Genetic Variability and Population Structure of Ethiopian Sesame (Sesamum indicum L.) Germplasm Assessed through Phenotypic Traits and Simple Sequence Repeats Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Phenotyping
2.2.1. Description of the Study Sites
2.2.2. Experimental Design and Trial Management
2.2.3. Phenotypic Data Collection
2.2.4. Phenotypic Data Analysis
2.3. Genotyping
2.3.1. DNA Extraction, Primer Selection, Polymerase Chain Reaction, and Electrophoresis
2.3.2. Genotypic Data Analysis
3. Results
3.1. Genetic Variation and Mean Performance of Sesame Accessions
3.2. Correlations of Yield and Yield Components
3.3. Principal Component Analysis
3.4. Genetic Polymorphism of the SSR Markers
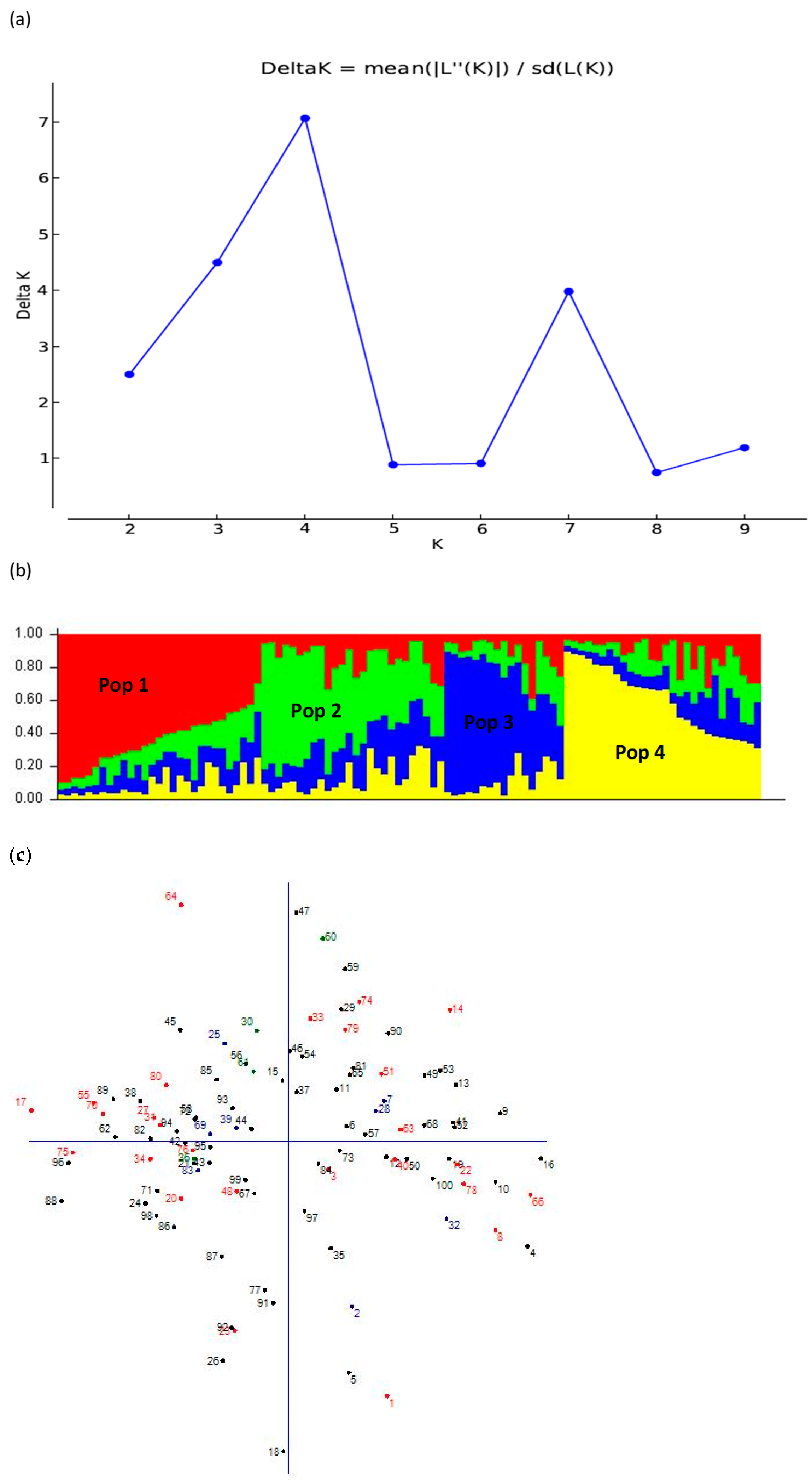
3.5. Population Structure Analysis
3.6. Cluster Analysis of 100 Sesame Accessions
4. Discussion
4.1. Genotypic Variation and Mean Performance for Seed and Oil Yields, and Yield-Component Traits
4.2. Traits Associations
4.3. Genetic Diversity and Population Structure of Sesame Germplasm Based on SSR Markers
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De la Vega, A.J.; Hall, A.J. Effect of planting date, genotype, and their interactions on sunflower yield: II. Components of oil yield. Crop Sci. 2002, 42, 1202–1210. [Google Scholar] [CrossRef]
- Zheljazkov, V.D.; Vick, B.A.; Ebelhar, M.W.; Buehring, N.; Baldwin, B.S.; Astatkie, T.; Mille, J.F. Yield, oil content, and composition of sunflower grown at multiple locations in Mississippi. Agron. J. 2008, 100, 635–639. [Google Scholar] [CrossRef]
- Wei, W.; Zhang, Y.; Lv, H.; Li, D.; Wang, L.; Zhang, X. Association analysis for quality traits in a diverse panel of Chinese sesame (Sesamum indicum L.) germplasm. J. Integr. Plant Biol. 2013, 55, 745–758. [Google Scholar] [CrossRef]
- Dossa, K.; Wei, X.; Niang, M.; Liu, P.; Zhang, Y.; Wang, L.; Liao, B.; Cissé, N.; Zhang, X.; Diouf, D. Near-infrared reflectance spectroscopy reveals wide variation in major components of sesame seeds from Africa and Asia. Crop J. 2018, 6, 202–206. [Google Scholar] [CrossRef]
- Uzun, B.; Ülger, S.; Çagirgan, M.I. Comparison of determinate and indeterminate types of sesame for oil content and fatty acid composition. Turk. J. Agric. For. 2002, 26, 269–274. [Google Scholar]
- Uzun, B.; Arslan, Ç.; Furat, Ş. Variation in fatty acid compositions, oil content and oil yield in a germplasm collection of sesame (Sesamum indicum L.). J. Am. Oil. Chem. Soc. 2008, 85, 1135–1142. [Google Scholar] [CrossRef]
- Anastasi, U.; Sortino, O.; Tuttobene, R. Agronomic performance and grain quality of sesame (Sesamum indicum L.) landraces and improved varieties grown in a Mediterranean environment. Genet. Resour. Crop Evol. 2017, 64, 127–137. [Google Scholar] [CrossRef]
- Were, B.A.; Onkware, A.O.; Gudu, S.; Welander, M.; Carlsson, A.S. Seed oil content and fatty acid composition in East African sesame (Sesamum indicum L.) accessions evaluated over 3 years. Field Crops Res. 2006, 97, 254–260. [Google Scholar] [CrossRef]
- Gharby, S.; Harhar, H.; Bouzoubaa, Z.; Asdadi, A.; El Yadini, A.; Charrouf, Z. Chemical characterization and oxidative stability of seeds and oil of sesame grown in Morocco. J. Saudi Soc. Agric. Sci. 2017, 16, 105–111. [Google Scholar] [CrossRef]
- Gebremedhn, M.B.; Tessema, W.; Gebre, G.G.; Mawcha, K.T.; Assefa, M.K. Value chain analysis of sesame (Sesamum indicum L.) in Humera district, Tigray, Ethiopia. Cogent Food Agric. 2019, 5, 1705741. [Google Scholar] [CrossRef]
- Ethiopian Agricultural Sample Enumeration: Report on the Primary Results of Area, Production and Yield of Temporary Crops of Private Peasant Holdings in Meher Season; Central Statistic Authority (CSA): Addis Ababa, Ethiopia, 2018.
- FAOSTAT. Food and Agriculture Organization of the United Nations; FAOSTAT Online Statistical Service: Rome, Italy, 2018; Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 14 April 2020).
- Woldesenbet, D.T.; Tesfaye, K.; Bekele, E. Genetic diversity of sesame germplasm collection (Sesamum indicum L.): Implication for conservation, improvement and use. Int. J. Biotechnol. Mol. Biol. Res. 2015, 6, 7–18. [Google Scholar] [CrossRef]
- Nyongesa, B.O.; Were, B.A.; Gudu, S.; Dangasuk, O.G.; Onkware, A.O. Genetic diversity in cultivated sesame (Sesamum indicum L.) and related wild species in East Africa. J. Crop Sci. Biotech. 2013, 16, 9–15. [Google Scholar] [CrossRef]
- Gidey, T.; Kebede, S.A.; Gashawbeza, G.T. Extent and pattern of the genetic diversity for morpho-agronomic traits in Ethiopian sesame landraces (Sesamum indicum L.). Asian J. Agric. 2012, 6, 118–128. [Google Scholar] [CrossRef][Green Version]
- Teklu, D.H.; Kebede, S.A.; Gebremichael, D.E. Assessment of genetic variability, genetic advance, correlation, and path analysis for morphological traits in sesame genotypes. Asian J. Agric. Res. 2014, 7, 118–128. [Google Scholar] [CrossRef]
- Hika, G.; Geleta, N.; Jaleta, Z. Correlation and divergence analysis for phenotypic traits in sesame (Sesamum indicum L.) Genotypes. Sci. Technol. Arts Res. J. 2014, 3, 01–09. [Google Scholar] [CrossRef]
- Hika, G.; Geleta, N.; Jaleta, Z. Genetic variability, heritability and genetic advance for the phenotypic sesame (Sesamum indicum L.) Populations from Ethiopia. Sci. Technol. Arts Res. J. 2015, 4, 20–26. [Google Scholar] [CrossRef]
- Gebremichael, D.E.; Parzies, H.K. Genetic variability among landraces of sesame in Ethiopia. Afr. Crop Sci. J. 2011, 19, 1–13. [Google Scholar] [CrossRef][Green Version]
- Wei, W.; Zhang, Y.; Wang, L.; Li, D.; Gao, Y.; Zhang, X. Genetic diversity, population structure, and association mapping of 10 agronomic traits in sesame. Crop Sci. 2016, 56, 331–343. [Google Scholar] [CrossRef]
- Asekova, S.; Kulkarni, K.P.; Oh, K.W.; Lee, M.H.; Oh, E.; Kim, J.I.; Yeo, U.; Pae, U.S.; Ha, T.J.; Kim, S.U. Analysis of molecular variance and population structure of sesame (Sesamum indicum L.) genotypes using SSR markers. Plant Breed. Biotech. 2018, 6, 321–336. [Google Scholar] [CrossRef]
- Araújo, E.D.S.; Arriel, N.H.C.; Santos, R.C.D.; Lima, L.M.D. Assessment of genetic variability in sesame accessions using SSR markers and morpho agronomic traits. Aust. J. Crop Sci. AJCS 2019, 13, 45–54. [Google Scholar] [CrossRef]
- Gupta, P.K.; Varshney, R. The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica 2000, 113, 163–185. [Google Scholar] [CrossRef]
- HuARC. Annual Research Report for the Period 2009/10; Humera Agricultural Research Centre: Tigray, Ethiopia, 2010. [Google Scholar]
- SAS Institute. Statistical Analysis Software; Version 9.4; SAS Institute Inc.: Cary, NC, USA, 2018. [Google Scholar]
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Computing: Vienna, Austria, 2020. [Google Scholar]
- Wei, X.; Wang, L.; Zhang, Y.; Qi, X.; Wang, X.; Ding, X.; Zhang, J.; Zhang, X. Development of Simple Sequence Repeat (SSR) Markers of Sesame (Sesamum indicum L.) from a Genome Survey. Molecules 2014, 19, 5150–5162. [Google Scholar] [CrossRef]
- Pritchard, J.; Stephens, M.; Rosenberg, N.; Donnelly, P. Association mapping in structured populations. Am. J. Hum. Genet. 2000, 67, 170–180. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Wijnands, J.H.M.; Biersteker, J.; Van Loo, E.N. Oilseeds business opportunities in Ethiopia. In Public Private Partnership in Oil Seed; Wageningen University and Research: Wageningen, The Netherlands, 2009. [Google Scholar]
- Adu-Gyamfi, R.; Prempeh, R.; Zakari, I. Diversity assessment of some sesame (Sesamum indicum L.) genotypes cultivated in northern Ghana using morphological and SSR markers. Adv. Agric. 2019, 6067891. [Google Scholar] [CrossRef]
- He, Q.; Li, X.W.; Liang, G.L.; Ji, K.; Guo, Q.G. Genetic diversity and identity of Chinese loquat cultivars/accessions (Eriobotrya japonica) using apple SSR markers. Plant Mol. Biol. Repo. 2011, 29, 197–208. [Google Scholar] [CrossRef]
- Baraket, G.; Chatti, K.; Saddoud, O.; Abdelkarim, A.B.; Mars, M. Comparative assessment of SSR and AFLP markers for evaluation of genetic diversity and conservation of Fig (Ficus carica L.) genetic resources in Tunisia. Plant Mol. Biol. Repo. 2011, 29, 171–184. [Google Scholar] [CrossRef]
- Jifar, H.; Tesfaye, K.; Dagne, K.; Assefa, K.; Tadele, Z. Genetic diversity and population structure of tef [Eragrostis tef (Zucc.) Trotter] as Revealed by SSR Markers. Adv. Crop Sci. Technol. 2020, 8, 438. [Google Scholar]
- Mulualem, T.; Mekbib, F.; Shimelis, H.; Gebre, E.; Amelework, B. Genetic diversity of yam (Dioscorea spp.) landrace collections from Ethiopia using simple sequence repeat markers. Aust. J. Crop Sci. 2018, 12, 1223–1230. [Google Scholar] [CrossRef]
- William, T.S.; Hussein, S.; Mark, L.; Isack, M.; Admire, I.T.S. Assessment of the genetic diversity and population structure of rice genotypes using SSR markers. Acta Agric. Scand.Sect. B Soil Plant Sci. 2019, 70, 76–86. [Google Scholar] [CrossRef]
- Zhang, H.; Wei, L.; Miao, H.; Zhang, T.; Wang, C. Development and validation of genetic SSR markers in sesame by RNA-seq. BioMed Central. 2012, 13, 316–317. [Google Scholar]
- Ganesamurthy, K.; Punitha, D.; Elangovan, M. Genetic diversity among the land races of sorghum collected in Tamil Nadu. Electron. J. Plant Breed. 2010, 1, 1375–1379. [Google Scholar]
- Barnaud, A.; Trigueros, G.; McKey, D.; Joly, H.I. High outcrossing rates in fields with mixed sorghum landraces: How are landraces maintained? Heredity 2008, 101, 445–452. [Google Scholar] [CrossRef] [PubMed]



| Entry Number | Entry Name or Designation | Description | Source (Regions or Research Center in Ethiopia) | Entry Number | Entry Name or Designation | Description | Source (Regions or Research Center in Ethiopia) |
|---|---|---|---|---|---|---|---|
| 1 | Hirhir Kebabo Hairless Sel-2 | Accession | Tigray | 51 | Hirhir Baeker-Sel-3 | Accession | Tigray |
| 2 | GXT = 85(28-2) | Accession | Afar | 52 | NN0068-3 | Accession | Amhara |
| 3 | Hirhir Kebabo Hairless-9 | Accession | Tigray | 53 | NN0074-3 | Accession | Amhara |
| 4 | NN-0068-1 | Accession | Amhara | 54 | NN0036-1 | Accession | Amhara |
| 5 | NN-0108-2 | Accession | Amhara | 55 | Hirhir Kebabo Hairless Sel-4 | Accession | Tigray |
| 6 | NN-034 | Accession | Amhara | 56 | NN0001-2 | Accession | Amhara |
| 7 | BCS-0041 | Accession | Afar | 57 | Bawnji Sel-2 | Accession | Amhara |
| 8 | ACC-031-5-14 | Accession | Tigray | 58 | NN0058-2 | Accession | Amhara |
| 9 | NN-0129-2 | Accession | Amhara | 59 | G-02 | Accession | Amhara |
| 10 | ACC-203-020 | Accession | Amhara | 60 | ACC-NS-007(2) | Accession | Oromia |
| 11 | NN-0038-2 | Accession | Amhara | 61 | GA-002(3) | Accession | Gambela |
| 12 | Bawnji Fiyel Kolet | Accession | Amhara | 62 | Endelemi Kirem Sel-2 | Accession | Amhara |
| 13 | Gojam Azene (Aleka) | Accession | Amhara | 63 | ACC-205-299 | Accession | Tigray |
| 14 | Hirhir Humera Sel-6 | Accession | Tigray | 64 | Hirhir Kebabo Early Sel-1 | Accession | Tigray |
| 15 | Bawnji Gobate | Accession | Amhara | 65 | NN0016-1 | Accession | Amhara |
| 16 | Shwarobit (83) | Accession | Amhara | 66 | Hirhir Adgeshu Sel -8 | Accession | Tigray |
| 17 | Humera-1 | Variety | HuARC † | 67 | NN0015 | Accession | Amhara |
| 18 | ACC-202-950 | Accession | Amhara | 68 | NN01-13 | Accession | Amhara |
| 19 | NN-0026 | Accession | Amhara | 69 | Bering Bawany | Accession | Afar |
| 20 | ACC-NO-041 | Accession | Tigray | 70 | Hirhir Nigara 1st Sel-2 | Accession | Tigray |
| 21 | ACC-203-612 | Accession | Amhara | 71 | Gojam Azene (Yohans Sel-1) | Accession | Amhara |
| 22 | ACC-200-064-1 | Accession | Tigray | 72 | NN0038-1 | Accession | Amhara |
| 23 | Setit-1 | Variety | HuARC | 73 | NN0104 | Accession | Amhara |
| 24 | Tejareb Kokit Sel-3 | Accession | Amhara | 74 | Hirhir Kebabo Hairless Sel-6 | Accession | Tigray |
| 25 | Orofalc ACC-2 | Accession | Afar | 75 | ACC 205-180 | Accession | Tigray |
| 26 | NN-0022 | Accession | Amhara | 76 | Hirhir | Landrace | Tigray |
| 27 | Setit-3 | Variety | HuARC | 77 | ACC 203-616 | Accession | Amhara |
| 28 | ABX = 2-01-2 | Accession | Afar | 78 | NN0025 | Accession | Amhara |
| 29 | NN-0020 | Accession | Amhara | 79 | NN-0183-3 | Accession | Amhara |
| 30 | ACC-NS-010 | Accession | Oromia | 80 | Hirhir Humera | Accession | Tigray |
| 31 | Hirhir Sel-2 | Accession | Tigray | 81 | NN0031 | Accession | Amhara |
| 32 | ABXT-85-SEL-2-1 | Accession | Afar | 82 | NN0061 | Accession | Amhara |
| 33 | Setit-2 | Variety | HuARC | 83 | ABXC-50402 | Accession | Afar |
| 34 | Hirhir Kebabo Hairless-Sel-7 | Accession | Tigray | 84 | NN0021 | Accession | Amhara |
| 35 | NN-0143 | Accession | Amhara | 85 | NN0079-1 | Accession | Amhara |
| 36 | ACC NS-031 | Accession | Oromia | 86 | ACC 202-333 | Accession | Amhara |
| 37 | NN-0029(2) | Accession | Amhara | 87 | NN-0052 | Accession | Amhara |
| 38 | NN-0054 | Accession | Amhara | 88 | NN-0029-1 | Accession | Amhara |
| 39 | Morgo-Sel-P = 13 | Accession | Afar | 89 | Teiahir Sanja Sel-6 | Accession | Amhara |
| 40 | Hirhir Humera Sel-8 | Accession | Tigray | 90 | ACC-202-358 | Accession | Amhara |
| 41 | Tejareb Girar | Accession | Amhara | 91 | NN0032 | Accession | Amhara |
| 42 | NN0027 | Accession | Amhara | 92 | NN0071 | Accession | Amhara |
| 43 | NN0009 | Accession | Amhara | 93 | NN0064-1 | Accession | Amhara |
| 44 | ACC-203-610 | Accession | Amhara | 94 | NN0056 | Accession | Amhara |
| 45 | NN-0146 | Accession | Amhara | 95 | NN-01-03 | Accession | Amhara |
| 46 | NN-0044-2 | Accession | Amhara | 96 | NN0032-2 | Accession | Amhara |
| 47 | NN-0018-2 | Accession | Amhara | 97 | Bawnji Maksegnt | Accession | Amhara |
| 48 | Hirhir Nigara 1st Sel-1 | Accession | Tigray | 98 | Bawnji Flwha Sel-2 | Accession | Amhara |
| 49 | NN00136-1 | Accession | Amhara | 99 | NN0068-2 | Accession | Amhara |
| 50 | NN-0088-2 | Accession | Amhara | 100 | Hirhir Filwha Large Seeded | Accession | Amhara |
| No | Primers | Forward Primer Sequence | Reverse Primer Sequence | Product Size (bp) |
|---|---|---|---|---|
| 1 | ID0046 | TCAACGTGATTGCTCCCATA | CAGCTGCCTGAAAGAAGAGG | 101 |
| 2 | ZMM1043 | CCCGAAAATAGGATTTCTAACCA | TTTTGGACTGCTATTGAGGGA | 184 |
| 3 | ZMM3261 | CGAAAGCATGAGACGAGTATG | AACTAGTGCGCAATTCATTCAA | 244 |
| 4 | ID0041 | AGGCTTTCACATCATCAAATG | CATGTAGGATGCAACTCTTCAAA | 280 |
| 5 | ZMM5015 | ATTTATTGGGTTGCTGGGAA | TGAAAATTAAGTCACCAGTACCACC | 151 |
| 6 | ZMM4664 | CCTTCACTTCAAATCCGTCAA | TTTGGTTTGCATAGATGCTCTT | 184 |
| 7 | ZMM1809 | TTAAGCCTCGTTGACTCCAA | ATTGTACGGCATGTTGTCCC | 256 |
| 8 | ZMM2321 | CAACACCACCAACGCATATC | AGCAACGATTCACGACATTG | 280 |
| 9 | ZMM5358 | TAGGATGCTTTGAATTGGGC | AGGAACAAACATACGGCGTC | 164 |
| 10 | ID0068 | TCTTCGGAGTTAACACCCTCA | TTGGATTTCCATGTATGCCA | 199 |
| 11 | ZMM3312 | GCAAAATCTTCTTTCCTCCG | GCAGCAAGGGAATTGAATGT | 264 |
| 12 | ZMM1033 | CGTAGTGGTTCCCCTCACAT | ATGCTTTCCCCCAAATAACC | 179 |
| 13 | ZMM1189 | TATCCAGGGGAAAACCAGAA | TTGGATTTTCCTTCTCACGC | 212 |
| 14 | ZMM2202 | TCAGGAAGAAAGAATTGCTGC | CAATTTAACCATCCTGACTC | 276 |
| 15 | ZMM1637 | GCGGTGACATATTAAGGGCA | ACCGGAATCCGAACATGTAA | 265 |
| 16 | ZMM4645 | TTGAGCGATTCATCGACTTG | TTCTCCGGCCATTTTAATCA | 179 |
| 17 | ZMM1700 | CATTAACACCATTACGCAAACA | TTTGGCAAAACTAGCAATGAA | 258 |
| 18 | ID0175 | CAATTTTGATTTCTTTATCTATTTTCG | TCGAGTGCCCGAATTTTAAG | 271 |
| 19 | ZMM1353 | GCCAAAACAAAGGATTCAAGA | TGAGCTTTGTGTGACCATGA | 169 |
| 20 | ID0145 | ACCCTCCCTCCATGAATTTT | CCTCCATCTCATCTCATCCC | 196 |
| 21 | ZMM4803 | TGCATGAGCTAAGGGAAAGG | TGGTGGCAATTTGCAAGTAA | 268 |
| 22 | ZMM6141 | AAAAAGCAAAATCCATAATTTGA | TTGCCCCCTCAACTATTTG | 167 |
| 23 | ZMM3013 | TGCCAGTTGGCATATACCATTA | GAGCCGGTCTGAAATTTATCC | 216 |
| 24 | ZMM2818 | CGTGTGCCCAATATTTGAGTT | TCAACCTCCTCCCTACACAA | 279 |
| 25 | ZMM3223 | CGATGGTTATTAAATTAAGTATTCGG | GACATTTGAAGCAAAGTGTATCG | 279 |
| 26 | ZMM1691 | CTTGACCTGGAGTGTACGGC | GGATCAAACAGACACGAGCA | 220 |
| 27 | ZMM1851 | TGACTCTTTCGATTTGGGCT | CGAAAAATACGGGCGTTACT | 280 |
| Traits | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source of Variation | d.f. | DF | DM | PH | INL | NPB | NSB | NCPP | NSPC | SHB | DFLBC | TSW | GYH | OYH |
| Rep (Loc) | 2 | 171.27 | 205.65 | 714.76 | 0.00 | 121.72 | 28.62 | 14,855.85 | 522.58 | 1730.39 | 374.90 | 17.61 | 0.00 | 0.06 |
| Block (Loc*Rep) | 36 | 5.05 | 16.04 | 240.28 | 1.16 | 1.15 | 0.14 | 276.21 | 249.16 | 70.61 | 111.98 | 6.36 | 0.05 | 0.07 |
| Entry (Gen) | 99 | 6.93 * | 16.46 * | 360.33 * | 2.87 ** | 0.61 ns | 1.24 ** | 145.00 ns | 189.08 ** | 70.52 ns | 122.26 ** | 5.43 ns | 0.08 ** | 0.07 ns |
| Location (Env) | 1 | 201.64 ** | 60.06 * | 423.33 ns | 852.35 ** | 39.06 ** | 35.40** | 116.64 ns | 2787.84 ** | 11,306.07 ** | 36,898.56 ** | 1.32 ns | 3.94 ** | 1.47 ** |
| Gen × Env | 99 | 5.05 ns | 14.03 ns | 336.30 * | 3.43 ** | 0.67 * | 0.68 * | 179.46 ns | 136.61 ns | 48.88 ns | 107.58 ** | 5.49 ns | 0.70 ** | 0.05 ns |
| Error | 162 | 4.69 | 11.08 | 243.30 | 1.54 | 0.47 | 0.40 | 143.65 | 112.01 | 52.94 | 65.88 | 5.37 | 0.03 | 0.05 |
| Traits | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No | Entry Name or Designation | DF | DM | PH | INL | NPB | NSB | NCPP | NSPC | SHB | DFLBC | TSW | GYH | OYH | OC |
| Top 10 entries | |||||||||||||||
| 1 | Hirhir Kebabo Hairless-9 | 41 bcd | 90 cd | 123.9 abc | 9.8 abc | 4 a | 1 d | 41 abcd | 58 abcd | 18.1 cd | 42.8 ab | 3.0 a | 1.01 a | 0.40 a | 50.9 |
| 2 | Setit-3 | 43 abc | 91 bcd | 131.6 a | 10.0 ab | 3 b | 2c | 52 a | 63 a | 18.0 cd | 26.5 cd | 3.1 a | 0.84 ab | 0.40 a | 49.1 |
| 3 | Orofalc ACC-2 | 42 abcd | 89 d | 103.9 cd | 8.3 bcd | 3 b | 2 c | 36 abcd | 56 abcd | 13.8 d | 22.4 d | 3.1 a | 0.80 ab | 0.40 a | 52.5 |
| 4 | Hirhir Humera Sel-6 | 43 abc | 92 bcd | 114.8 abcd | 9.7 abc | 4 a | 4 a | 48 abc | 61 abc | 27.2 abc | 38.1 ab | 2.9 a | 0.78 ab | 0.39 ab | 53.9 |
| 5 | ABX = 2-01-2 | 44 ab | 95 ab | 133.2 a | 8.7 bcd | 4 a | 3 b | 40 abcd | 65 a | 18.9 bcd | 40.9 ab | 2.7 a | 0.74 b | 0.36 abc | 48.9 |
| 6 | Setit-1 | 40 cd | 88 d | 125.6 ab | 7.9 d | 4 a | 2 c | 39 abcd | 56 abcd | 21.9 bcd | 38.9 ab | 3.3 a | 0.73 b | 0.39 ab | 53.8 |
| 7 | ACC 205-180 | 45 a | 97 a | 126.9 ab | 10.5 a | 3 b | 2 c | 40 abcd | 55 abcd | 32.5 a | 42.0 ab | 3.1 a | 0.72 b | 0.38 ab | 53.1 |
| 8 | ACC 203-616 | 45 a | 94 abc | 106.8 bcd | 8.5 bcd | 3 b | 1 d | 37 abcd | 54 abcd | 25.5 abc | 39.0 ab | 2.6 a | 0.69 b | 0.35 abc | 51.7 |
| 9 | NN-0029(2) | 40 cd | 88 d | 118.5 abcd | 9.5 abcd | 4 a | 1 d | 44 abcd | 65 a | 28.2 abc | 41.9 ab | 3.4 a | 0.68 b | 0.36 abc | 50.7 |
| 10 | GA-002(3) | 42 abcd | 90 cd | 119 abcd | 10.3 ab | 4 a | 2 c | 49 ab | 63 a | 27.7 abc | 34.8 bc | 2.8 a | 0.67 b | 0.33 abc | 49.3 |
| Bottom 5 entries | |||||||||||||||
| 1 | NN-0183-3 | 43 abc | 92 bcd | 117.1 abcd | 8.1 cd | 3 b | 2 c | 34 bcd | 48 bcd | 20.8 bcd | 42.6 ab | 2.3 a | 0.17 c | 0.07 bc | 45.8 |
| 2 | NN-0020 | 43 abc | 98 a | 125.8 ab | 8.2 cd | 4 a | 2 c | 34 bcd | 62 ab | 29.0 ab | 44.4 ab | 2.6 a | 0.24 c | 0.12 abc | 49.3 |
| 3 | NN-0108-2 | 39 d | 90 cd | 135.2 a | 10.2 ab | 4 a | 0 e | 32 cd | 47 cd | 20.6bcd | 47.2 a | 2.9 a | 0.26 c | 0.04 c | 47.3 |
| 4 | NN00136-1 | 43 abc | 97 a | 99.6 d | 10.2 ab | 3 b | 2 c | 42 abcd | 44 d | 26.1 abc | 47.4 a | 3.0 a | 0.26 c | 0.13 abc | 47.5 |
| 5 | NN-0143 | 42 abcd | 94 abc | 132.2 a | 10.3 ab | 4 a | 3 b | 30 d | 55 abcd | 27.2 abc | 39.7 ab | 3.2 a | 0.28 c | 0.15 abc | 48.3 |
| Mean | 43 | 92 | 119.6 | 9.2 | 3.5 | 2 | 41 | 51 | 24.4 | 39.8 | 2.9 | 0.48 | 0.24 | 49.7 | |
| CV (%) | 5.03 | 3.61 | 13.30 | 13.52 | 19.21 | 38.58 | 29.42 | 20.92 | 29.74 | 20.44 | 78.06 | 35.26 | 90.13 | NA | |
| R2 (%) | 72.87 | 70.33 | 68.59 | 86.27 | 85.16 | 82.97 | 77.76 | 70.65 | 77.22 | 85.95 | 60.98 | 82.15 | 66.42 | NA | |
| Traits | DM | PH | INL | NPB | NSB | NCPP | NSPC | SHB | DFLBC | TSW | OC | OYH | GYH |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DF | 0.31 ** | 0.05 ns | 0.14 ** | 0.09 ns | 0.09 ns | 0.16 ** | −0.21 ** | 0.05 ns | −0.13 ** | −0.02 ns | −0.11 * | 0.08 ns | 0.09 ns |
| DM | 1.00 | 0.07 ns | 0.08 ns | −0.20 ns | 0.07 ns | −0.10 * | −0.12 * | 0.09 ns | 0.00 ns | −0.15 ** | −0.06 ns | −0.10 * | −0.09 ns |
| PH | 1.00 | 0.09 ns | 0.06 ns | 0.12 * | 0.07 ns | 0.05 ns | 0.01 ns | 0.07 ns | 0.10 * | −0.00 ns | 0.05 ns | 0.05 ns | |
| INL | 1.00 | −0.17 ** | 0.21 ** | 0.08 ns | −0.08 ns | 0.38 ** | −0.45 ** | 0.00 ns | 0.03 ns | 0.35 ** | 0.35 ** | ||
| NPB | 1.00 | 0.06 ns | 0.30 ** | 0.08 ns | −0.14 ** | 0.25 ** | 0.20 ** | 0.05 ns | −0.05 ns | −0.06 ns | |||
| NSB | 1.00 | −0.05 ns | 0.04 ns | 0.34 ** | −0.166 ** | −0.122 * | 0.051 ns | 0.214 ** | 0.207 ** | ||||
| NCPP | 1.00 | 0.119 * | −0.00 ns | −0.01 ns | 0.16 ** | −0.01 ns | 0.17 ** | 0.18 ** | |||||
| NSPC | 1.00 | −0.07 ns | 0.21 ** | 0.04 ns | −0.08 ns | 0.15 ** | 0.17 ** | ||||||
| SHB | 1.00 | −0.26 ** | −0.13 ** | 0.02 ns | 0.16 ** | 0.16 ** | |||||||
| DFLBC | 1.00 | −0.01 ns | −0.02 ns | −0.29 ** | −0.29 ** | ||||||||
| TSW | 1.00 | 0.05 ns | 0.23 ** | 0.23 ** | |||||||||
| OC | 1.00 | 0.16 ** | 0.06 ns | ||||||||||
| OYH | 1.00 | 0.99 ** |
| Locus | Product Size (bp) | Genetic Parameter | |||
|---|---|---|---|---|---|
| MAF | He | Ho | PIC | ||
| ID0046 | 101 | 0.72 | 0.40 | 0.56 | 0.32 |
| ZMM1043 | 184 | 0.75 | 0.38 | 0.51 | 0.31 |
| ZMM3261 | 244 | 0.59 | 0.48 | 0.82 | 0.37 |
| ID0041 | 280 | 0.96 | 0.08 | 0.08 | 0.07 |
| ZMM5015 | 151 | 0.79 | 0.34 | 0.43 | 0.28 |
| ZMM4664 | 184 | 0.60 | 0.48 | 0.80 | 0.36 |
| ZMM1809 | 256 | 0.86 | 0.24 | 0.28 | 0.21 |
| ZMM2321 | 280 | 0.90 | 0.18 | 0.20 | 0.16 |
| ZMM5358 | 164 | 0.62 | 0.47 | 0.77 | 0.36 |
| ID0068 | 199 | 0.86 | 0.25 | 0.29 | 0.22 |
| ZMM3312 | 264 | 0.56 | 0.49 | 0.89 | 0.37 |
| ZMM1033 | 179 | 0.76 | 0.37 | 0.49 | 0.30 |
| ZMM1189 | 212 | 0.52 | 0.50 | 0.96 | 0.37 |
| ZMM2202 | 276 | 0.89 | 0.20 | 0.22 | 0.18 |
| ZMM1637 | 265 | 0.68 | 0.44 | 0.65 | 0.34 |
| ZMM4645 | 179 | 0.81 | 0.31 | 0.39 | 0.26 |
| ZMM1700 | 258 | 0.95 | 0.10 | 0.10 | 0.09 |
| ID0175 | 271 | 0.96 | 0.08 | 0.08 | 0.07 |
| ZMM1353 | 169 | 0.94 | 0.12 | 0.13 | 0.11 |
| ID0145 | 196 | 0.77 | 0.36 | 0.47 | 0.29 |
| ZMM4803 | 268 | 0.95 | 0.10 | 0.11 | 0.10 |
| ZMM6141 | 167 | 0.75 | 0.38 | 0.51 | 0.31 |
| ZMM3013 | 216 | 0.69 | 0.43 | 0.63 | 0.34 |
| ZMM2818 | 279 | 0.96 | 0.08 | 0.08 | 0.07 |
| ZMM3223 | 279 | 0.82 | 0.30 | 0.36 | 0.25 |
| ZMM1691 | 220 | 0.73 | 0.39 | 0.54 | 0.32 |
| ZMM1851 | 280 | 0.90 | 0.18 | 0.20 | 0.16 |
| Mean | 221 | 0.78 | 0.30 | 0.43 | 0.25 |
| Population | Entries | Membership % | Expected Heterozygosity | Mean Fixation Index |
|---|---|---|---|---|
| I | ACC NS-031E63, Hirhir Nigara 1st Sel-2, Hirhir Kebabo Hairless Sel-6, NN-0183-3, GXT = 85(28 − 2), ABXT-85-Sel-2-1, ABXC-50402, NN-0129-2, ACC-203-020, Gonjam Azene (Aleka), NN-0026, Tejareb Kokit Sel-3, NN0027, NN0009, NN-0088-2 Bawnji Sel-2, G-02, Endelemi kirem sel-2, NN0016-1, NN0038-1, NN-0052, NN0071, NN0064-1 | 24 | 0.15 | 0.39 |
| II | Hirhir Baeker-Sel-3, NN0025, BCS-0041, Orofalc ACC-2, ABX = 2-01-2, Bering Bawany, ACC-203-612, NN-0143, NN-0146, NN-0044-2, NN-0018-2, NN-0029-1, NN0056 | 13 | 0.22 | 0.23 |
| III | Hirhir kebabo hairless sel-2, Hirhir kebabo hairless-9, ACC-200-064-1, HIRHIR NIGARA 1ST SEL-1, Bawnji Fiyel Kolet, NN0104, Bawnji Maksegnt, Bawnji Flwha Sel-2, Hirhir Filwha Large Seeded | 9 | 0.29 | 0.02 |
| IV | Hirhir Humera Sel-6, Humera-1, Setit-1, Setit-3, Hirhir Sel-2, Hirhir Kebabo Hairless-Sel-7, Hirhir Humera Sel-8, NN0036-1, Hirhir Kebabo Early Sel-1, Hirhir Adgeshu Sel -8, Morgo-Sel-P = 13, NN-0020 ACC-203-610, NN0001-2, NN01-13, Gojam Azene(Yohans Sel-1), NN0031 | 17 | 0.20 | 0.29 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teklu, D.H.; Shimelis, H.; Tesfaye, A.; Mashilo, J.; Zhang, X.; Zhang, Y.; Dossa, K.; Shayanowako, A.I.T. Genetic Variability and Population Structure of Ethiopian Sesame (Sesamum indicum L.) Germplasm Assessed through Phenotypic Traits and Simple Sequence Repeats Markers. Plants 2021, 10, 1129. https://doi.org/10.3390/plants10061129
Teklu DH, Shimelis H, Tesfaye A, Mashilo J, Zhang X, Zhang Y, Dossa K, Shayanowako AIT. Genetic Variability and Population Structure of Ethiopian Sesame (Sesamum indicum L.) Germplasm Assessed through Phenotypic Traits and Simple Sequence Repeats Markers. Plants. 2021; 10(6):1129. https://doi.org/10.3390/plants10061129
Chicago/Turabian StyleTeklu, Desawi Hdru, Hussein Shimelis, Abush Tesfaye, Jacob Mashilo, Xiurong Zhang, Yanxin Zhang, Komivi Dossa, and Admire Isaac Tichafa Shayanowako. 2021. "Genetic Variability and Population Structure of Ethiopian Sesame (Sesamum indicum L.) Germplasm Assessed through Phenotypic Traits and Simple Sequence Repeats Markers" Plants 10, no. 6: 1129. https://doi.org/10.3390/plants10061129
APA StyleTeklu, D. H., Shimelis, H., Tesfaye, A., Mashilo, J., Zhang, X., Zhang, Y., Dossa, K., & Shayanowako, A. I. T. (2021). Genetic Variability and Population Structure of Ethiopian Sesame (Sesamum indicum L.) Germplasm Assessed through Phenotypic Traits and Simple Sequence Repeats Markers. Plants, 10(6), 1129. https://doi.org/10.3390/plants10061129









