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antoni, R.; Gonzalez-Guzman, M.; Rodriguez, L.; Peirats-Llobet, M.; Pizzio, G.A.; Fernandez, M.A.; De Winne, N.; De Jaeger, G.; Dietrich, D.; Bennett, M.J.; et al. PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol. 2013, 161, 931–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, L.; Cheng, J.; Zhu, Y.; Ding, Y.; Meng, J.; Chen, Z.; Xie, Q.; Guo, Y.; Li, J.; Yang, S.; et al. Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 2015, 6, 8630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Q.; Zhang, X.; Peirats-Llobet, M.; Belda-Palazon, B.; Wang, X.; Cui, S.; Yu, X.; Rodriguez, P.L.; An, C. Ubiquitin Ligases RGLG1 and RGLG5 Regulate Abscisic Acid Signaling by Controlling the Turnover of Phosphatase PP2CA. Plant Cell 2016, 28, 2178–2196. [Google Scholar] [CrossRef] [Green Version]
- Baek, W.; Lim, C.W.; Luan, S.; Lee, S.C. The RING finger E3 ligases PIR1 and PIR2 mediate PP2CA degradation to enhance abscisic acid response in Arabidopsis. Plant J. 2019, 100, 473–486. [Google Scholar] [CrossRef]
- Julian, J.; Coego, A.; Lozano-Juste, J.; Lechner, E.; Wu, Q.; Zhang, X.; Merilo, E.; Belda-Palazon, B.; Park, S.Y.; Cutler, S.R.; et al. The MATH-BTB BPM3 and BPM5 subunits of Cullin3-RING E3 ubiquitin ligases target PP2CA and other clade A PP2Cs for degradation. Proc. Natl. Acad. Sci. USA 2019, 116, 15725–15734. [Google Scholar] [CrossRef] [Green Version]
- Nishimura, N.; Tsuchiya, W.; Moresco, J.J.; Hayashi, Y.; Satoh, K.; Kaiwa, N.; Irisa, T.; Kinoshita, T.; Schroeder, J.I.; Yates, J.R.; et al. Control of seed dormancy and germination by DOG1-AHG1 PP2C phosphatase complex via binding to heme. Nat. Commun. 2018, 9, 2132. [Google Scholar] [CrossRef]
- Née, G.; Kramer, K.; Nakabayashi, K.; Yuan, B.; Xiang, Y.; Miatton, E.; Finkemeier, I.; Soppe, W.J.J. DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nat. Commun. 2017, 8, 72. [Google Scholar] [CrossRef]
- Yatusevich, R.; Fedak, H.; Ciesielski, A.; Krzyczmonik, K.; Kulik, A.; Dobrowolska, G.; Swiezewski, S. Antisense transcription represses. EMBO Rep. 2017, 18, 2186–2196. [Google Scholar] [CrossRef]
- Zhang, L.; Li, X.; Li, D.; Sun, Y.; Li, Y.; Luo, Q.; Liu, Z.; Wang, J.; Zhang, H.; Lou, Z.; et al. CARK1 mediates ABA signaling by phosphorylation of ABA receptors. Cell Discov. 2018, 4, 30. [Google Scholar] [CrossRef]
- Chen, H.H.; Qu, L.; Xu, Z.H.; Zhu, J.K.; Xue, H.W. EL1-like Casein Kinases Suppress ABA Signaling and Responses by Phosphorylating and Destabilizing the ABA Receptors PYR/PYLs in Arabidopsis. Mol. Plant 2018, 11, 706–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mega, R.; Abe, F.; Kim, J.S.; Tsuboi, Y.; Tanaka, K.; Kobayashi, H.; Sakata, Y.; Hanada, K.; Tsujimoto, H.; Kikuchi, J.; et al. Tuning water-use efficiency and drought tolerance in wheat using abscisic acid receptors. Nat. Plants 2019, 5, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhao, Y.; Li, Z.; Hsu, C.C.; Liu, X.; Fu, L.; Hou, Y.J.; Du, Y.; Xie, S.; Zhang, C.; et al. Reciprocal Regulation of the TOR Kinase and ABA Receptor Balances Plant Growth and Stress Response. Mol. Cell 2018, 69, 100–112.e106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, C.; Xiao, L.; Hua, K.; Zou, C.; Zhao, Y.; Bressan, R.A.; Zhu, J.K. Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc. Natl. Acad. Sci. USA 2018, 115, 6058–6063. [Google Scholar] [CrossRef] [Green Version]
- Kulik, A.; Anielska-Mazur, A.; Bucholc, M.; Koen, E.; Szymanska, K.; Zmienko, A.; Krzywinska, E.; Wawer, I.; McLoughlin, F.; Ruszkowski, D.; et al. SNF1-related protein kinases type 2 are involved in plant responses to cadmium stress. Plant Physiol. 2012, 160, 868–883. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Chua, N.H. Dehydration stress activates Arabidopsis MPK6 to signal DCP1 phosphorylation. EMBO J. 2012, 31, 1975–1984. [Google Scholar] [CrossRef] [Green Version]
- Maszkowska, J.; Dębski, J.; Kulik, A.; Kistowski, M.; Bucholc, M.; Lichocka, M.; Klimecka, M.; Sztatelman, O.; Szymańska, K.P.; Dadlez, M.; et al. Phosphoproteomic Analysis Reveals that Dehydrins ERD10 and ERD14 are Phosphorylated by SNF1-related Protein Kinase 2.10 in Response to Osmotic Stress. Plant Cell Environ. 2018, 42, 931–946. [Google Scholar] [CrossRef]
- Szymańska, K.P.; Polkowska-Kowalczyk, L.; Lichocka, M.; Maszkowska, J.; Dobrowolska, G. SNF1-Related Protein Kinases SnRK2.4 and SnRK2.10 Modulate ROS Homeostasis in Plant Response to Salt Stress. Int. J. Mol. Sci. 2019, 20, 143. [Google Scholar] [CrossRef] [Green Version]
- Stecker, K.E.; Minkoff, B.B.; Sussman, M.R. Phosphoproteomic Analyses Reveal Early Signaling Events in the Osmotic Stress Response. Plant Physiol. 2014, 165, 1171–1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krzywińska, E.; Kulik, A.; Bucholc, M.; Fernandez, M.A.; Rodriguez, P.L.; Dobrowolska, G. Protein phosphatase type 2C PP2CA together with ABI1 inhibits SnRK2.4 activity and regulates plant responses to salinity. Plant Signal. Behav. 2016, 11, e1253647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Julkowska, M.M.; McLoughlin, F.; Galvan-Ampudia, C.S.; Rankenberg, J.M.; Kawa, D.; Klimecka, M.; Haring, M.A.; Munnik, T.; Kooijman, E.E.; Testerink, C. Identification and functional characterization of the Arabidopsis Snf1-related protein kinase SnRK2.4 phosphatidic acid-binding domain. Plant Cell Environ. 2015, 38, 614–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Song, T.; Wallrad, L.; Kudla, J.; Wang, X.; Zhang, W. Tissue-specific accumulation of pH-sensing phosphatidic acid determines plant stress tolerance. Nat. Plants 2019, 5, 1012–1021. [Google Scholar] [CrossRef]
- Takahashi, F.; Kuromori, T.; Urano, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Drought Stress Responses and Resistance in Plants: From Cellular Responses to Long-Distance Intercellular Communication. Front. Plant Sci. 2020, 11, 556972. [Google Scholar] [CrossRef]
- Yoshida, T.; Fernie, A.R.; Shinozaki, K.; Takahashi, F. Long-distance stress and developmental signals associated with abscisic acid signaling in environmental responses. Plant J. 2021, 105, 477–488. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 1994, 6, 251–264. [Google Scholar] [CrossRef] [Green Version]
- Maruyama, K.; Todaka, D.; Mizoi, J.; Yoshida, T.; Kidokoro, S.; Matsukura, S.; Takasaki, H.; Sakurai, T.; Yamamoto, Y.Y.; Yoshiwara, K.; et al. Identification of cis-acting promoter elements in cold- and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 2012, 19, 37–49. [Google Scholar] [CrossRef]
- Fujita, Y.; Fujita, M.; Satoh, R.; Maruyama, K.; Parvez, M.M.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 2005, 17, 3470–3488. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef]
- Shen, Q.; Zhang, P.; Ho, T.H. Modular nature of abscisic acid (ABA) response complexes: Composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell 1996, 8, 1107–1119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [Green Version]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 2006, 18, 1292–1309. [Google Scholar] [CrossRef] [Green Version]
- Engels, C.; Fuganti-Pagliarini, R.; Marin, S.R.; Marcelino-Guimaraes, F.C.; Oliveira, M.C.; Kanamori, N.; Mizoi, J.; Nakashima, K.; Yamaguchi-Shinozaki, K.; Nepomuceno, A.L. Introduction of the rd29A:AtDREB2A CA gene into soybean (Glycine max L. Merril) and its molecular characterization in leaves and roots during dehydration. Genet. Mol. Biol. 2013, 36, 556–565. [Google Scholar] [CrossRef]
- Qin, F.; Sakuma, Y.; Tran, L.S.; Maruyama, K.; Kidokoro, S.; Fujita, Y.; Fujita, M.; Umezawa, T.; Sawano, Y.; Miyazono, K.; et al. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 2008, 20, 1693–1707. [Google Scholar] [CrossRef] [Green Version]
- Sakuma, Y.; Maruyama, K.; Qin, F.; Osakabe, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. USA 2006, 103, 18822–18827. [Google Scholar] [CrossRef] [Green Version]
- Morimoto, K.; Ohama, N.; Kidokoro, S.; Mizoi, J.; Takahashi, F.; Todaka, D.; Mogami, J.; Sato, H.; Qin, F.; Kim, J.S.; et al. BPM-CUL3 E3 ligase modulates thermotolerance by facilitating negative regulatory domain-mediated degradation of DREB2A in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E8528–E8536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, H.; Mizoi, J.; Tanaka, H.; Maruyama, K.; Qin, F.; Osakabe, Y.; Morimoto, K.; Ohori, T.; Kusakabe, K.; Nagata, M.; et al. Arabidopsis DPB3-1, a DREB2A interactor, specifically enhances heat stress-induced gene expression by forming a heat stress-specific transcriptional complex with NF-Y subunits. Plant Cell 2014, 26, 4954–4973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizoi, J.; Kanazawa, N.; Kidokoro, S.; Takahashi, F.; Qin, F.; Morimoto, K.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Heat-induced inhibition of phosphorylation of the stress-protective transcription factor DREB2A promotes thermotolerance of Arabidopsis thaliana. J. Biol. Chem. 2019, 294, 902–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.S.; Mizoi, J.; Yoshida, T.; Fujita, Y.; Nakajima, J.; Ohori, T.; Todaka, D.; Nakashima, K.; Hirayama, T.; Shinozaki, K.; et al. An ABRE Promoter Sequence is Involved in Osmotic Stress-Responsive Expression of the DREB2A Gene, Which Encodes a Transcription Factor Regulating Drought-Inducible Genes in Arabidopsis. Plant Cell Physiol. 2011, 52, 2136–2146. [Google Scholar] [CrossRef]
- Himmelbach, A.; Hoffmann, T.; Leube, M.; Hohener, B.; Grill, E. Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 2002, 21, 3029–3038. [Google Scholar] [CrossRef] [Green Version]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef]
- Tran, L.S.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takasaki, H.; Maruyama, K.; Kidokoro, S.; Ito, Y.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K.; Nakashima, K. The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol. Genet. Genom. 2010, 284, 173–183. [Google Scholar] [CrossRef]
- Xu, Z.Y.; Kim, S.Y.; Hyeon do, Y.; Kim, D.H.; Dong, T.; Park, Y.; Jin, J.B.; Joo, S.H.; Kim, S.K.; Hong, J.C.; et al. The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 2013, 25, 4708–4724. [Google Scholar] [CrossRef] [Green Version]
- Sakuraba, Y.; Kim, Y.S.; Han, S.H.; Lee, B.D.; Paek, N.C. The Arabidopsis Transcription Factor NAC016 Promotes Drought Stress Responses by Repressing AREB1 Transcription through a Trifurcate Feed-Forward Regulatory Loop Involving NAP. Plant Cell 2015, 27, 1771–1787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takasaki, H.; Maruyama, K.; Takahashi, F.; Fujita, M.; Yoshida, T.; Nakashima, K.; Myouga, F.; Toyooka, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J. 2015, 84, 1114–1123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abe, H.; Yamaguchi-Shinozaki, K.; Urao, T.; Iwasaki, T.; Hosokawa, D.; Shinozaki, K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 1997, 9, 1859–1868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [Green Version]
- Kazan, K.; Manners, J.M. MYC2: The master in action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Duan, G.; Li, C.; Liu, L.; Han, G.; Zhang, Y.; Wang, C. The Crosstalks Between Jasmonic Acid and Other Plant Hormone Signaling Highlight the Involvement of Jasmonic Acid as a Core Component in Plant Response to Biotic and Abiotic Stresses. Front. Plant Sci. 2019, 10, 1349. [Google Scholar] [CrossRef] [Green Version]
- Seo, P.J.; Xiang, F.; Qiao, M.; Park, J.Y.; Lee, Y.N.; Kim, S.G.; Lee, Y.H.; Park, W.J.; Park, C.M. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009, 151, 275–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, Y.; Sunarti, S.; Kissoudis, C.; Visser, R.G.F.; van der Linden, C.G. The Role of Tomato WRKY Genes in Plant Responses to Combined Abiotic and Biotic Stresses. Front. Plant Sci. 2018, 9, 801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rushton, D.L.; Tripathi, P.; Rabara, R.C.; Lin, J.; Ringler, P.; Boken, A.K.; Langum, T.J.; Smidt, L.; Boomsma, D.D.; Emme, N.J.; et al. WRKY transcription factors: Key components in abscisic acid signalling. Plant. Biotechnol. J. 2012, 10, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Yan, L.; Liu, Z.Q.; Cao, Z.; Mei, C.; Xin, Q.; Wu, F.Q.; Wang, X.F.; Du, S.Y.; Jiang, T.; et al. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 2010, 22, 1909–1935. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Lai, Z.; Shi, J.; Xiao, Y.; Chen, Z.; Xu, X. Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 2010, 10, 281. [Google Scholar] [CrossRef] [Green Version]
- Ren, X.; Chen, Z.; Liu, Y.; Zhang, H.; Zhang, M.; Liu, Q.; Hong, X.; Zhu, J.K.; Gong, Z. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010, 63, 417–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soma, F.; Takahashi, F.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Cellular Phosphorylation Signaling and Gene Expression in Drought Stress Responses: ABA-Dependent and ABA-Independent Regulatory Systems. Plants 2021, 10, 756. https://doi.org/10.3390/plants10040756
Soma F, Takahashi F, Yamaguchi-Shinozaki K, Shinozaki K. Cellular Phosphorylation Signaling and Gene Expression in Drought Stress Responses: ABA-Dependent and ABA-Independent Regulatory Systems. Plants. 2021; 10(4):756. https://doi.org/10.3390/plants10040756
Chicago/Turabian StyleSoma, Fumiyuki, Fuminori Takahashi, Kazuko Yamaguchi-Shinozaki, and Kazuo Shinozaki. 2021. "Cellular Phosphorylation Signaling and Gene Expression in Drought Stress Responses: ABA-Dependent and ABA-Independent Regulatory Systems" Plants 10, no. 4: 756. https://doi.org/10.3390/plants10040756
APA StyleSoma, F., Takahashi, F., Yamaguchi-Shinozaki, K., & Shinozaki, K. (2021). Cellular Phosphorylation Signaling and Gene Expression in Drought Stress Responses: ABA-Dependent and ABA-Independent Regulatory Systems. Plants, 10(4), 756. https://doi.org/10.3390/plants10040756





