Overexpression of Cassava MeAnn2 Enhances the Salt and IAA Tolerance of Transgenic Arabidopsis
Abstract
1. Introduction
2. Results
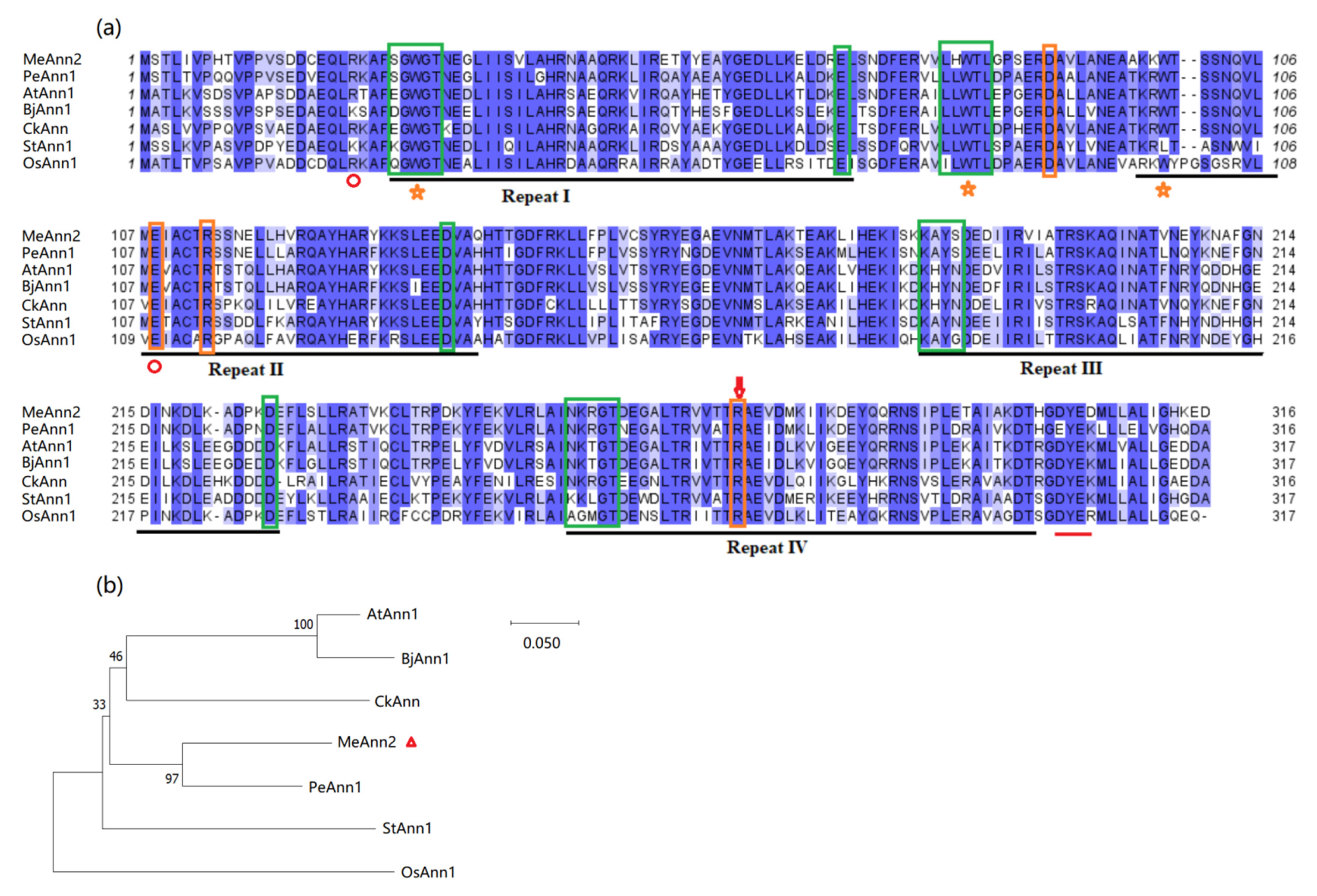
2.1. Cloning and Characterization of MeAnn2 in Cassava
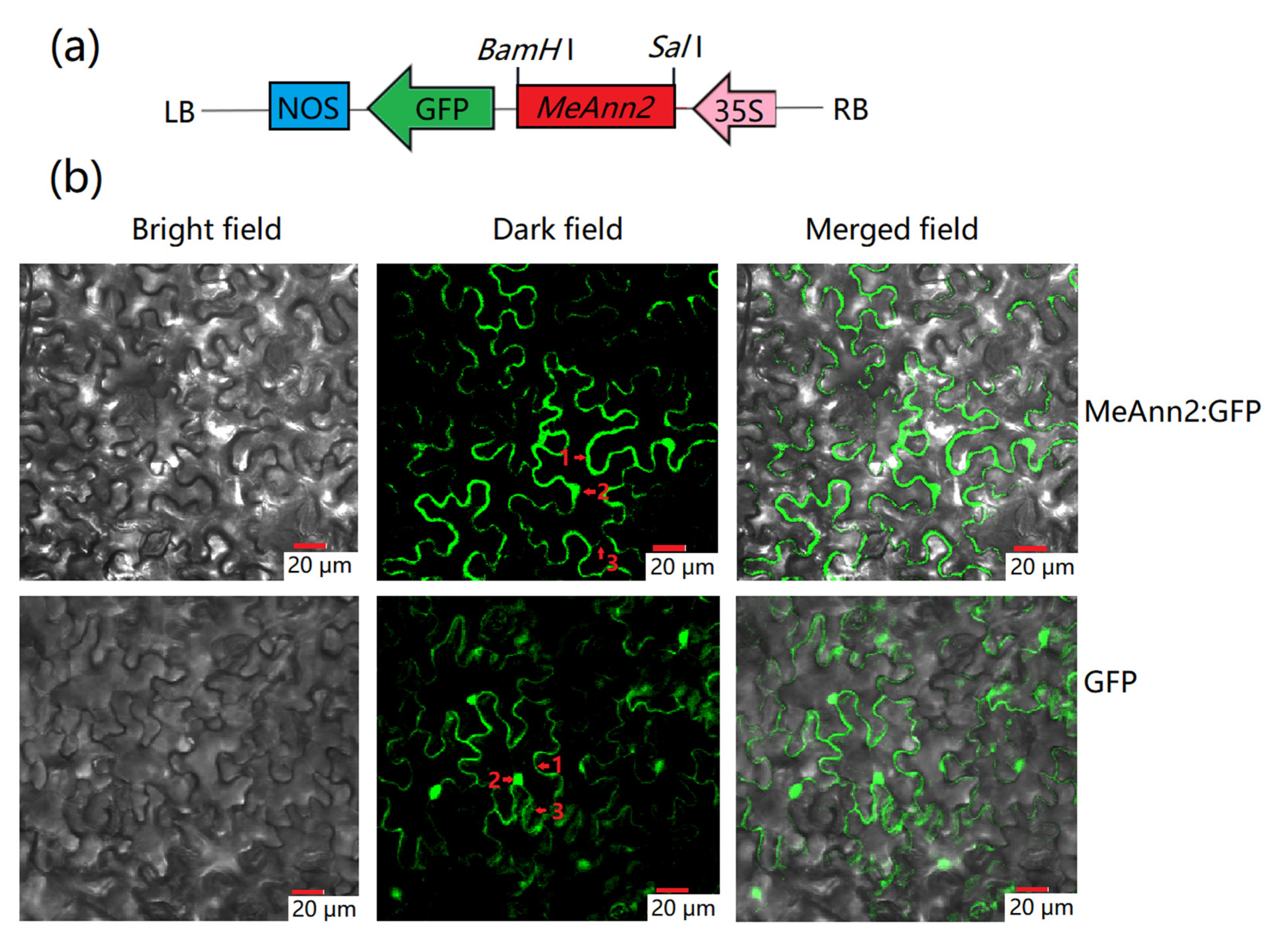
2.2. Subcellular Localization of the MeAnn2 Protein
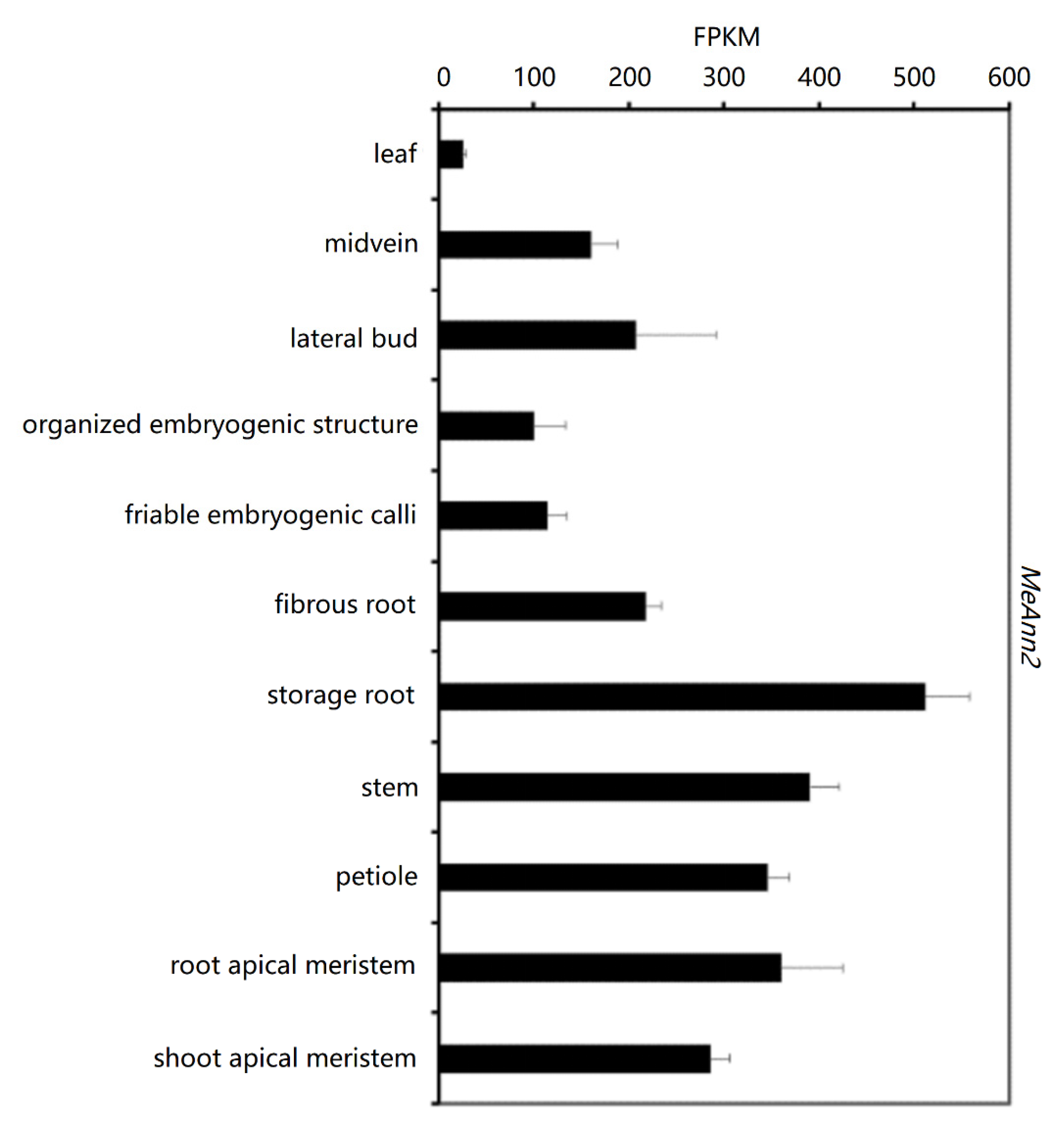
2.3. Analysis of MeAnn2 Expression in Cassava
2.4. Transformation of MeAnn2 in Arabidopsis
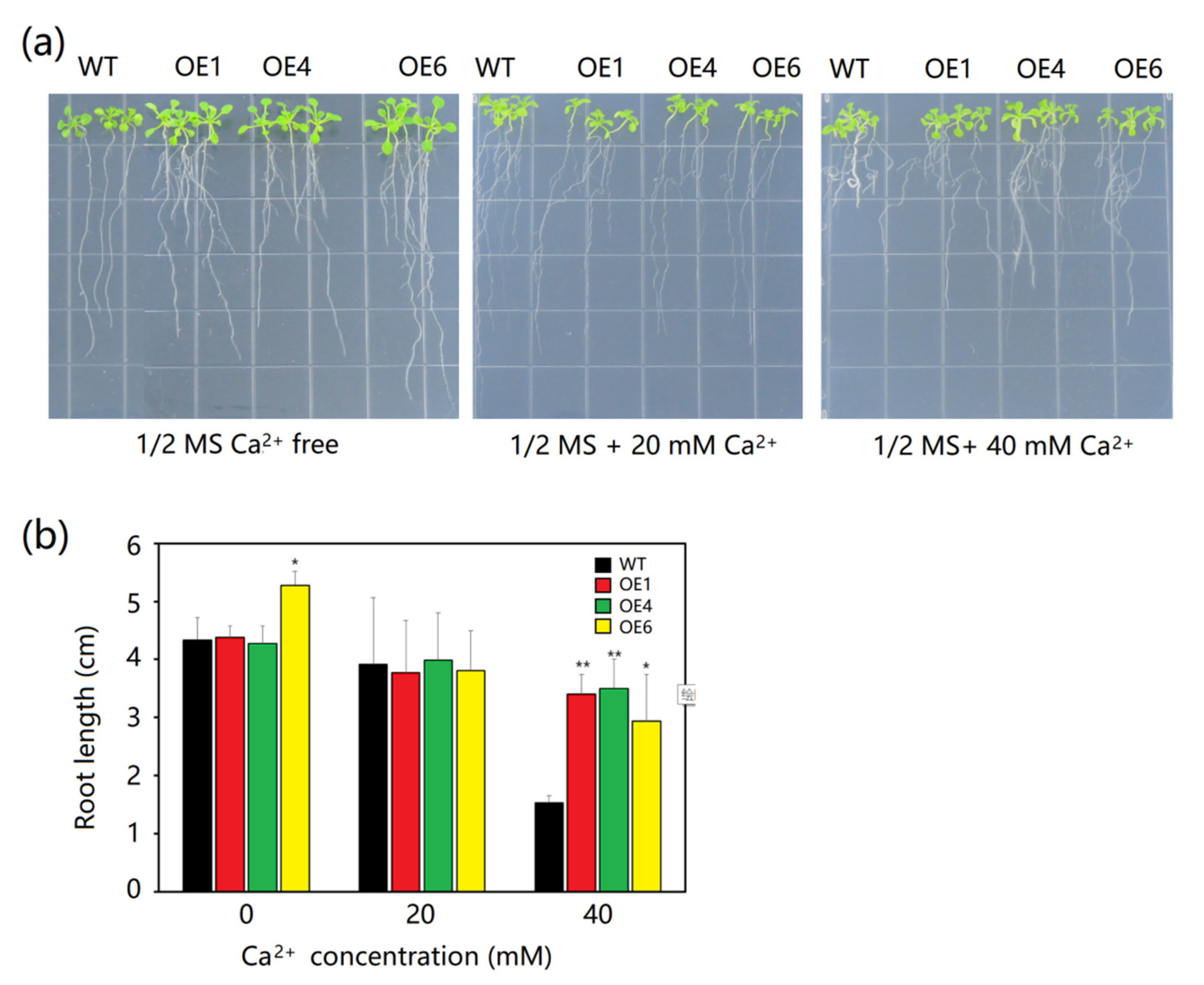
2.5. Effect of Different Ca2+ Concentrations on Arabidopsis Seedlings
2.6. Effects of NaCl Stress on Growth in MeAnn2-Overexpressing Arabidopsis
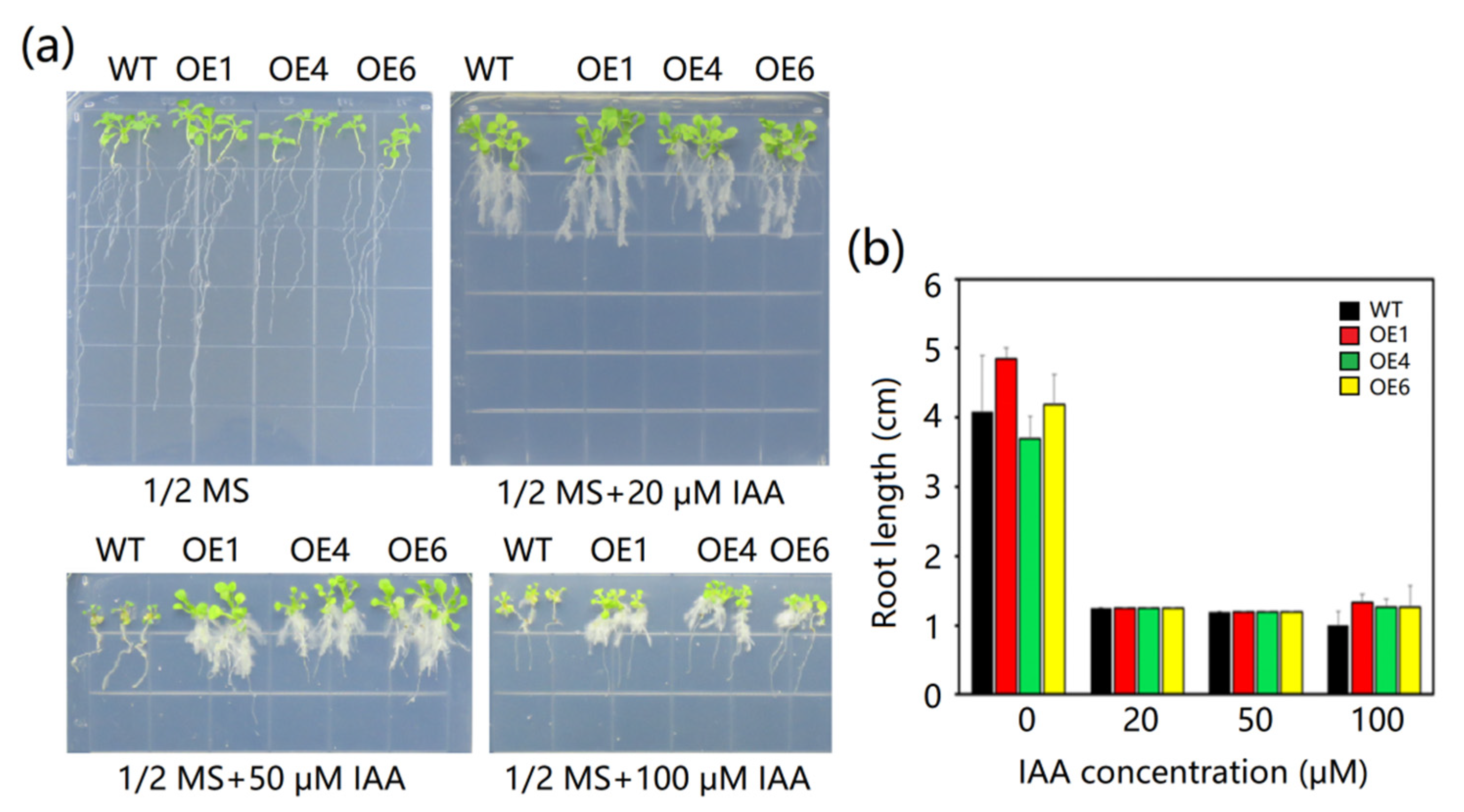
2.7. The Effects of IAA on MeAnn2 Overexpressing Arabidopsis
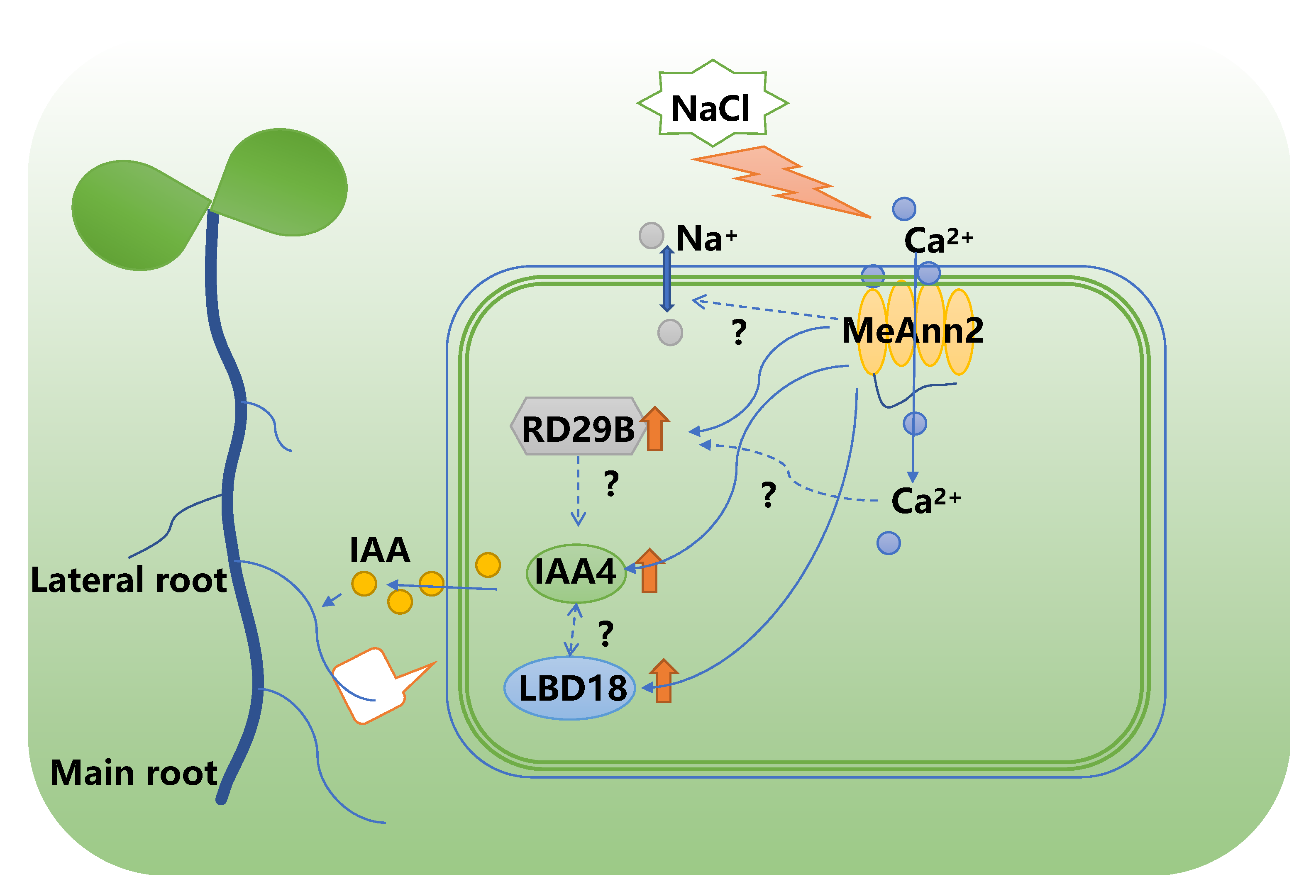
3. Discussion
4. Materials and Methods
4.1. Total RNA Extraction and PCR Amplification of the MeAnn2 Gene
4.2. MeAnn2 Gene Bioinformatics Analysis and Gene Expression
4.3. Subcellular Localization of MeAnn2:GFP
4.4. Genetic Transformation and Identification of MeAnn2 Overexpression Transgenic Lines
4.5. Transgenic Arabidopsis Stress Treatment
4.6. Quantitative RT-PCR Analysis of Stress Resistance-Related Genes
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Saad, R.B.; Ben Romdhane, W.; Ben Hsouna, A.; Mihoubi, W.; Harbaoui, M.; Brini, F. Insights into plant annexins function in abiotic and biotic stress tolerance. Plant Signal Behav. 2020, 15, 1699264. [Google Scholar] [CrossRef]
- Moss, S.E.; Morgan, R.O. The annexins. Genome Biol. 2004, 5, 219. [Google Scholar] [CrossRef]
- Konopka-Postupolska, D.; Clark, G.; Hofmann, A. Structure, function and membrane interactions of plant annexins: An update. Plant Sci. 2011, 181, 230–241. [Google Scholar] [CrossRef]
- Gerke, V.; Moss, S.E. Annexins: From structure to function. Physiol. Rev. 2002, 82, 331–371. [Google Scholar] [CrossRef]
- Mortimer, J.C.; Laohavisit, A.; Macpherson, N.; Webb, A.; Brownlee, C.; Battey, N.H.; Davies, J.M. Annexins: Multifunctional components of growth and adaptation. J. Exp. Bot. 2008, 59, 533–544. [Google Scholar] [CrossRef]
- De Carvalho-Niebel, F.; Timmers, A.C.; Chabaud, M.; Defaux-Petras, A.; Barker, D.G. The Nod factor-elicited annexin MtAnn1 is preferentially localised at the nuclear periphery in symbiotically activated root tissues of Medicago truncatula. Plant J. 2002, 32, 343–352. [Google Scholar] [CrossRef]
- Clark, G.B.; Dauwalder, M.; Roux, S.J. Immunological and biochemical evidence for nuclear localization of annexin in peas. Plant Physiol. Biochem. 1998, 36, 621–627. [Google Scholar] [CrossRef]
- Lee, S.; Lee, E.J.; Yang, E.J.; Lee, J.E.; Park, A.R.; Song, W.H.; Park, O.K. Proteomic identification of annexins, calcium-dependent membrane binding proteins that mediate osmotic stress and abscisic acid signal transduction in Arabidopsis. Plant Cell 2004, 16, 1378–1391. [Google Scholar] [CrossRef]
- Peltier, J.B.; Emanuelsson, O.; Kalume, D.E.; Ytterberg, J.; Friso, G.; Rudella, A.; Liberles, D.A.; Soderberg, L.; Roepstorff, P.; von Heijne, G.; et al. Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 2002, 14, 211–236. [Google Scholar] [CrossRef]
- Peltier, J.B.; Cai, Y.; Sun, Q.; Zabrouskov, V.; Giacomelli, L.; Rudella, A.; Ytterberg, A.J.; Rutschow, H.; van Wijk, K.J. The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Mol. Cell. Proteom. 2006, 5, 114–133. [Google Scholar] [CrossRef]
- Wang, F.; Chen, S.; Liang, D.; Qu, G.Z.; Chen, S.; Zhao, X. Transcriptomic analyses of Pinus koraiensis under different cold stresses. BMC Genom. 2020, 21, 10. [Google Scholar] [CrossRef]
- Ticha, M.; Richter, H.; Ovecka, M.; Maghelli, N.; Hrbackova, M.; Dvorak, P.; Samaj, J.; Samajova, O. Advanced Microscopy Reveals Complex Developmental and Subcellular Localization Patterns of ANNEXIN 1 in Arabidopsis. Front. Plant Sci. 2020, 11, 1153. [Google Scholar] [CrossRef] [PubMed]
- Lichocka, M.; Rymaszewski, W.; Morgiewicz, K.; Barymow-Filoniuk, I.; Chlebowski, A.; Sobczak, M.; Samuel, M.A.; Schmelzer, E.; Krzymowska, M.; Hennig, J. Nucleus- and plastid-targeted annexin 5 promotes reproductive development in Arabidopsis and is essential for pollen and embryo formation. BMC Plant Biol. 2018, 18, 183. [Google Scholar] [CrossRef] [PubMed]
- Laohavisit, A.; Shang, Z.; Rubio, L.; Cuin, T.A.; Very, A.A.; Wang, A.; Mortimer, J.C.; Macpherson, N.; Coxon, K.M.; Battey, N.H.; et al. Arabidopsis annexin1 mediates the radical-activated plasma membrane Ca2+- and K+-permeable conductance in root cells. Plant Cell 2012, 24, 1522–1533. [Google Scholar] [CrossRef] [PubMed]
- Laohavisit, A.; Richards, S.L.; Shabala, L.; Chen, C.; Colaco, R.D.; Swarbreck, S.M.; Shaw, E.; Dark, A.; Shabala, S.; Shang, Z.; et al. Salinity-induced calcium signaling and root adaptation in Arabidopsis require the calcium regulatory protein annexin1. Plant Physiol. 2013, 163, 253–262. [Google Scholar] [CrossRef]
- Mohammad-Sidik, A.; Sun, J.; Shin, R.; Song, Z.; Ning, Y.; Matthus, E.; Wilkins, K.A.; Davies, J.M. Annexin 1 is a component of eATP-induced cytosolic calcium elevation in Arabidopsis thaliana roots. Int. J. Mol. Sci. 2021, 22, 494. [Google Scholar] [CrossRef]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Kiegle, E.; Moore, C.A.; Haseloff, J.; Tester, M.A.; Knight, M.R. Cell-type-specific calcium responses to drought, salt and cold in the Arabidopsis root. Plant J. 2000, 23, 267–278. [Google Scholar] [CrossRef]
- Pan, Z.; Zhao, Y.; Zheng, Y.; Liu, J.; Jiang, X.; Guo, Y. A high-throughput method for screening Arabidopsis mutants with disordered abiotic stress-induced calcium signal. J. Genet. Genom. 2012, 39, 225–235. [Google Scholar] [CrossRef]
- Choi, W.G.; Toyota, M.; Kim, S.H.; Hilleary, R.; Gilroy, S. Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. USA 2014, 111, 6497–6502. [Google Scholar] [CrossRef]
- Cantero, A.; Barthakur, S.; Bushart, T.J.; Chou, S.; Morgan, R.O.; Fernandez, M.P.; Clark, G.B.; Roux, S.J. Expression profiling of the Arabidopsis annexin gene family during germination, de-etiolation and abiotic stress. Plant Physiol. Biochem. 2006, 44, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Ye, J.; Yang, Y.; Lin, H.; Yue, L.; Luo, J.; Long, Y.; Fu, H.; Liu, X.; Zhang, Y.; et al. The SOS2-SCaBP8 complex generates and fine-tunes an AtANN4-dependent calcium signature under salt stress. Dev. Cell 2019, 48, 697–709.e5. [Google Scholar] [CrossRef] [PubMed]
- Huh, S.M.; Noh, E.K.; Kim, H.G.; Jeon, B.W.; Bae, K.; Hu, H.C.; Kwak, J.M.; Park, O.K. Arabidopsis annexins AnnAt1 and AnnAt4 interact with each other and regulate drought and salt stress responses. Plant Cell Physiol. 2010, 51, 1499–1514. [Google Scholar] [CrossRef] [PubMed]
- Jami, S.K.; Hill, R.D.; Kirti, P.B. Transcriptional regulation of annexins in Indian Mustard, Brassica juncea and detoxification of ROS in transgenic tobacco plants constitutively expressing AnnBj1. Plant Signal Behav. 2010, 5, 618–621. [Google Scholar] [CrossRef][Green Version]
- Jami, S.K.; Clark, G.B.; Turlapati, S.A.; Handley, C.; Roux, S.J.; Kirti, P.B. Ectopic expression of an annexin from Brassica juncea confers tolerance to abiotic and biotic stress treatments in transgenic tobacco. Plant Physiol. Biochem. 2008, 46, 1019–1030. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.; Yadav, D.; Shukla, P.; Kirti, P.B. Heterologous expression of Brassica juncea annexin, AnnBj2 confers salt tolerance and ABA insensitivity in transgenic tobacco seedlings. Funct. Integr. Genom. 2018, 18, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Harbaoui, M.; Ben Saad, R.; Ben Halima, N.; Choura, M.; Brini, F. Structural and functional characterisation of two novel durum wheat annexin genes in response to abiotic stress. Funct. Plant Biol. 2018, 45, 542–552. [Google Scholar] [CrossRef] [PubMed]
- Harbaoui, M.; Ben Romdhane, W.; Ben Hsouna, A.; Brini, F.; Ben Saad, R. The durum wheat annexin, TdAnn6, improves salt and osmotic stress tolerance in Arabidopsis via modulation of antioxidant machinery. Protoplasma 2021. [Google Scholar] [CrossRef] [PubMed]
- Mu, C.; Zhou, L.; Shan, L.; Li, F.; Li, Z. Phosphatase GhDsPTP3a interacts with annexin protein GhANN8b to reversely regulate salt tolerance in cotton (Gossypium spp.). New Phytol. 2019, 223, 1856–1872. [Google Scholar] [CrossRef]
- Koevoets, I.T.; Venema, J.H.; Elzenga, J.T.; Testerink, C. Roots Withstanding their Environment: Exploiting root system architecture responses to abiotic stress to improve crop tolerance. Front. Plant Sci. 2016, 7, 1335. [Google Scholar] [CrossRef]
- Liu, W.; Li, R.J.; Han, T.T.; Cai, W.; Fu, Z.W.; Lu, Y.T. Salt stress reduces root meristem size by nitric oxide-mediated modulation of auxin accumulation and signaling in Arabidopsis. Plant Physiol. 2015, 168, 343–356. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Kim, J. EXPANSINA17 up-regulated by LBD18/ASL20 promotes lateral root formation during the auxin response. Plant Cell Physiol. 2013, 54, 1600–1611. [Google Scholar] [CrossRef]
- Goh, T.; Kasahara, H.; Mimura, T.; Kamiya, Y.; Fukaki, H. Multiple AUX/IAA-ARF modules regulate lateral root formation: The role of Arabidopsis SHY2/IAA3-mediated auxin signalling. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 1461–1468. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Cho, C.; Pandey, S.K.; Park, Y.; Kim, M.J.; Kim, J. LBD16 and LBD18 acting downstream of ARF7 and ARF19 are involved in adventitious root formation in Arabidopsis. BMC Plant Biol. 2019, 19, 46. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wang, L.Y.; Dai, J.X.; Wang, Y.; Lin, D. The NAC-type transcription factor CaNAC46 regulates the salt and drought tolerance of transgenic Arabidopsis thaliana. BMC Plant Biol. 2021, 21, 11. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yuan, S.; He, Y.; Fan, J.; Zhou, Y.; Qiu, T.; Lin, X.; Yao, Y.; Liu, J.; Fu, S.J.A. Genome-wide identification and expression profiling analysis of the galactinol synthase gene family in cassava (Manihot esculenta Crantz). Agronomy 2018, 8, 250. [Google Scholar] [CrossRef]
- Ding, Z.; Fu, L.; Yan, Y.; Tie, W.; Xia, Z.; Wang, W.; Peng, M.; Hu, W.; Zhang, J. Genome-wide characterization and expression profiling of HD-Zip gene family related to abiotic stress in cassava. PLoS ONE 2017, 12, e0173043. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Ouyang, B.; Zhang, J.; Wang, T.; Lu, C.; Han, Q.; Zhao, S.; Ye, Z.; Li, H. Genomic organization, phylogenetic comparison and expression profiles of annexin gene family in tomato (Solanum lycopersicum). Gene 2012, 499, 14–24. [Google Scholar] [CrossRef]
- Huang, Y.; Deng, T.; Zuo, K. Cotton annexin proteins participate in the establishment of fiber cell elongation scaffold. Plant Signal Behav. 2013, 8, e66160. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xu, L.; Tang, Y.; Gao, S.; Su, S.; Hong, L.; Wang, W.; Fang, Z.; Li, X.; Ma, J.; Quan, W.; et al. Comprehensive analyses of the annexin gene family in wheat. BMC Genom. 2016, 17, 415. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.L.; Laohavisit, A.; Mortimer, J.C.; Shabala, L.; Swarbreck, S.M.; Shabala, S.; Davies, J.M. Annexin 1 regulates the H2O2-induced calcium signature in Arabidopsis thaliana roots. Plant J. 2014, 77, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Ding, Y.; Shi, Y.; Ma, L.; Wang, Y.; Song, C.; Wilkins, K.A.; Davies, J.M.; Knight, H.; Knight, M.R.; et al. The calcium transporter ANNEXIN1 mediates cold-induced calcium signaling and freezing tolerance in plants. EMBO J. 2021, 40, e104559. [Google Scholar] [CrossRef] [PubMed]
- Konopka-Postupolska, D.; Clark, G.; Goch, G.; Debski, J.; Floras, K.; Cantero, A.; Fijolek, B.; Roux, S.; Hennig, J. The role of annexin 1 in drought stress in Arabidopsis. Plant Physiol. 2009, 150, 1394–1410. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.X.; Xu, Y.J.; Lei, Y.; Li, Q.; Zhao, J.Q.; Li, Y.; Fan, J.; Xiao, S.; Wang, W.M. ANNEXIN 8 negatively regulates RPW8.1-mediated cell death and disease resistance in Arabidopsis. J. Integr. Plant Biol. 2021, 63, 378–392. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, A.; Proust, J.; Dorowski, A.; Schantz, R.; Huber, R. Annexin 24 from Capsicum annuum. X-ray structure and biochemical characterization. J. Biol. Chem. 2000, 275, 8072–8082. [Google Scholar] [CrossRef]
- Zhang, Y.; Sa, G.; Zhang, Y.; Hou, S.; Wu, X.; Zhao, N.; Zhang, Y.; Deng, S.; Deng, C.; Deng, J.; et al. Populus euphratica annexin1 facilitates cadmium enrichment in transgenic Arabidopsis. J. Hazard. Mater. 2021, 405, 124063. [Google Scholar] [CrossRef]
- Gao, S.; Song, T.; Han, J.; He, M.; Zhang, Q.; Zhu, Y.; Zhu, Z. A calcium-dependent lipid binding protein, OsANN10, is a negative regulator of osmotic stress tolerance in rice. Plant Sci. 2020, 293, 110420. [Google Scholar] [CrossRef]
- Qiao, B.; Zhang, Q.; Liu, D.; Wang, H.; Yin, J.; Wang, R.; He, M.; Cui, M.; Shang, Z.; Wang, D.; et al. A calcium-binding protein, rice annexin OsANN1, enhances heat stress tolerance by modulating the production of H2O2. J. Exp. Bot. 2015, 66, 5853–5866. [Google Scholar] [CrossRef]
- Kovacs, I.; Ayaydin, F.; Oberschall, A.; Ipacs, I.; Bottka, S.; Pongor, S.; Dudits, D.; Toth, E.C. Immunolocalization of a novel annexin-like protein encoded by a stress and abscisic acid responsive gene in alfalfa. Plant J. 1998, 15, 185–197. [Google Scholar] [CrossRef]
- Vandeputte, O.; Lowe, Y.O.; Burssens, S.; Van Raemdonck, D.; Hutin, D.; Boniver, D.; Geelen, D.; El Jaziri, M.; Baucher, M. The tobacco NtAnn12 gene, encoding an annexin, is induced upon Rhodoccocus fascians infection and during leafy gall development. Mol. Plant Pathol. 2007, 8, 185–194. [Google Scholar] [CrossRef]
- Li, Q.; Zhao, H.; Wang, X.; Kang, J.; Lv, B.; Dong, Q.; Li, C.; Chen, H.; Wu, Q. Tartary Buckwheat Transcription Factor FtbZIP5, Regulated by FtSnRK2.6, Can Improve Salt/Drought Resistance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2020, 21, 1123. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Magwanga, R.O.; Kirungu, J.N.; Hu, Y.; Dong, Q.; Cai, X.; Zhou, Z.; Wang, X.; Zhang, Z.; Hou, Y.; et al. Overexpression of cotton a DTX/MATE gene enhances drought, salt, and cold stress tolerance in transgenic Arabidopsis. Front. Plant Sci. 2019, 10, 299. [Google Scholar] [CrossRef] [PubMed]
- Zolla, G.; Heimer, Y.M.; Barak, S. Mild salinity stimulates a stress-induced morphogenic response in Arabidopsis thaliana roots. J. Exp. Bot. 2010, 61, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.C.; Mutka, A.M.; Hummel, A.W.; Berry, J.; Chauhan, R.D.; Vijayaraghavan, A.; Taylor, N.J.; Voytas, D.F.; Chitwood, D.H.; Bart, R.S. Gene expression atlas for the food security crop cassava. New Phytol. 2017, 213, 1632–1641. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, X.; Li, R.; Zhou, Y.; Tang, F.; Wang, Y.; Lu, X.; Wang, S.; Yao, Y.; Liu, J.; Hu, X.; et al. Overexpression of Cassava MeAnn2 Enhances the Salt and IAA Tolerance of Transgenic Arabidopsis. Plants 2021, 10, 941. https://doi.org/10.3390/plants10050941
Lin X, Li R, Zhou Y, Tang F, Wang Y, Lu X, Wang S, Yao Y, Liu J, Hu X, et al. Overexpression of Cassava MeAnn2 Enhances the Salt and IAA Tolerance of Transgenic Arabidopsis. Plants. 2021; 10(5):941. https://doi.org/10.3390/plants10050941
Chicago/Turabian StyleLin, Xuejun, Ruimei Li, Yangjiao Zhou, Fenlian Tang, Yajie Wang, Xiaohua Lu, Shijia Wang, Yuan Yao, Jiao Liu, Xinwen Hu, and et al. 2021. "Overexpression of Cassava MeAnn2 Enhances the Salt and IAA Tolerance of Transgenic Arabidopsis" Plants 10, no. 5: 941. https://doi.org/10.3390/plants10050941
APA StyleLin, X., Li, R., Zhou, Y., Tang, F., Wang, Y., Lu, X., Wang, S., Yao, Y., Liu, J., Hu, X., & Guo, J. (2021). Overexpression of Cassava MeAnn2 Enhances the Salt and IAA Tolerance of Transgenic Arabidopsis. Plants, 10(5), 941. https://doi.org/10.3390/plants10050941





