Morpho-Physiological Testing of NaCl Sensitivity of Tobacco Plants Overexpressing Choline Oxidase Gene
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. In Vitro Cultivation, Agrobacterial Transformation, and Plant Regeneration
2.3. Segregation Analysis
2.4. Determination of Salt Stress Sensitivity
2.5. Biochemical Analyses
2.6. Molecular Analysis of Regenerants
2.7. Statistical Data Processing
3. Results
3.1. Agrobacterial Transformation and Transformation Efficiency Assessment
3.2. Screening of Regenerants for the Integration of Heterologous codA Gene Expression
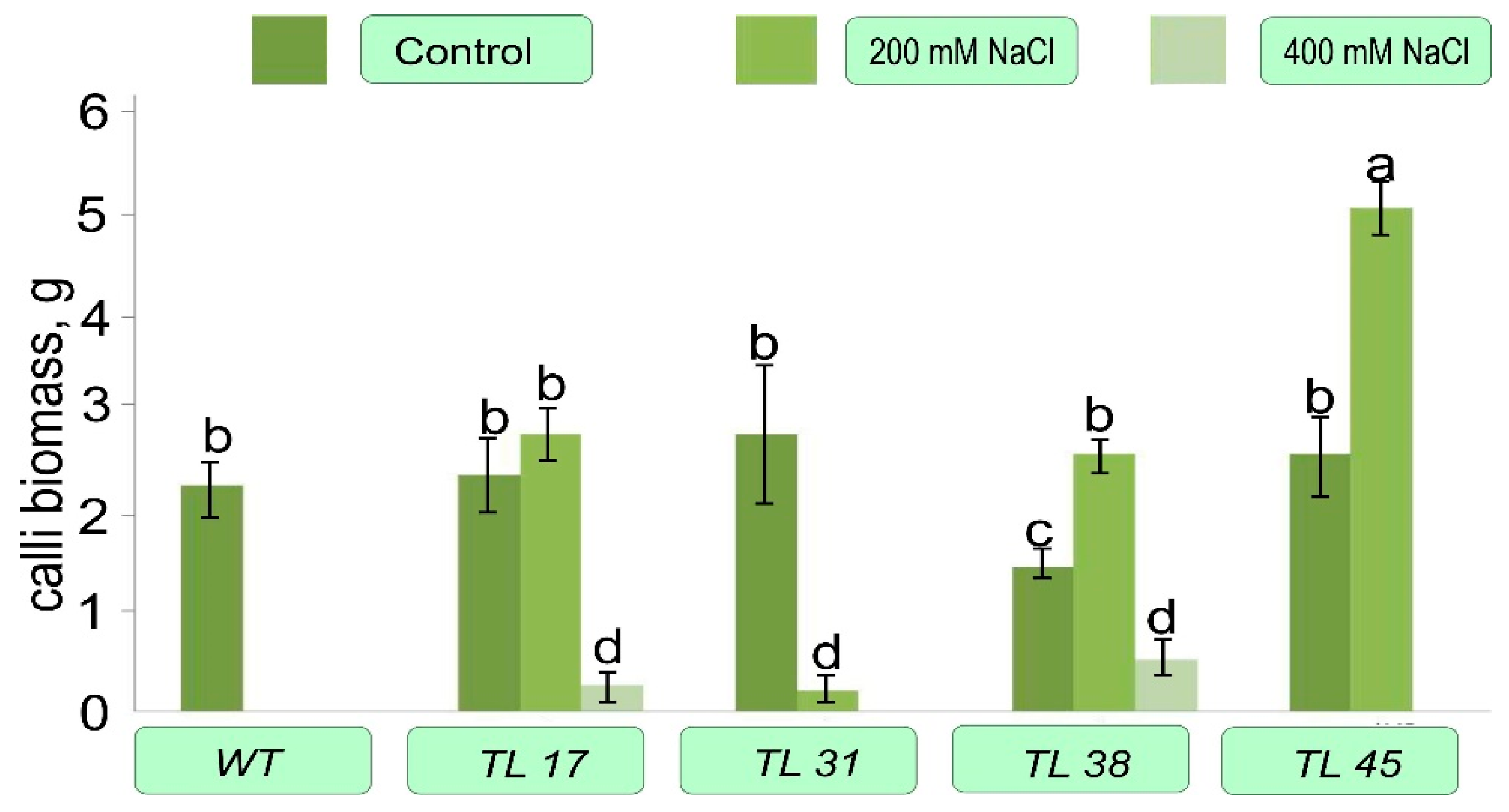
3.3. Evaluation of the Regeneration Potential of codA Calli of Transgenic Lines during Selection for NaCl
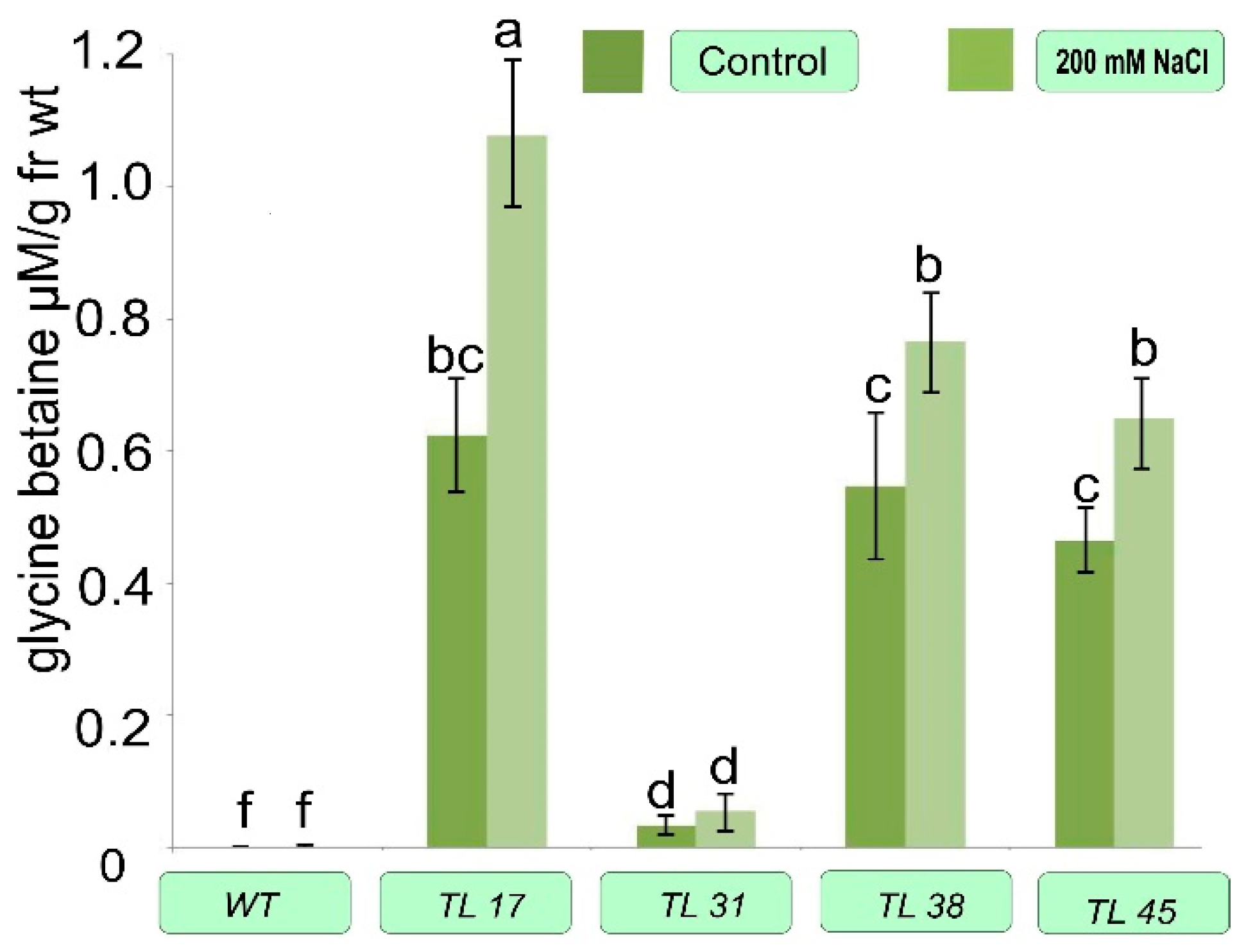
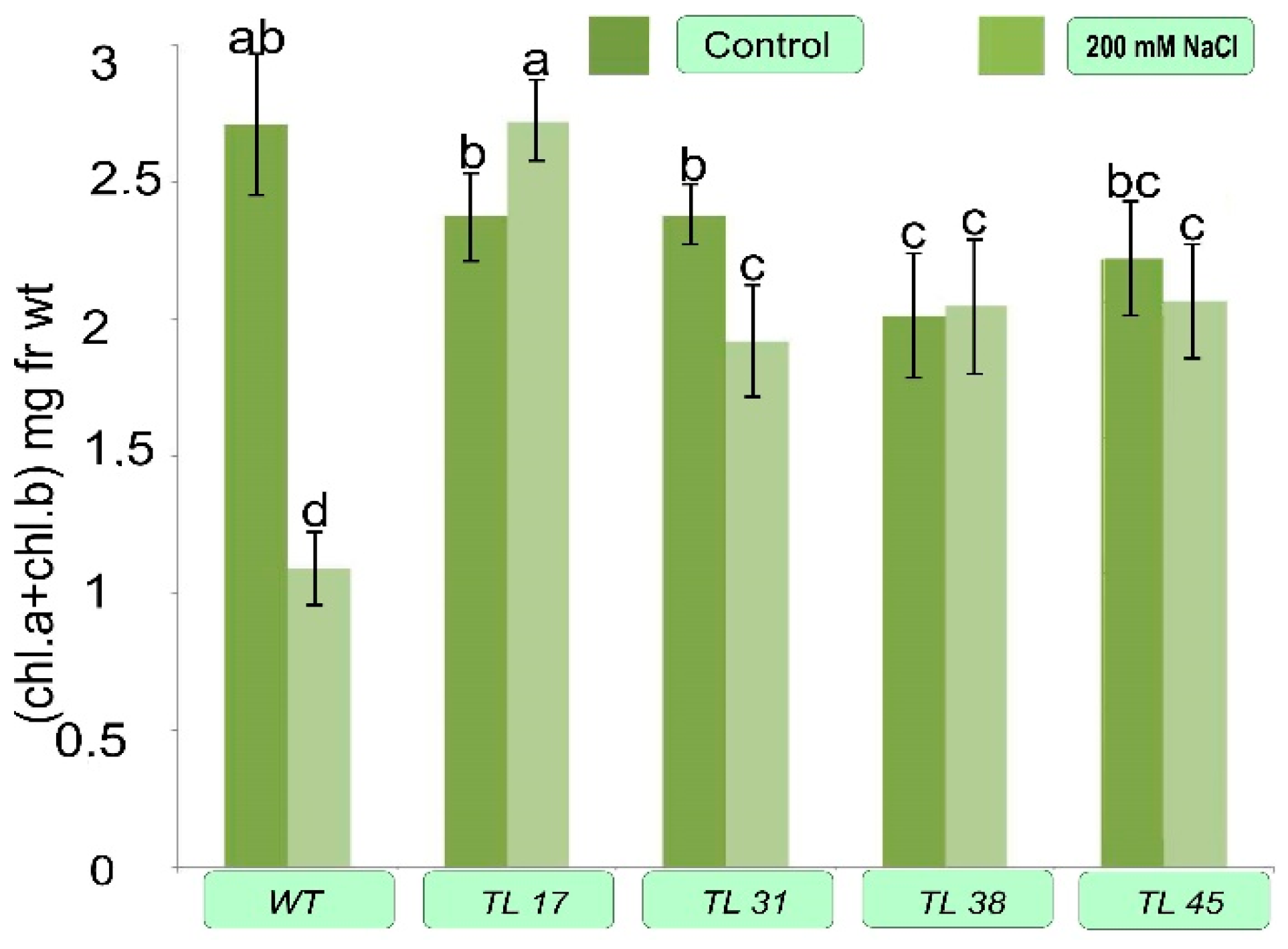
3.4. Biochemical Features of Transformed Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Golkar, P.; Taghizadeh, M. In vitro evaluation of phenolic and osmolite compounds, ionic content, and antioxidant activity in safflower (Carthamus tinctorius L.) under salinity stress. Plant Cell Tissue Organ Cult. 2018, 134, 357–368. [Google Scholar] [CrossRef]
- Ashraf, M.; Foolad, M.R. Roles of GB and proline in improving plant abiotic-stress resistance. Environ. Exp. Bot. 2007, 59, 206–216. [Google Scholar] [CrossRef]
- Rhodes, D.; Hanson, A.D. Quaternary ammonium and tertiary sulfonium compounds in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1993, 44, 357–384. [Google Scholar] [CrossRef]
- Chen, T.H.H.; Murata, N. GB: An effective protectant against abiotic stress in plants. Trends Plant Sci. 2008, 13, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Papageorgiou, G.C.; Murata, N. The unusually strong stabilizing effects of GB on the structure and function of the oxygen-evolving photosystem-II complex. Photosynth. Res. 1995, 44, 243–252. [Google Scholar] [CrossRef]
- He, Y.; Wang, Q.; Zeng, J.; Sun, T.; Yang, G.-X.; He, G.-Y. Current status and trends of wheat genetic transformation studies in China. J. Integr. Agric. 2015, 14, 438–452. [Google Scholar] [CrossRef] [Green Version]
- Su, J.; Hirji, R.; Zhang, L.; He, C.; Selvaraj, G.; Wu, R. Evaluation of the stress-inducible production of choline oxidase in transgenic rice as a strategy for producing the stress-protectant GB. J. Exp. Bot. 2006, 57, 1129–1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNeil, S.D.; Rhodes, D.; Russell, B.L.; Nuccio, M.L.; Shachar-Hill, Y.; Hanson, A.D. Metabolic modeling identifies key constraints on an engineered GB synthesis pathway in tobacco. Plant Physiol. 2000, 124, 153–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Dhingra, A.; Daniell, H. Stable transformation of the cotton plastid genome and maternal inheritance of transgenes. Plant Mol. Biol. 2004, 56, 203–216. [Google Scholar] [CrossRef]
- Gulevich, A.A.; Kurenina, L.V.; Baranova, E.N. Application of a system for targeting Fe-dependent superoxide dismutase and choline oxidase enzymes to chloroplast as a strategy for effective plant resistance to abiotic stresses. Russ. Agric. Sci. 2018, 44, 118–123. [Google Scholar] [CrossRef]
- Fan, F.; Ghanem, M.; Gadda, G. Cloning, sequence analysis, and purification of choline oxidase from Arthrobacter globiformis: A bacterial enzyme involved in osmotic stress tolerance. Arch. Biochem. Biophys. 2004, 421, 149–158. [Google Scholar] [CrossRef]
- Deshnium, P.; Los, D.A.; Hayashi, H.; Mustardy, L.; Murata, N. Transformation of Synechococcus with a gene for choline oxidase enhances tolerance to salt stress. Plant Mol. Biol. 1995, 29, 897–907. [Google Scholar] [CrossRef]
- Hayashi, H.; Mustardy, L.; Deshnium, P.; Ida, M.; Murata, N. Transformation of Arabidopsis thaliana with the codA gene for choline oxidase; accumulation of glycinebetaine and enhanced tolerance to salt and cold stress. Plant J. 1997, 12, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, R.; Kim, M.D.; Back, K.H.; Kim, H.S.; Lee, H.S.; Kwon, S.Y.; Murata, N.; Chung, W.I.; Kwak, S.S. Stress-induced expression of choline oxidase in potato plant chloroplasts confers enhanced tolerance to oxidative, salt, and drought stresses. Plant Cell Rep. 2008, 27, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Gulevich, A.A.; Baranova, E.N. Creation of a genetic construct with the choline oxidase gene for improved expression in plant. Russ. Agric. Sci. 2008, 34, 145–148. [Google Scholar] [CrossRef]
- Danilova, S.A.; Kusnetsov, V.V.; Dolgikh, Y.I. A novel efficient method for maize genetic transformation: Usage of agrobacterial monolayer. Russ. J. Plant Physiol. 2009, 56, 258. [Google Scholar] [CrossRef]
- Grieve, C.M.; Grattan, R. Rapid assay for determination of water soluble quaternary-amino compounds. Plant Soil 1983, 70, 303–307. [Google Scholar] [CrossRef]
- Zhang, F.; Yang, Y.L.; He, W.L.; Zhao, X.; Zhang, L.X. Effects of salinity on growth and compatible solutes of callus induced from Populus euphratica. In Vitro Cell Dev. Biol. Plant 2004, 40, 491–494. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Shlyk, A.A. Definition of a Chlorophyll and Carotenoids in Extracts of Green Leaves. Biochemical Methods in Physiology of Plants; Nauka: Moscow, Russia, 1971. [Google Scholar]
- Ahmad, R.; Lim, C.J.; Kwon, S.Y. GB: A versatile compound with great potential for gene pyramiding to improve crop plant performance against environmental stresses. Plant Biotechnol. Rep. 2013, 7, 49–57. [Google Scholar] [CrossRef]
- Nakamura, T.; Yokota, S.; Muramoto, Y.; Tsutsui, K.; Oguri, Y.; Fukui, K.; Takabe, T. Expression of a betaine aldehyde dehydrogenase gene in rice, a glycinebetaine nonaccumulator, and possible localization of its protein in peroxisomes. Plant J. 1997, 11, 1115–1120. [Google Scholar] [CrossRef] [Green Version]
- You, L.; Song, Q.; Wu, Y.; Li, S.; Jiang, C.; Chang, L.; Yang, X.; Zhang, J. Accumulation of GB in transplastomic potato plants expressing choline oxidase confers improved drought tolerance. Planta 2019, 249, 1963–1975. [Google Scholar] [CrossRef]
- Park, E.J.; Jeknić, Z.; Sakamoto, A.; DeNoma, J.; Yuwansiri, R.; Murata, N.; Chen, T.H. Genetic engineering of GB synthesis in tomato protects seeds, plants, and flowers from chilling damage. Plant J. 2004, 40, 474–487. [Google Scholar] [CrossRef]
- Tran, N.H.T.; Oguchi, T.; Matsunaga, E.; Kawaoka, A.; Watanabe, K.N.; Kikuchi, A. Transcriptional enhancement of a bacterial choline oxidase A gene by an HSP terminator improves the GB production and salinity stress tolerance of Eucalyptus camaldulensis trees. Plant Biotechnol. 2018, 35, 215–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmström, K.O.; Somersalo, S.; Mandal, A.; Palva, T.E.; Welin, B. Improved tolerance to salinity and low temperature in transgenic tobacco producing GB. J. Exp. Bot. 2000, 51, 177–185. [Google Scholar] [CrossRef] [Green Version]
- Sakamoto, A.; Murata, N. Genetic engineering of GB synthesis in plants: Current status and implications for enhancement of stress tolerance. J. Exp. Bot. 2000, 51, 81–88. [Google Scholar] [CrossRef]
- Park, E.J.; Jeknić, Z.; Pino, M.T.; Murata, N.; Chen, T.H.H. GB accumulation is more effective in chloroplasts than in the cytosol for protecting transgenic tomato plants against abiotic stress. Plant Cell Environ. 2007, 30, 994–1005. [Google Scholar] [CrossRef]
- Srivastava, T.P.; Gupta, S.C.; Lal, P.; Muralia, P.N.; Kumar, A. The effect of salt stress on physiological and biochemical parameters of wheat. Ann. Arid Zone 1998, 27, 197–204. [Google Scholar]
- Hayashi, H.; Sakamoto, A.; Nonaka, H.; Chen, T.H.H.; Murata, N. Enhanced germination under high-salt conditions of seeds of transgenic Arabidopsis with a bacterial gene (codA) for choline oxidase. J. Plant Res. 1998, 111, 357–362. [Google Scholar] [CrossRef]
- Bohnert, H.J.; Nelson, D.E.; Jensen, R.G. Adaptations to environmental stresses. Plant Cell 1995, 7, 1099–1111. [Google Scholar] [CrossRef] [PubMed]
- Yancey, P.H. Compatible and counteracting solutes. In Cellular and Molecular Physiology of Cell Volume Regulation; Strange, K., Ed.; CRC Press: Boca Raton, FL, USA, 1994; pp. 81–109. [Google Scholar]
- Rontein, D.; Basset, G.; Hanson, A.D. Metabolic engineering of osmoprotectant accumulation in plants. Metab. Eng. 2002, 4, 49–56. [Google Scholar] [CrossRef] [Green Version]
- Shabala, S.; Cuin, T.A. Osmoregulation versus osmoprotection: Reevaluating the role of compatible solutes. In Floriculture, Ornamental and Plant Biotechnology—Advances and Topical Issues; Teixeira, S.J., Ed.; Global Science Books: Tokyo, Japan, 2006; pp. 405–416. [Google Scholar]
- Wani, S.H.; Singh, N.B.; Haribhushan, A.; Mir, J.I. Compatible solute engineering in plants for abiotic stress tolerance—Role of GB. Curr. Genom. 2013, 14, 157. [Google Scholar] [CrossRef] [Green Version]
- Fan, W.; Wang, H.; Zhang, P. Engineering GB metabolism for enhanced drought stress tolerance in plants. In Drought Stress Tolerance in Plants; Hossain, M., Wani, S., Bhattacharjee, S., Burritt, D., Tran, L.S., Eds.; Springer: Cham, Switzerland, 2016; Volume 2. [Google Scholar]
- Sable, A.D.; Kardile, P.B.; Sable, A.D.; Kharde, A.V. Studies on effect of different concentration of NaCI on bacoside production from brahmi (Bacopa monnieri) under in vitro condition. J. Pharmacogn. Phytochem. 2018, 7, 1386–1389. [Google Scholar]
- Dogan, M. Effect of salt stress on in vitro organogenesis from nodal explant of Limnophila aromatica (Lamk.) Merr. and Bacopa monnieri (L.) Wettst. and their physio-morphological and biochemical responses. Physiol. Mol. Biol. Plants 2020, 26, 803–816. [Google Scholar] [CrossRef]
- Burssens, S.; Himanen, K.; Van de Cotte, B.; Beeckman, T.; Van Montagu, M.; Inzé, D.; Verbruggen, N. Expression of cell cycle regulatory genes and morphological alterations in response to salt stress in Arabidopsis thaliana. Planta 2000, 211, 632–640. [Google Scholar] [CrossRef]
- Bogoutdinova, L.R.; Lazareva, E.M.; Chaban, I.A.; Kononenko, N.V.; Dilovarova, T.; Khaliluev, M.R.; Kurenina, L.V.; Gulevich, A.A.; Smirnova, E.A.; Baranova, E.N. Salt stress-induced structural changes are mitigated in transgenic tomato plants over-expressing superoxide dismutase. Biology 2020, 9, 297. [Google Scholar] [CrossRef]
- Radi, A.A.; Farghaly, F.A.; Hamada, A.M. Physiological and biochemical responses of salt-tolerant and salt-sensitive wheat and bean cultivars to salinity. J. Biol. Earth Sci. 2013, 3, 72–88. [Google Scholar]
- Kato, M.; Shimizu, S. Chlorophyll metabolism in higher plants VI. Involvement of peroxidase in chlorophyll degradation. Plant Cell Physiol. 1985, 26, 1291–1301. [Google Scholar]
- Akcin, A.; Yalcin, E. Effect of salinity stress on chlorophyll, carotenoid content, and proline in Salicornia prostrata Pall. and Suaeda prostrata Pall. subsp. prostrata (Amaranthaceae). Braz. J. Bot. 2016, 39, 101–106. [Google Scholar] [CrossRef]
- Perbea, M.; Petcu, E. The effect of soil water content on sunflower seedlings. Rom. Agric. Res. 2000, 13, 43–47. [Google Scholar]
- Higbie, S.M.; Wang, F.; Stewart, J.M.; Sterling, T.M.; Lindemann, W.C.; Hughs, E.; Zhang, J. Physiological response to salt (NaCl) stress in selected cultivated tetraploid cottons. Int. J. Agron. 2010, 10, 1155–1167. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Sharma, M.K.; Sengar, R.S. Osmolytes: Proline metabolism in plants as sensors of abiotic stress. J. Appl. Nat. Sci. 2017, 9, 2079–2092. [Google Scholar] [CrossRef]
- Kavi Kishor, P.B.; Sangam, S.; Amrutha, R.N.; Laxmi, P.S.; Naidu, K.R.; Rao, K.R.S.; Rao, S.; Reddy, K.J.; Theriappan, P.; Sreenivasulu, N. Regulation of proline biosynthesis, degradation, uptake and transport in higher plants: Its implications in plant growth and abiotic stress tolerance. Curr. Sci. 2005, 88, 424–438. [Google Scholar]
- Khan, M.S.; Yu, X.; Kikuchi, A.; Asahina, M.; Watanabe, K.N. Genetic engineering of GB biosynthesis to enhance abiotic stress tolerance in plants. Plant Biotechnol. 2009, 26, 125–134. [Google Scholar] [CrossRef] [Green Version]
- Hussain, S.J.; Masood, A.; Anjum, N.A.; Khan, N.A. Sulfur-mediated control of salinity impact on photosynthesis and growth in mungbean cultivars screened for salt tolerance involves glutathione and proline metabolism, and glucose sensitivity. Acta Physiol. Plant. 2019, 41, 1–13. [Google Scholar] [CrossRef]
- Gomes, F.P.; Oliva, M.A.; Mielke, M.S.; Almeida, A.A.F.; Aquino, L.A. Osmotic adjustment, proline accumulation and cell membrane stability in leaves of Cocos nucifera submitted to drought stress. Sci. Hortic. 2010, 126, 379–384. [Google Scholar] [CrossRef]
- Sakamoto, A.; Valverde, R.; Chen, T.H.H.; Murata, N. Transformation of Arabidopsis with the codA gene for choline oxidase enhances freezing tolerance of plants. Plant J. 2000, 22, 449–453. [Google Scholar] [CrossRef] [PubMed]
- Takabe, T.; Rai, V.; Hibino, T. Metabolic engineering of GB. In Abiotic Stress Tolerance in Plants; Springer: Dordrecht, The Netherlands, 2006; pp. 137–151. [Google Scholar]
- Chen, T.H.H.; Murata, N. GB protects plants against abiotic stress: Mechanisms and biotechnological applications. Plant Cell Environ. 2011, 34, 1–20. [Google Scholar] [CrossRef]
- Huang, J.; Hirji, R.; Adam, L.; Rozwadowski, K.L.; Hammerlindl, J.K.; Keller, W.A.; Selvaraj, G. Genetic engineering of GB production toward enhancing stress tolerance in plants: Metabolic limitations. Plant Physiol. 2000, 122, 747–756. [Google Scholar] [CrossRef] [Green Version]
- Lazareva, E.M.; Baranova, E.N.; Smirnova, E.A. Reorganization of interphase microtubules in root cells of Medicago sativa L. during acclimation to osmotic and salt stress. Cell Tissue Biol. 2017, 11, 324–334. [Google Scholar] [CrossRef]
- Baranova, E.N.; Gulevich, A.A.; Polyakov, V.Y. Effect of NaCl, Na2SO4, and mannitol on utilization of storage starch and formation of plastids in the cotyledons and roots of alfalfa seedlings. Russ. J. Plant Physiol. 2007, 54, 50–57. [Google Scholar] [CrossRef]
- Morohashi, K.; Okamoto, M.; Yamazaki, C.; Fujii, N.; Miyazawa, Y.; Kamada, M.; Takahashi, H. Gravitropism interferes with hydrotropism via counteracting auxin dynamics in cucumber roots: Clinorotation and spaceflight experiments. New Phytol. 2017, 215, 1476–1489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baranova, E.N.; Christov, N.K.; Kurenina, L.V.; Khaliluev, M.R.; Todorovska, E.G.; Smirnova, E.A. Formation of atypical tubulin structures in plant cells as a nonspecific response to abiotic stress. Bulg. J. Agric. Sci. 2016, 22, 987–992. [Google Scholar]
- Gorham, J. Betaines in higher plants-biosynthesis and role in stress metabolism. In Amino Acids and their Derivatives in Higher Plants; Wallsgrove, R.M., Ed.; Cambridge University Press: Cambridge, UK, 2010; pp. 173–204. [Google Scholar]
- Guo, B.H.; Zhang, Y.M.; Li, H.J.; Du, L.Q. Transformation of wheat with a gene encoding for the betaine aldehyde dehydrogenase (BADH). Acta Bot. Sin. 2000, 42, 279–283. [Google Scholar]
- Quan, R.; Shang, M.; Zhang, H.; Zhao, Y.; Zhang, J. Engineering of enhanced GB synthesis improves drought tolerance in maize. Plant Biotech. J. 2004, 2, 477–486. [Google Scholar] [CrossRef]
- Yousefi, A.R.; Rashidi, S.; Moradi, P.; Mastinu, A. Germination and seedling growth responses of Zygophyllum fabago, Salsola kali L. and Atriplex canescens to PEG-induced drought stress. Environments 2020, 7, 107. [Google Scholar] [CrossRef]


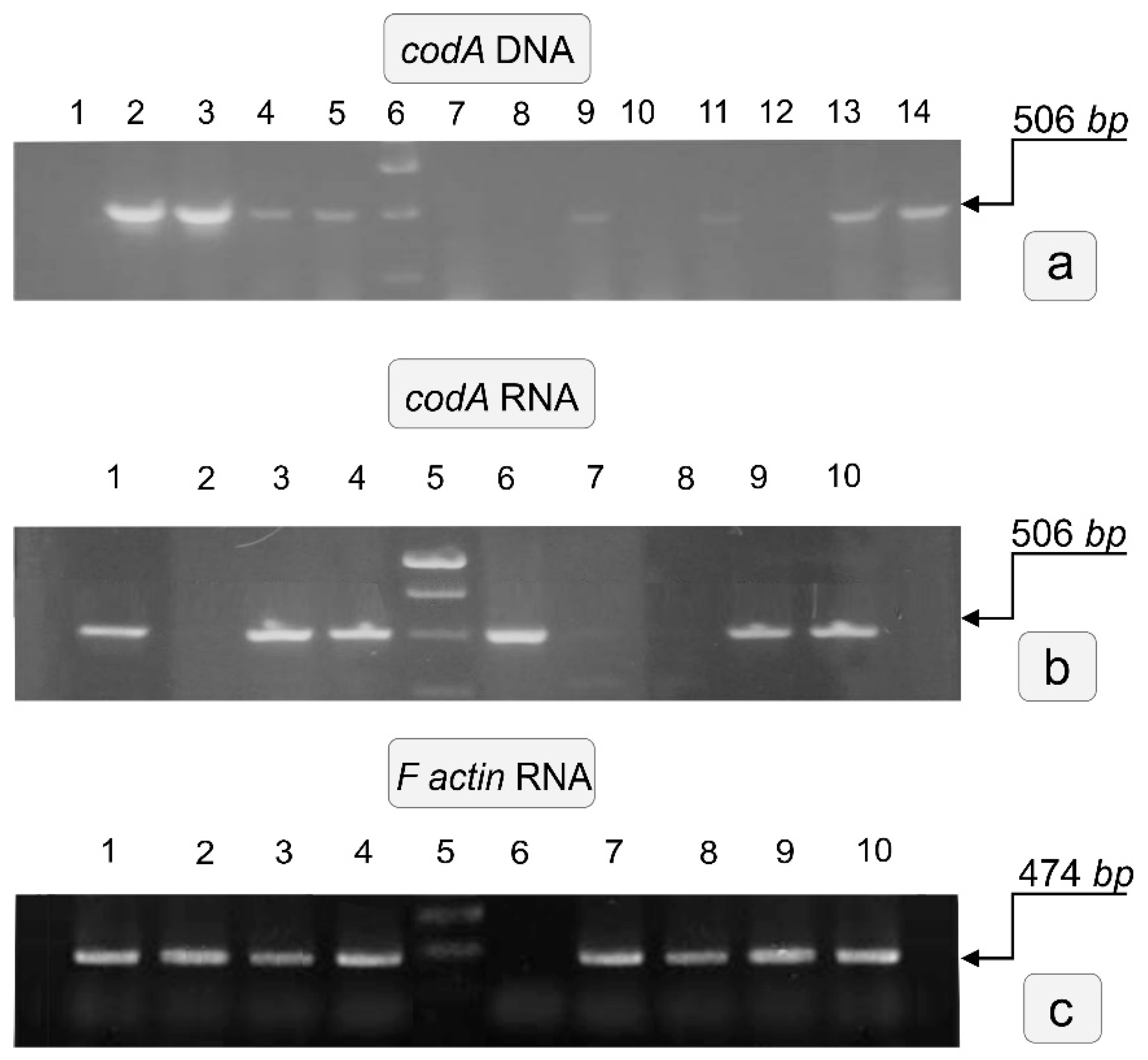


| Forward Primer | Reverse Primer | ||
|---|---|---|---|
| codAf | 5′-CGCCAACTTCTTCCAGATCAA-3′ | codAr | 5′-GGGTGTTCATGTCGAACG-3′ |
| actinf | 5′-CCTCCCACATGCTATTCTCC-3′ | actinr | 5′-AGAGCCTCCAATCCAGACAC-3′ |
| nptIIf | 5′-GTGGAGAAGGCTATTCGGCTA-3′ | nptIIr | 5′-CCACCATGATATTCGGCAAG-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raldugina, G.N.; Evsukov, S.V.; Bogoutdinova, L.R.; Gulevich, A.A.; Baranova, E.N. Morpho-Physiological Testing of NaCl Sensitivity of Tobacco Plants Overexpressing Choline Oxidase Gene. Plants 2021, 10, 1102. https://doi.org/10.3390/plants10061102
Raldugina GN, Evsukov SV, Bogoutdinova LR, Gulevich AA, Baranova EN. Morpho-Physiological Testing of NaCl Sensitivity of Tobacco Plants Overexpressing Choline Oxidase Gene. Plants. 2021; 10(6):1102. https://doi.org/10.3390/plants10061102
Chicago/Turabian StyleRaldugina, Galina N., Sergey V. Evsukov, Liliya R. Bogoutdinova, Alexander A. Gulevich, and Ekaterina N. Baranova. 2021. "Morpho-Physiological Testing of NaCl Sensitivity of Tobacco Plants Overexpressing Choline Oxidase Gene" Plants 10, no. 6: 1102. https://doi.org/10.3390/plants10061102







