Olive Cultivars Susceptible or Tolerant to Xylella fastidiosa Subsp. pauca Exhibit Mid-Term Different Metabolomes upon Natural Infection or a Curative Treatment
Abstract
:1. Introduction
2. Results and Discussion
2.1. Selection of Leccino, Ogliarola Salentina and Cellina Di Nardò Metabolic Profiles in a Mid-Term Treatment Study
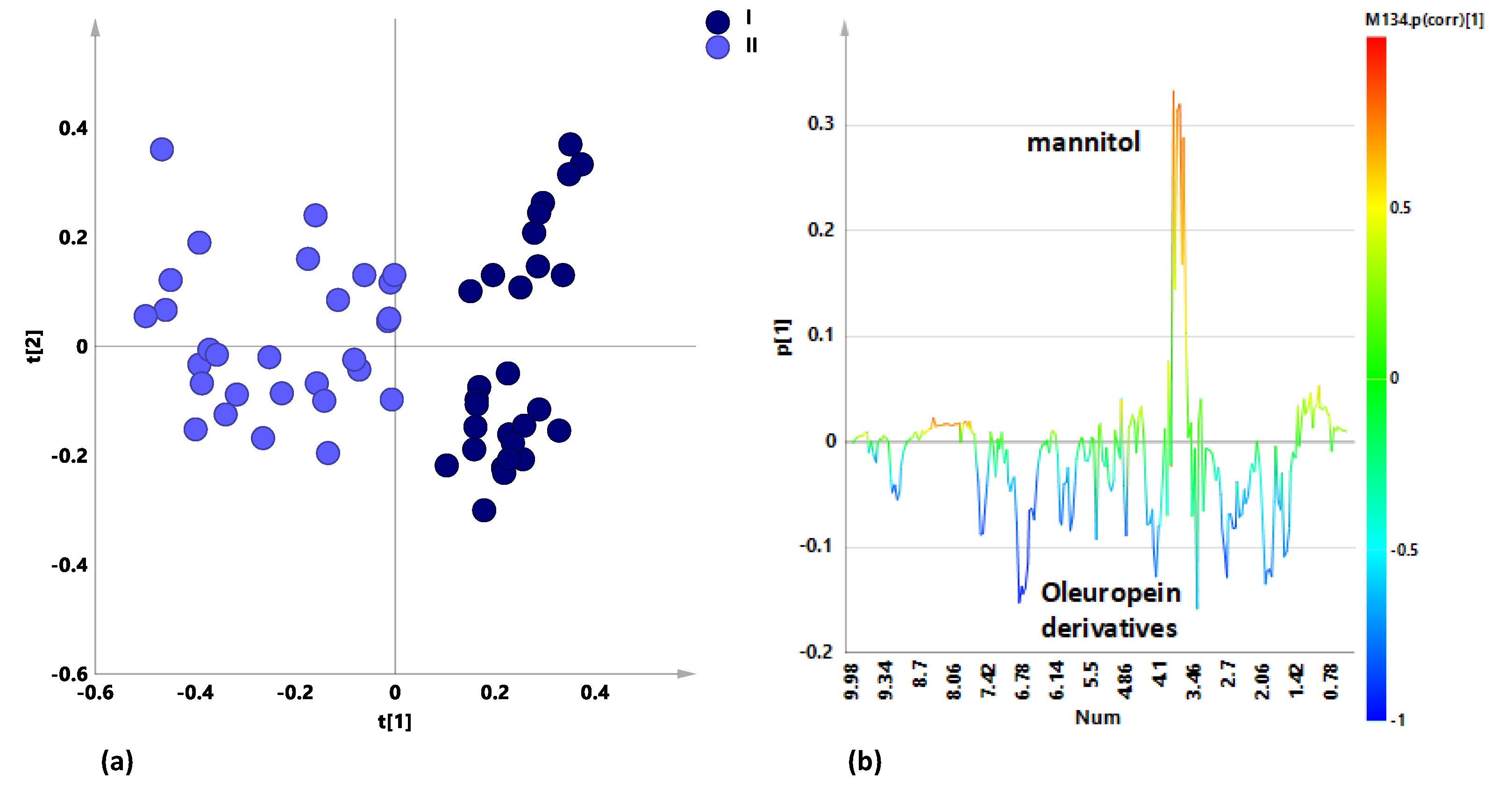
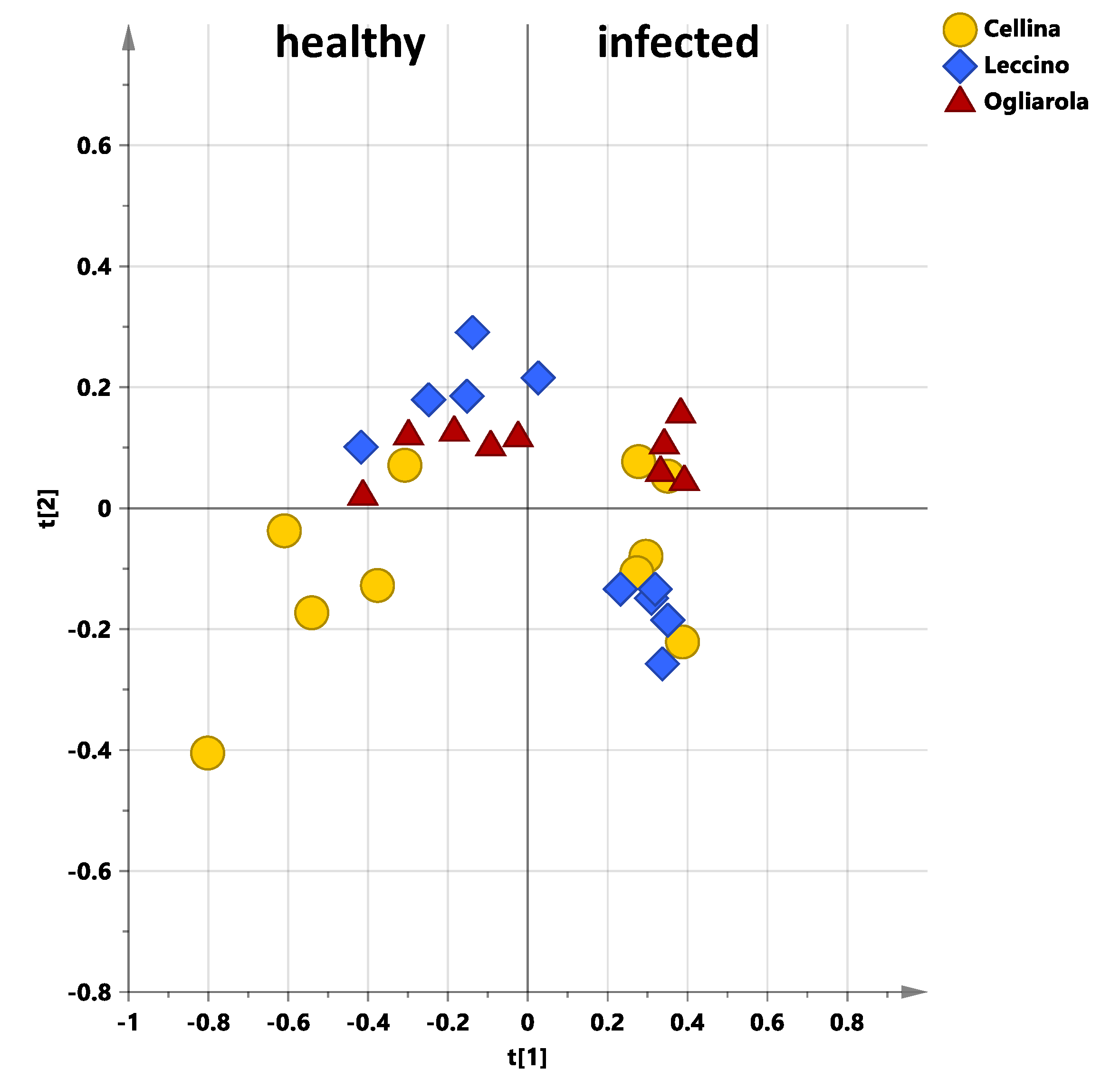
2.2. General Effects of Sampling Period and Treatment on Leaf Extracts Metabolic Profiles for Naturally Infected Leccino, Cellina Di Nardò and Ogliarola Salentina
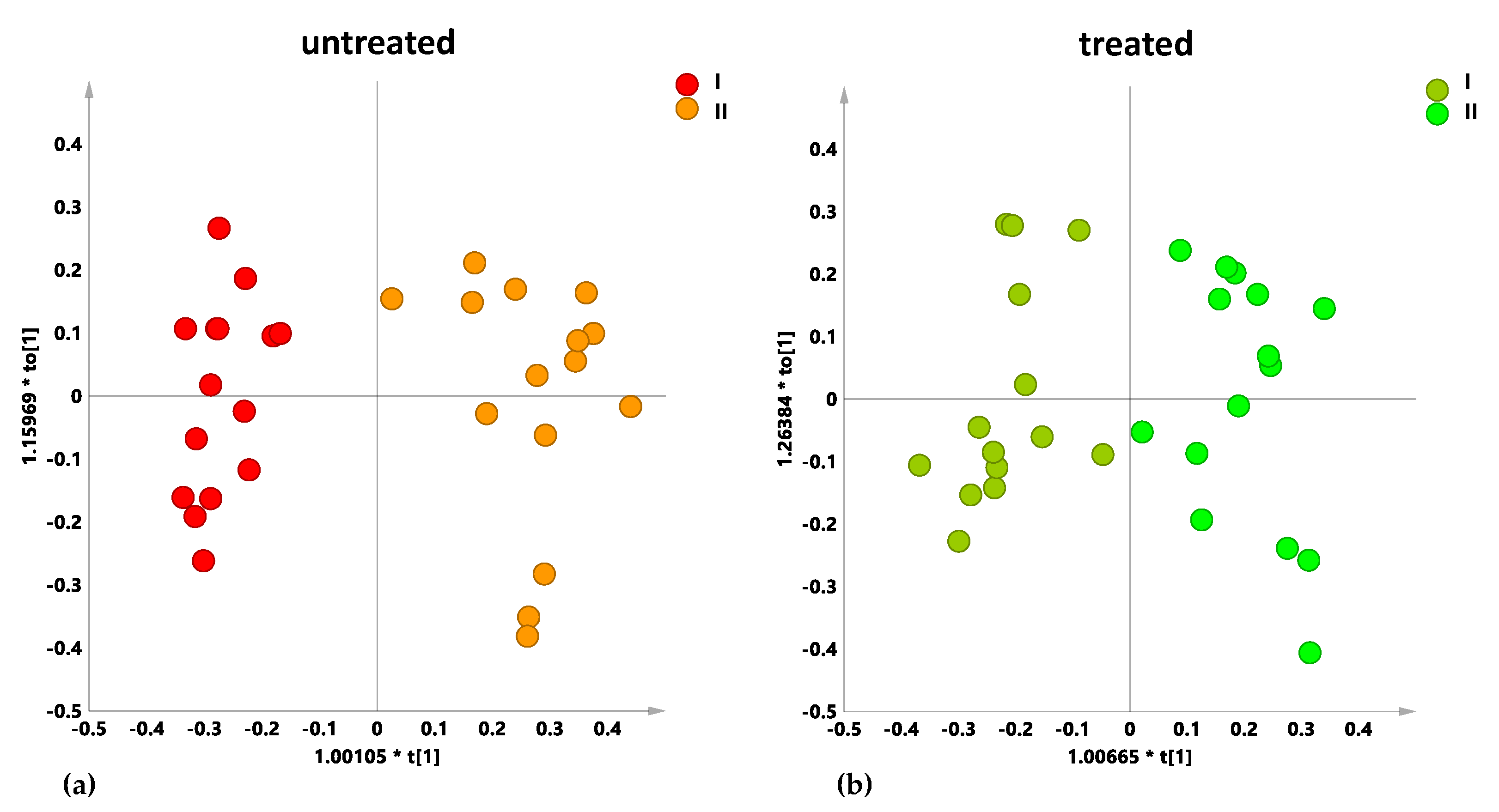
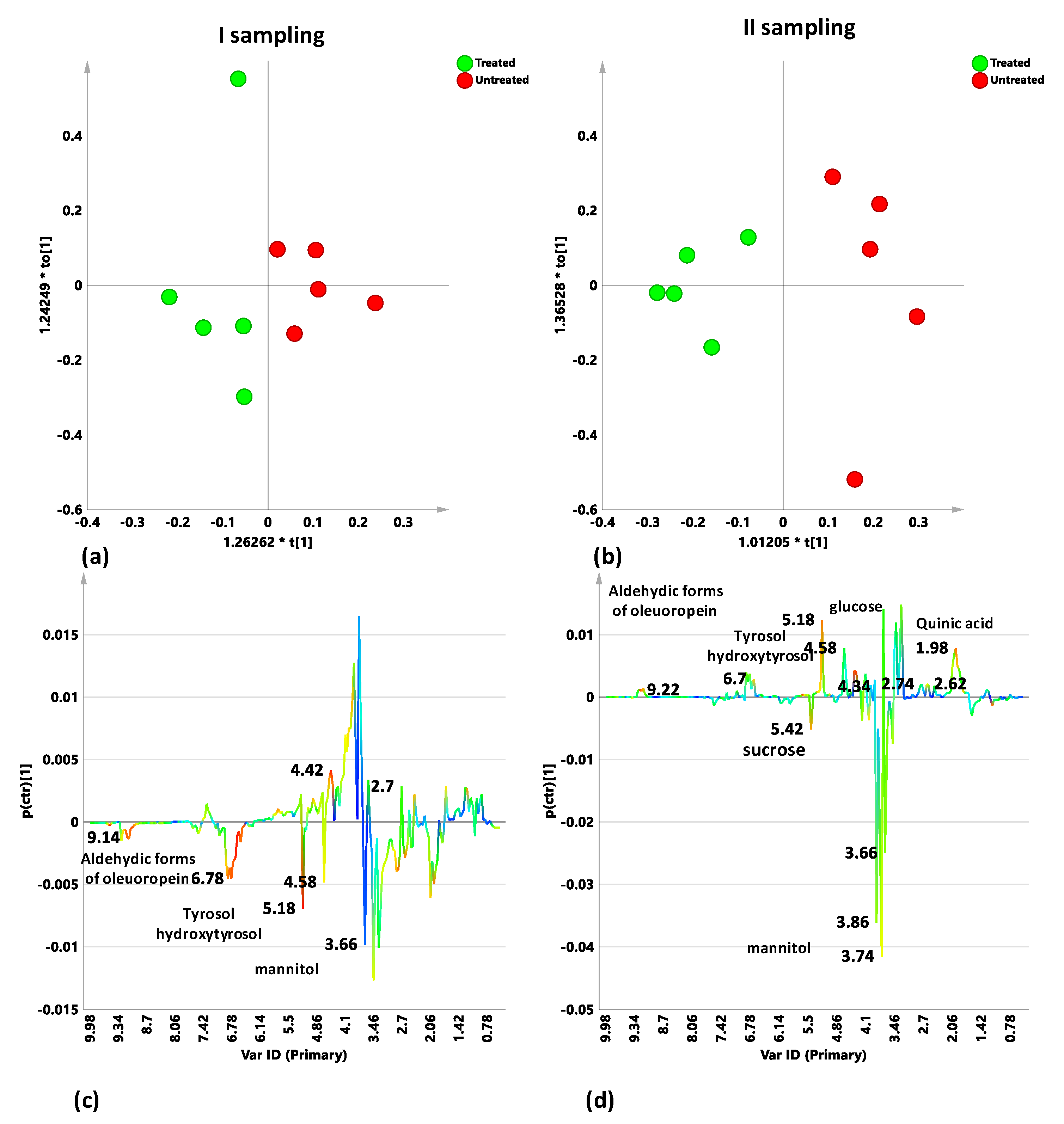
2.3. Sampling Period Effects on Leaf Extract Metabolic Profiles for Treated and Untreated Naturally Infected Leccino, Cellina Di Nardò and Ogliarola Salentina
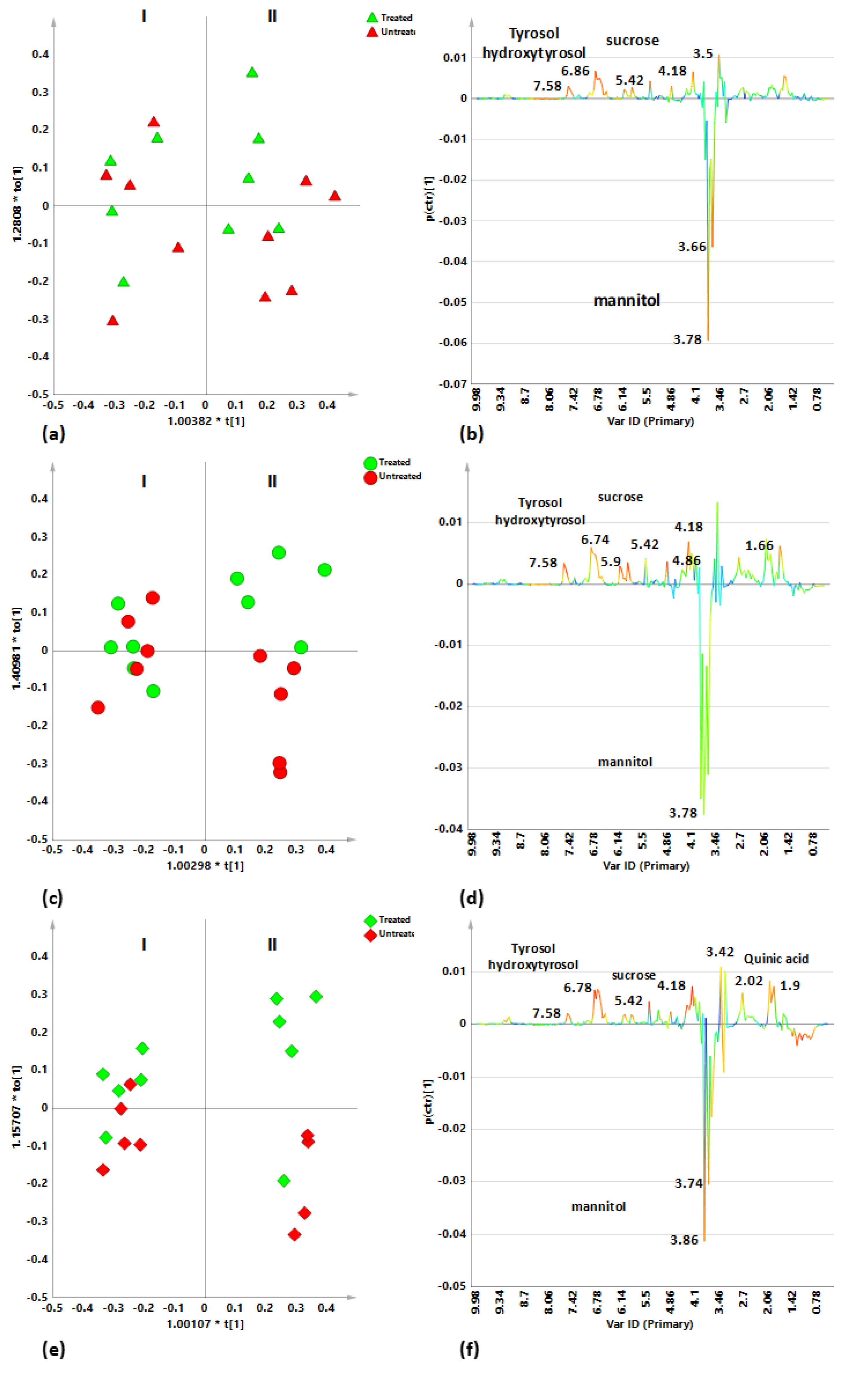
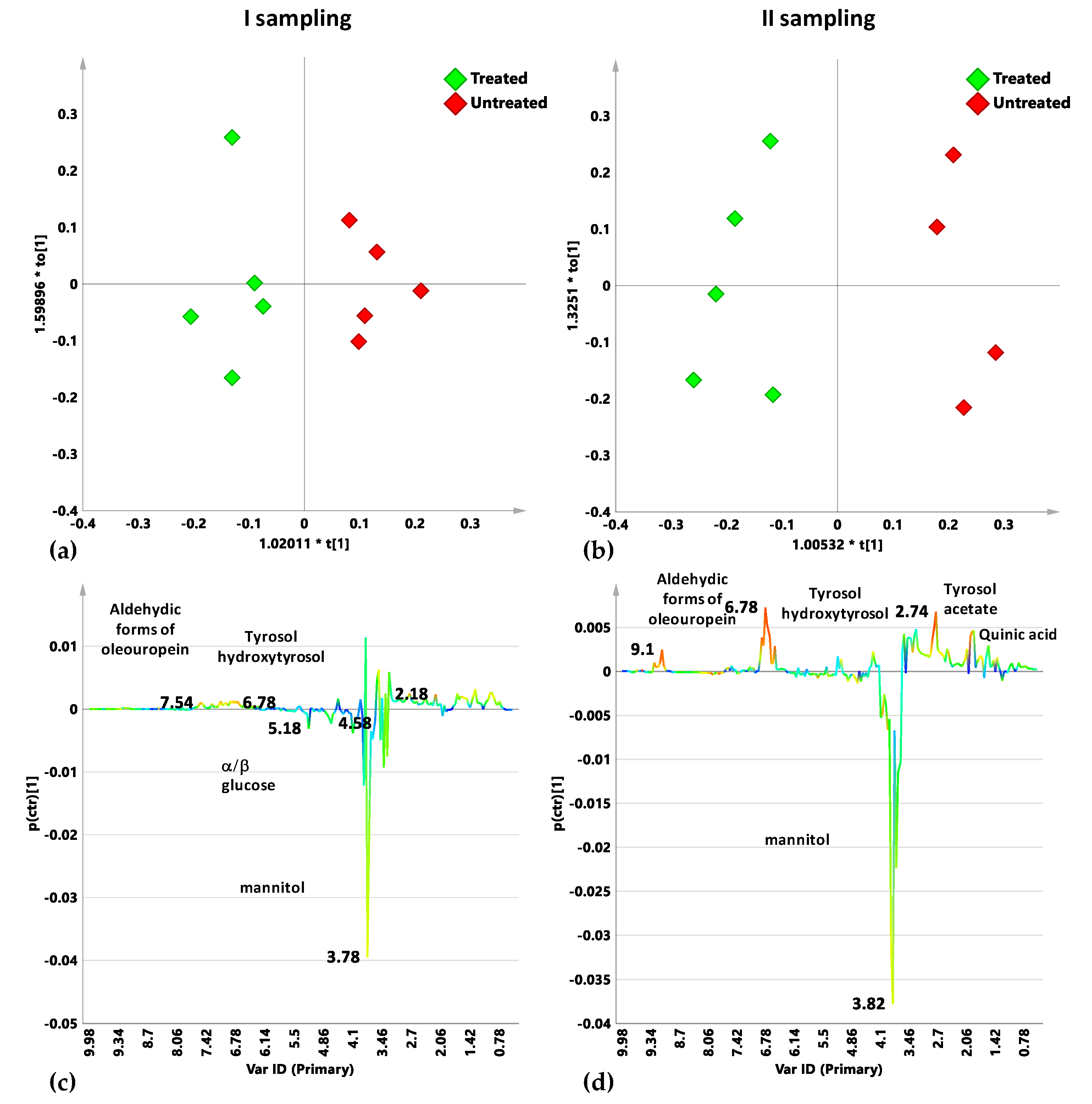
2.4. Cultivar-Related Treatment Effects for Naturally Infected Leccino, Cellina Di Nardò and Ogliarola Salentina over the Two Leaf Samplings
2.5. Leccino vs. Susceptible Cultivars Differences in Infected Trees
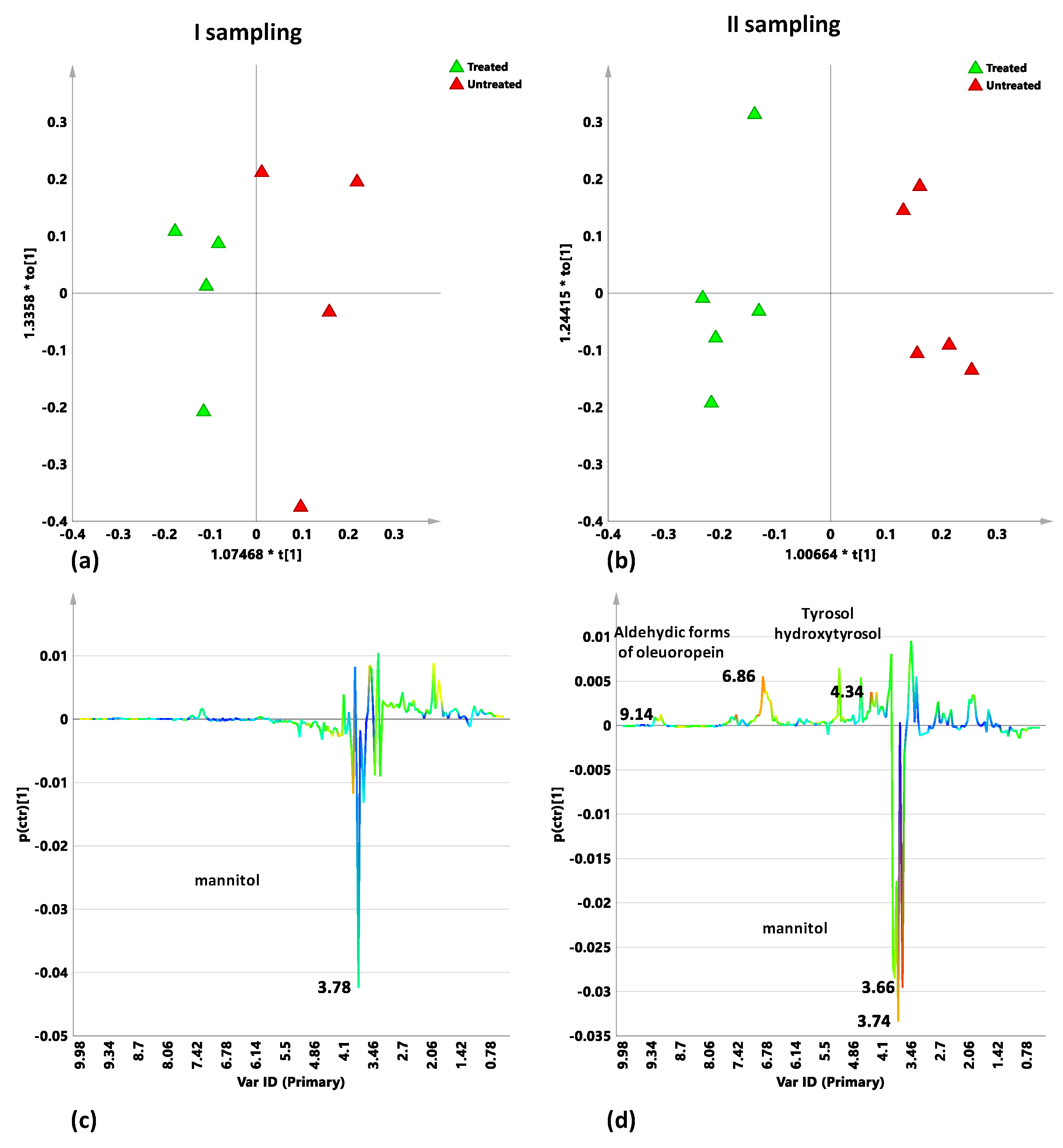
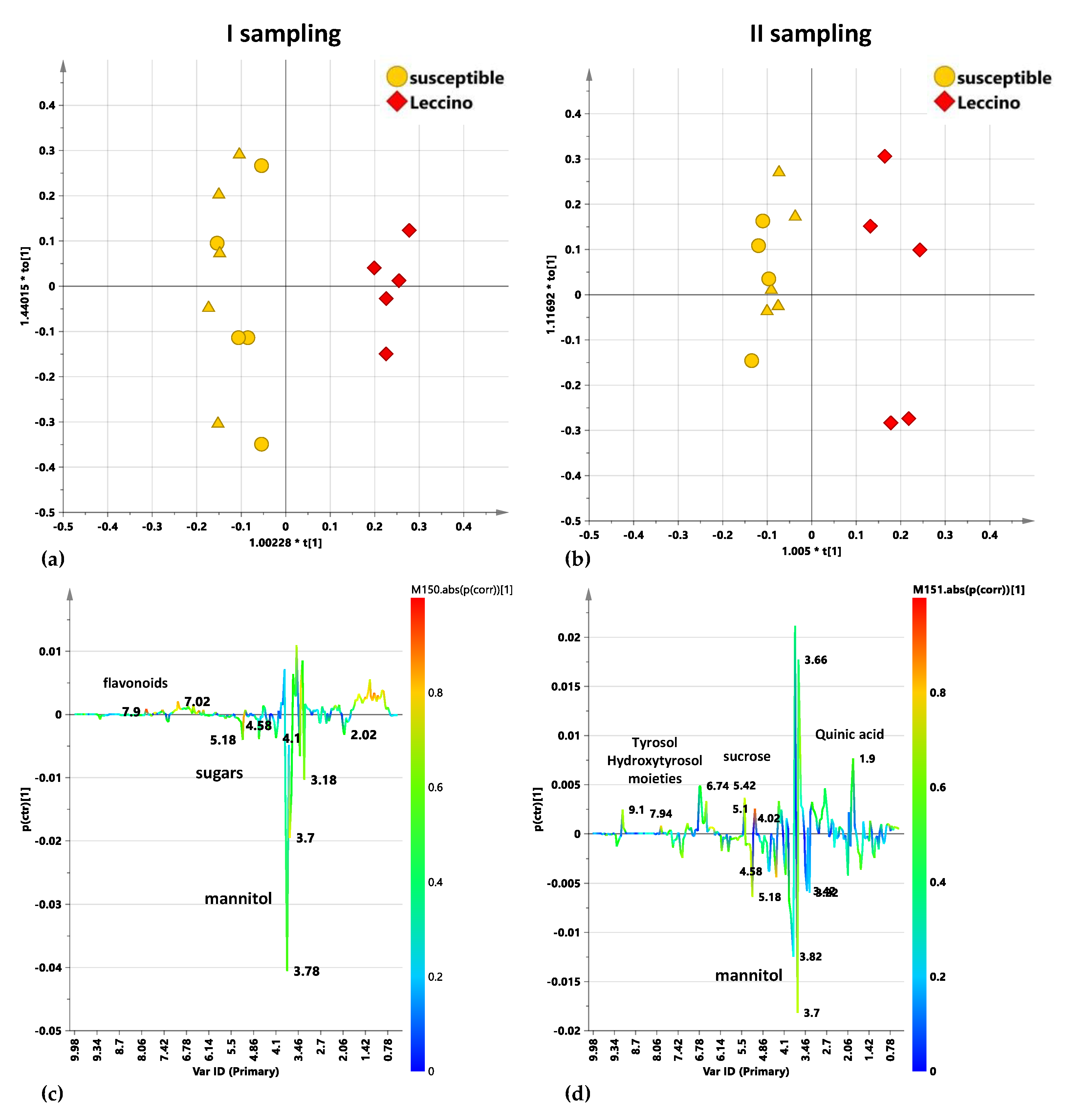
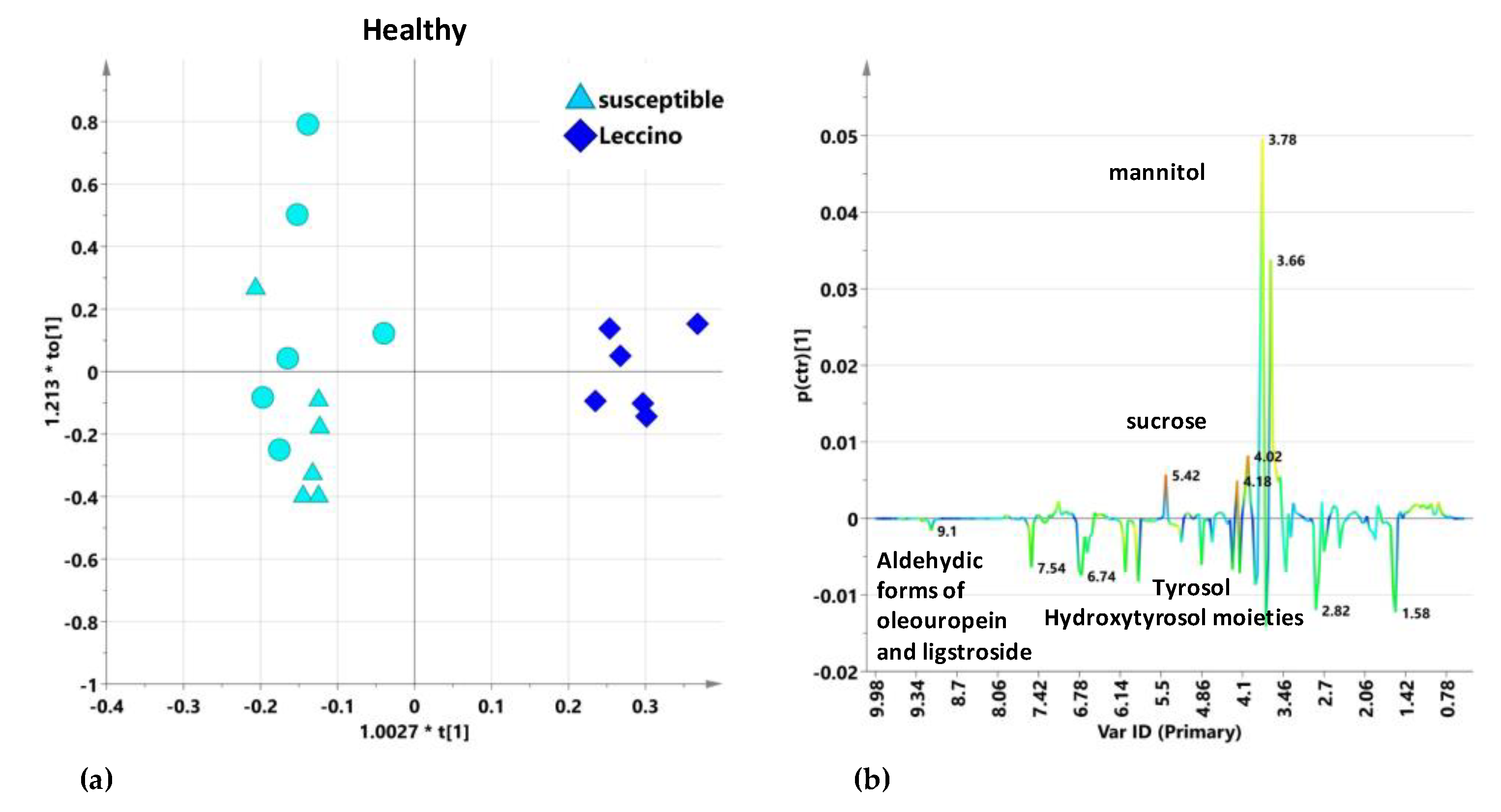
2.5.1. Untreated Leccino vs. Susceptible Cultivars Trees: I and II Sampling
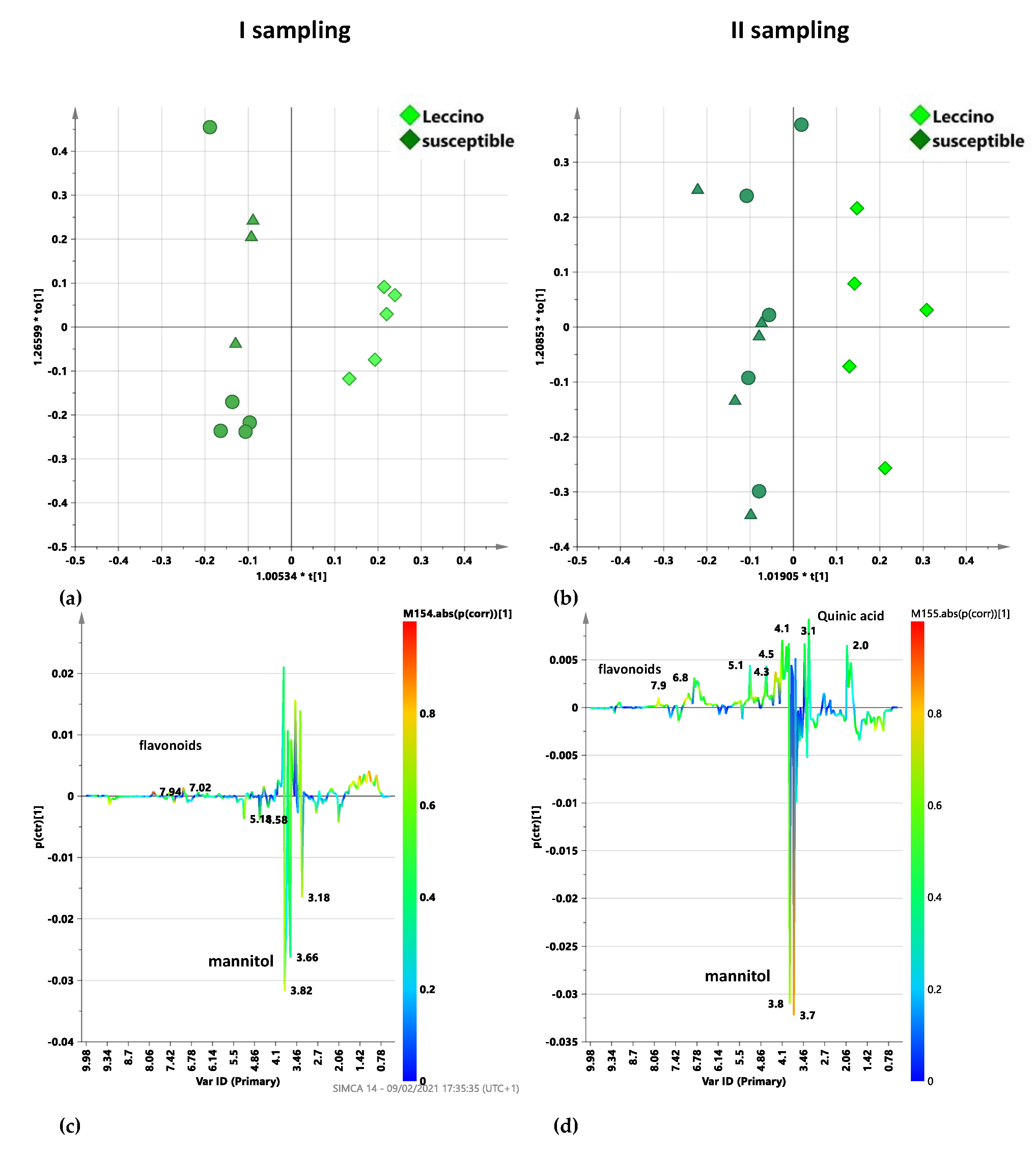
2.5.2. Dentamet®-Treated Leccino vs. Susceptible Cultivars Trees: I and II Sampling
2.5.3. Leccino vs. Susceptible Cultivars in Infected and Healthy Trees: General Remarks
3. Materials and Methods
3.1. Sampling
3.2. Sample Preparation for 1H-NMR Analysis
3.3. H-NMR Spectra Acquisition and Processing
3.4. Multivariate Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sardaro, R.; Fucilli, V.; Acciani, C. Measuring the value of rural landscape in support of preservation policies. Sci. Reg. 2015, 125–138. [Google Scholar] [CrossRef]
- ISTAT. Available online: http://dati.istat.it/ (accessed on 25 September 2020).
- Martelli, G.P. The current status of the quick decline syndrome of olive in southern Italy. Phytoparasitica 2016, 44, 1–10. [Google Scholar] [CrossRef]
- Cimato, A.; Sani, G.; De Rinaldis, G.; Calogiuri, A. Il germoplasma olivicolo in provincia di Lecce. In Recupero, Conservazione, Selezione e Caratterizzazione delle Varietà Autoctone; Camera di Commercio, Industria, Artigianato e Agricoltura: Lecce, Italy, 2001. [Google Scholar]
- Del Coco, L.; De Pascali, S.A.; Fanizzi, F.P. NMR-metabolomic study on monocultivar and blend salento EVOOs including some from secular olive trees. Food Nutr. Sci. 2014, 5, 89–95. [Google Scholar]
- Saponari, M.; Boscia, D.; Altamura, G.; Loconsole, G.; Zicca, S.; D’Attoma, G.; Morelli, M.; Palmisano, F.; Saponari, A.; Tavano, D. Isolation and pathogenicity of Xylella fastidiosa associated to the olive quick decline syndrome in southern Italy. Sci. Rep. 2017, 7, 17723. [Google Scholar] [CrossRef]
- Petruccelli, R.; Beghe, D.; Ganino, T.; Bartolini, G.; Ciaccheri, L.; Bernardi, R.; Durante, M. Evaluation of intra-cultivar variability in ‘Olea europaea’ L. cv. Leccino using morphological, biochemical and molecular markers. Aust. J. Crop Sci. 2020, 14, 588. [Google Scholar] [CrossRef]
- Giampetruzzi, A.; Morelli, M.; Saponari, M.; Loconsole, G.; Chiumenti, M.; Boscia, D.; Savino, V.N.; Martelli, G.P.; Saldarelli, P. Transcriptome profiling of two olive cultivars in response to infection by the CoDiRO strain of Xylella fastidiosa subsp. pauca. BMC Genom. 2016, 17, 475. [Google Scholar] [CrossRef]
- Sabella, E.; Luvisi, A.; Aprile, A.; Negro, C.; Vergine, M.; Nicolì, F.; Miceli, A.; De Bellis, L. Xylella fastidiosa induces differential expression of lignification related-genes and lignin accumulation in tolerant olive trees cv. Leccino. J. Plant Physiol. 2018, 220, 60–68. [Google Scholar] [CrossRef]
- Sabella, E.; Aprile, A.; Genga, A.; Siciliano, T.; Nutricati, E.; Nicolì, F.; Vergine, M.; Negro, C.; De Bellis, L.; Luvisi, A. Xylem cavitation susceptibility and refilling mechanisms in olive trees infected by Xylella fastidiosa. Sci. Rep. 2019, 9, 9602. [Google Scholar] [CrossRef]
- Vergine, M.; Meyer, J.B.; Cardinale, M.; Sabella, E.; Hartmann, M.; Cherubini, P.; De Bellis, L.; Luvisi, A. The Xylella fastidiosa-Resistant Olive Cultivar “Leccino” Has Stable Endophytic Microbiota during the Olive Quick Decline Syndrome (OQDS). Pathogens 2020, 9, 35. [Google Scholar] [CrossRef]
- Krishnan, P.; Kruger, N.; Ratcliffe, R. Metabolite fingerprinting and profiling in plants using NMR. J. Exp. Bot. 2005, 56, 255–265. [Google Scholar] [CrossRef]
- Deborde, C.; Moing, A.; Roch, L.; Jacob, D.; Rolin, D.; Giraudeau, P. Plant metabolism as studied by NMR spectroscopy. Prog. Nucl. Magn. Reson. Spectrosc. 2017, 102, 61–97. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Choi, Y.H.; Verpoorte, R. NMR-based metabolomic analysis of plants. Nat. Protoc. 2010, 5, 536–549. [Google Scholar] [CrossRef]
- Girelli, C.R.; Angilè, F.; Del Coco, L.; Migoni, D.; Zampella, L.; Marcelletti, S.; Cristella, N.; Marangi, P.; Scortichini, M.; Fanizzi, F.P. 1H-NMR metabolite fingerprinting analysis reveals a disease biomarker and a field treatment response in Xylella fastidiosa subsp. pauca-Infected Olive Trees. Plants 2019, 8, 115. [Google Scholar]
- Girelli, C.R.; Del Coco, L.; Scortichini, M.; Petriccione, M.; Zampella, L.; Mastrobuoni, F.; Cesari, G.; Bertaccini, A.; D’amico, G.; Contaldo, N. Xylella fastidiosa and olive quick decline syndrome (CoDiRO) in Salento (southern Italy): A chemometric 1H NMR-based preliminary study on Ogliarola salentina and Cellina di Nardò cultivars. Chem. Biol. Technol. Agric. 2017, 4, 25. [Google Scholar] [CrossRef]
- Girelli, C.R.; Del Coco, L.; Fanizzi, F.P. 1H NMR spectroscopy and multivariate analysis as possible tool to assess cultivars, from specific geographical areas, in EVOOs. Eur. J. Lipid Sci. Technol. 2016, 118, 1380–1388. [Google Scholar] [CrossRef]
- Girelli, C.R.; Schiavone, R.; Vilella, S.; Fanizzi, F.P. Salento Honey (Apulia, South-East Italy): A Preliminary Characterization by 1H-NMR Metabolomic Fingerprinting. Sustainability 2020, 12, 5009. [Google Scholar] [CrossRef]
- Luvisi, A.; Aprile, A.; Sabella, E.; Vergine, M.; Nicoli, F.; Nutricati, E.; Miceli, A.; Negro, C.; De Bellis, L. Xylella fastidiosa subsp. pauca (CoDiRO strain) infection in four olive (Olea europaea L.) cultivars: Profile of phenolic compounds in leaves and progression of leaf scorch symptoms. Phytopathol. Mediterr. 2017, 56, 259–273. [Google Scholar]
- Scortichini, M.; Jianchi, C.; De Caroli, M.; Dalessandro, G.; Pucci, N.; Modesti, V.; L’Aurora, A.; Petriccione, M.; Zampella, L.; Mastrobuoni, F. A zinc, copper and citric acid biocomplex shows promise for control of Xylella fastidiosa subsp. pauca in olive trees in Apulia region (southern Italy). Phytopathol. Mediterr. 2018, 57, 48–72. [Google Scholar]
- Tatulli, G.; Modesti, V.; Pucci, N.; Scala, V.; L’Aurora, A.; Lucchesi, S.; Salustri, M.; Scortichini, M.; Loreti, S. Further in Vitro Assessment and Mid-Term Evaluation of Control Strategy of Xylella fastidiosa subsp. pauca in Olive Groves of Salento (Apulia, Italy). Pathogens 2021, 10, 85. [Google Scholar]
- Oddo, E.; Saiano, F.; Alonzo, G.; Bellini, E. An investigation of the seasonal pattern of mannitol content in deciduous and evergreen species of the Oleaceae growing in northern Sicily. Ann. Bot. 2002, 90, 239–243. [Google Scholar] [CrossRef]
- Jlilat, A.; Ragone, R.; Gualano, S.; Santoro, F.; Gallo, V.; Varvaro, L.; Mastrorilli, P.; Saponari, M.; Nigro, F.; D’Onghia, A.M. A non-targeted metabolomics study on Xylella fastidiosa infected olive plants grown under controlled conditions. Sci. Rep. 2021, 11, 11. [Google Scholar] [CrossRef]
- Barros, J.; Serk, H.; Granlund, I.; Pesquet, E. The cell biology of lignification in higher plants. Ann. Bot. 2015, 115, 1053–1074. [Google Scholar] [CrossRef]
- Vergine, M.; Nicolì, F.; Sabella, E.; Aprile, A.; Bellis, L.D.; Luvisi, A. Secondary Metabolites in Xylella fastidiosa—Plant Interaction. Pathogens 2020, 9, 675. [Google Scholar] [CrossRef] [PubMed]
- Dichio, B.; Margiotta, G.; Xiloyannis, C.; Bufo, S.A.; Sofo, A.; Cataldi, T.R. Changes in water status and osmolyte contents in leaves and roots of olive plants (Olea europaea L.) subjected to water deficit. Trees 2009, 23, 247–256. [Google Scholar] [CrossRef]
- Mechri, B.; Tekaya, M.; Hammami, M.; Chehab, H. Effects of drought stress on phenolic accumulation in greenhouse-grown olive trees (Olea europaea). Biochem. Syst. Ecol. 2020, 92, 104112. [Google Scholar] [CrossRef]
- Tekaya, M.; El-Gharbi, S.; Mechri, B.; Chehab, H.; Bchir, A.; Chraief, I.; Ayachi, M.; Boujnah, D.; Attia, F.; Hammami, M. Improving performance of olive trees by the enhancement of key physiological parameters of olive leaves in response to foliar fertilization. Acta Physiol. Plant. 2016, 38, 101. [Google Scholar] [CrossRef]
- Jensen, S.R.; Franzyk, H.; Wallander, E. Chemotaxonomy of the Oleaceae: Iridoids as taxonomic markers. Phytochemistry 2002, 60, 213–231. [Google Scholar] [CrossRef]
- Castejón, M.L.; Montoya, T.; Alarcón-de-la-Lastra, C.; Sánchez-Hidalgo, M. Potential Protective Role Exerted by Secoiridoids from Olea europaea L. in Cancer, Cardiovascular, Neurodegenerative, Aging-Related, and Immunoinflammatory Diseases. Antioxidants 2020, 9, 149. [Google Scholar] [CrossRef]
- Emma, M.R.; Augello, G.; Di Stefano, V.; Azzolina, A.; Giannitrapani, L.; Montalto, G.; Cervello, M.; Cusimano, A. Potential Uses of Olive Oil Secoiridoids for the Prevention and Treatment of Cancer: A Narrative Review of Preclinical Studies. Int. J. Mol. Sci. 2021, 22, 1234. [Google Scholar] [CrossRef]
- Novelli, S.; Gismondi, A.; Di Marco, G.; Canuti, L.; Nanni, V.; Canini, A. Plant defense factors involved in Olea europaea resistance against Xylella fastidiosa infection. J. Plant Res. 2019, 132, 439–455. [Google Scholar] [CrossRef]
- Bartolozzi, F.; Rocchi, P.; Camerini, F.; Fontanazza, G. Changes of biochemical parameters in olive (Olea europaea L.) leaves during an entire vegetative season, and their correlation with frost resistance. Acta Hortic. 1999, 474, 435–440. [Google Scholar] [CrossRef]
- Harper, S.; Ward, L.; Clover, G. Development of LAMP and real-time PCR methods for the rapid detection of Xylella fastidiosa for quarantine and field applications. Phytopathology 2010, 100, 1282–1288. [Google Scholar] [CrossRef]
- Health, E.; Panel, O.P.; Bragard, C.; Dehnen-Schmutz, K.; Di Serio, F.; Gonthier, P.; Jacques, M.-A.; Jaques Miret, J.A.; Justesen, A.F.; MacLeod, A.; et al. Effectiveness of in planta control measures for Xylella fastidiosa. EFSA J. 2019, 17, e05666. [Google Scholar] [CrossRef]
- Van den Berg, R.A.; Hoefsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef]
- Sundekilde, U.; Larsen, L.; Bertram, H. NMR-based milk metabolomics. Metabolites 2013, 3, 204–222. [Google Scholar] [CrossRef]
- Kettaneh, N.; Berglund, A.; Wold, S. PCA and PLS with very large data sets. Comput. Stat. Data Anal. 2005, 48, 69–85. [Google Scholar] [CrossRef]
- Jackson, J.E. A User’s Guide to Principal Components; John Wiley & Sons: Hoboken, NJ, USA, 2005; Volume 587. [Google Scholar]
- Trygg, J.; Wold, S. Orthogonal projections to latent structures (O-PLS). J. Chemom. 2002, 16, 119–128. [Google Scholar] [CrossRef]
- Wold, S.; Eriksson, L.; Trygg, J.; Kettaneh, N. The PLS Method–Partial Least Squares Projections to Latent Structures—And Its Applications in Industrial RDP (Research, Development, and Production); Umeå University: Umeå, Sweden, 2004. [Google Scholar]
- Eriksson, L.; Byrne, T.; Johansson, E.; Trygg, J.; Vikström, C. Multi- and Megavariate Data Analysis Basic Principles and Applications; Umetrics Academy: Umeå, Sweden, 2013; Volume 1. [Google Scholar]
- Wheelock, Å.M.; Wheelock, C.E. Trials and tribulations of omics data analysis: Assessing quality of SIMCA-based multivariate models using examples from pulmonary medicine. Mol. Biosyst. 2013, 9, 2589–2596. [Google Scholar] [CrossRef] [PubMed]
| J | F | M | A | M | J | J | A | S | O | N | D | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2017 | T | T | T | T | T | T | ||||||
| 2018 | T | T | T | T | T | T | ||||||
| 2019 | S | T | T | T | T | T | T | S |
| I vs. II Sampling OPLS-DA (1 + 1 + 0) | R2X | R2Y | Q2 |
|---|---|---|---|
| Ogliarola salentina | 0.567 | 0874 | 0.698 |
| Cellina di Nardò | 0.488 | 0.924 | 0.813 |
| Leccino | 0.691 | 0.974 | 0.958 |
| Untreated vs. Treated OPLS-DA (1 + 1 + 0) | Sampling | R2X | R2Y | Q2 |
|---|---|---|---|---|
| Ogliarola salentina | I | 0.489 | 0.803 | −0.57 |
| II | 0.465 | 0.947 | 0.695 | |
| Cellina di Nardò | I | 0.536 | 0.704 | −1.01 |
| II | 0.596 | 0.895 | 0.763 | |
| Leccino | I | 0.39 | 0.887 | 0.323 |
| II | 0.563 | 0.945 | 0.554 |
| Leccino vs. Susceptible Cultivars OPLS-DA (1 + 1 + 0) | Sampling | R2X | R2Y | Q2 |
|---|---|---|---|---|
| Untreated samples | I | 0.668 | 0.953 | 0.811 |
| II | 0.645 | 0.948 | 0.684 | |
| Treated samples | I | 0.65 | 0.953 | 0.518 |
| II | 0.696 | 0.827 | 0.398 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Girelli, C.R.; Del Coco, L.; Angilè, F.; Scortichini, M.; Fanizzi, F.P. Olive Cultivars Susceptible or Tolerant to Xylella fastidiosa Subsp. pauca Exhibit Mid-Term Different Metabolomes upon Natural Infection or a Curative Treatment. Plants 2021, 10, 772. https://doi.org/10.3390/plants10040772
Girelli CR, Del Coco L, Angilè F, Scortichini M, Fanizzi FP. Olive Cultivars Susceptible or Tolerant to Xylella fastidiosa Subsp. pauca Exhibit Mid-Term Different Metabolomes upon Natural Infection or a Curative Treatment. Plants. 2021; 10(4):772. https://doi.org/10.3390/plants10040772
Chicago/Turabian StyleGirelli, Chiara Roberta, Laura Del Coco, Federica Angilè, Marco Scortichini, and Francesco Paolo Fanizzi. 2021. "Olive Cultivars Susceptible or Tolerant to Xylella fastidiosa Subsp. pauca Exhibit Mid-Term Different Metabolomes upon Natural Infection or a Curative Treatment" Plants 10, no. 4: 772. https://doi.org/10.3390/plants10040772
APA StyleGirelli, C. R., Del Coco, L., Angilè, F., Scortichini, M., & Fanizzi, F. P. (2021). Olive Cultivars Susceptible or Tolerant to Xylella fastidiosa Subsp. pauca Exhibit Mid-Term Different Metabolomes upon Natural Infection or a Curative Treatment. Plants, 10(4), 772. https://doi.org/10.3390/plants10040772









