Genome-Wide Differential Methylation Profiles from Two Terpene-Rich Medicinal Plant Extracts Administered in Osteoarthritis Rats
Abstract
1. Introduction
2. Results
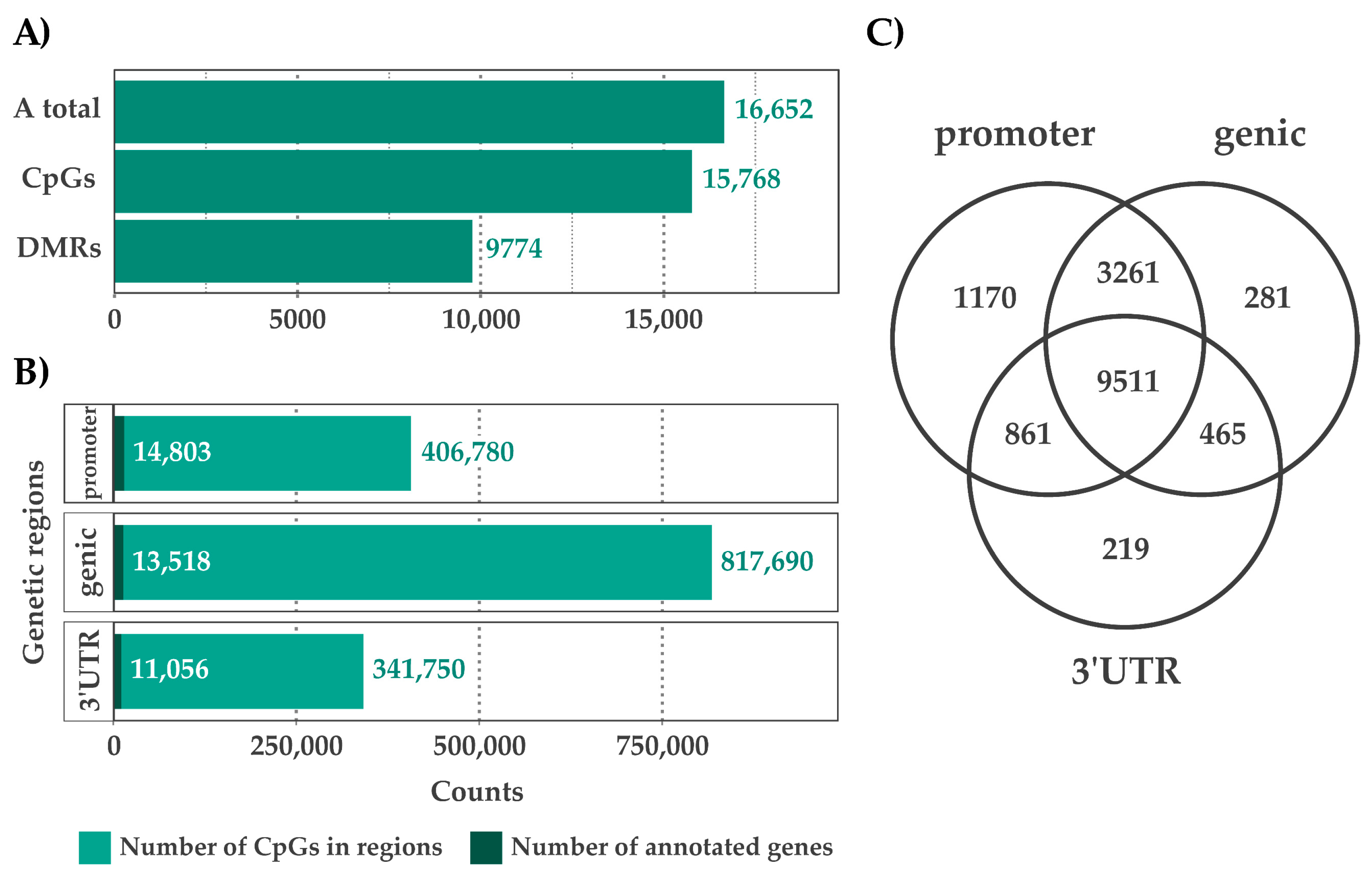
2.1. CpG Profiles and DMRs
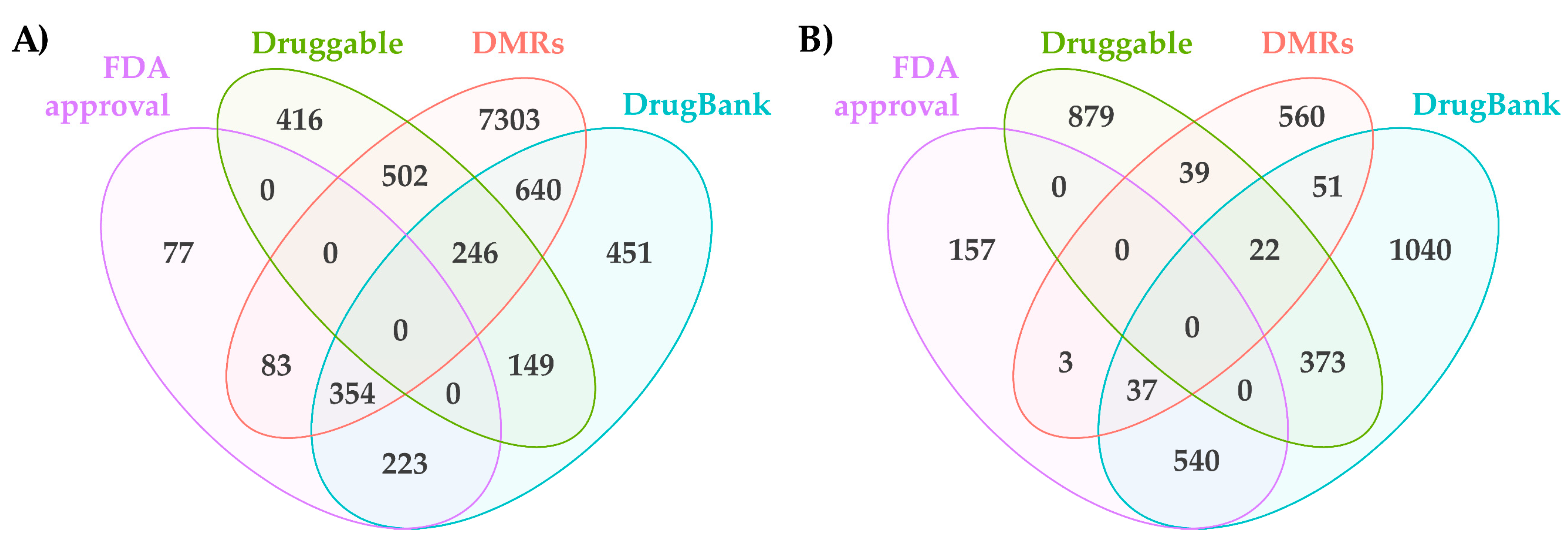
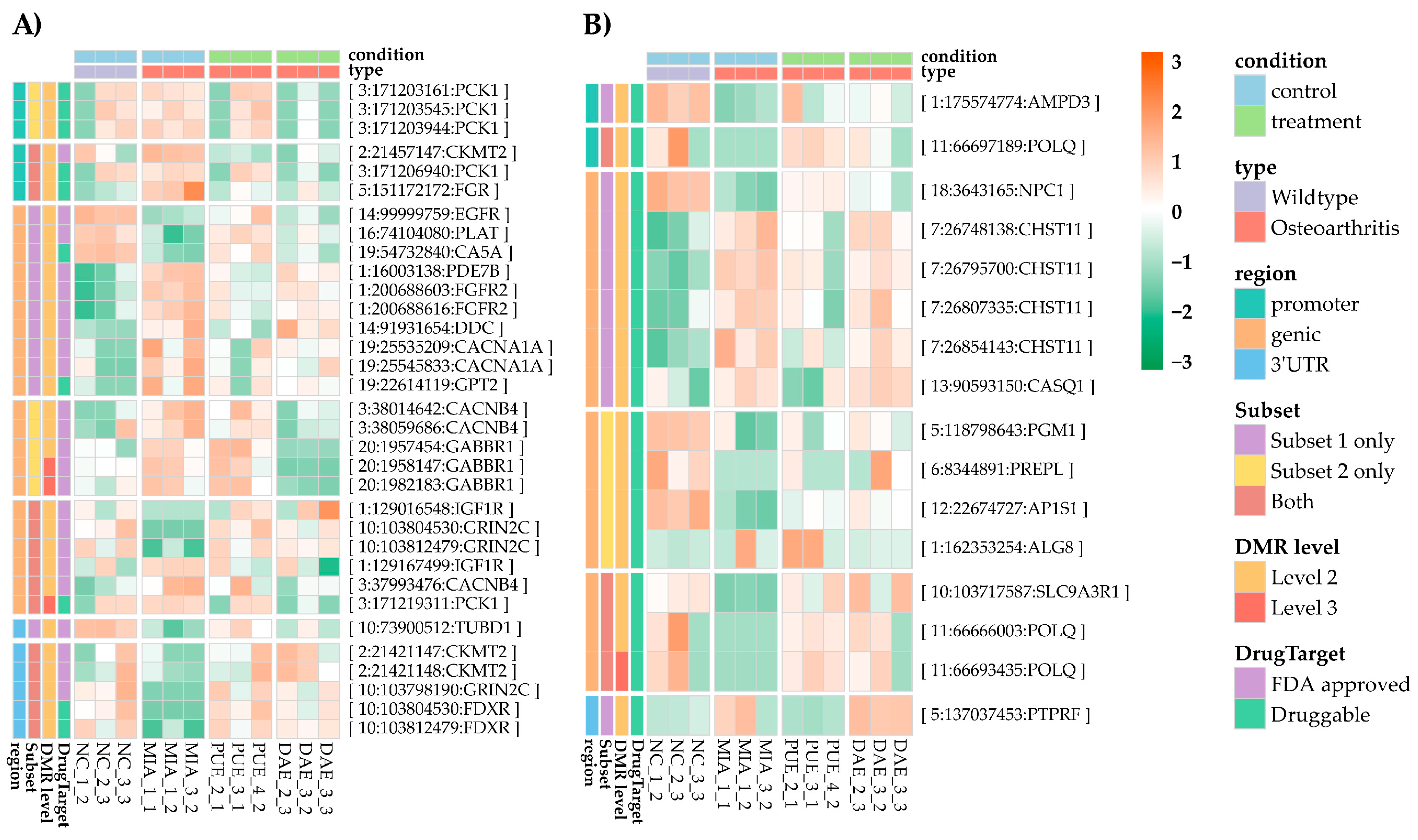
2.2. DMRs Common between Plant Treatments and Experimentally-Induced OA
2.3. Functional Annotation of CpGs
2.4. PI3K/AKT Signaling Pathway
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Sample Collection and Experimental Design
4.3. Library Preparation and Sequencing
4.4. Genome Wide Methylation Patterns from Bisulfite-Seq Data
4.5. Gene Set Enrichment Analysis among Union Genes in DMRs
4.6. Functional Database
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kulesa, A.; Kehe, J.; Hurtado, J.E.; Tawde, P.; Blainey, P.C. Combinatorial drug discovery in nanoliter droplets. Proc. Natl. Acad. Sci. USA 2018, 115, 6685–6690. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, W.; Zou, B.; Wang, J.; Deng, Y.; Deng, L. DrugCombDB: A comprehensive database of drug combinations toward the discovery of combinatorial therapy. Nucleic Acids Res. 2019, 48, D871–D881. [Google Scholar] [CrossRef]
- Posadzki, P.; Watson, L.K.; Ernst, E. Adverse effects of herbal medicines: An overview of systematic reviews. Clin. Med. 2013, 13, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.M.; Lee, A.Y.; Nam, J.Y.; Lee, M.Y.; Choe, M.S.; Lim, K.S.; Kim, C.; Kim, J.-S. Protective effects of Phlomis umbrosa extract on a monosodium iodoacetate–induced osteoarthritis model and prediction of molecular mechanisms using transcriptomics. Phytomedicine 2021, 81, 153429. [Google Scholar] [CrossRef]
- Chun, J.M.; Lee, A.Y.; Nam, J.Y.; Lim, K.S.; Choe, M.S.; Lee, M.Y.; Kim, C.; Kim, J.-S. Effects of Dipsacus asperoides Extract on Monosodium Iodoacetate–Induced Osteoarthritis in Rats Based on Gene Expression Profiling. Front. Pharmacol. 2021, 12, 293. [Google Scholar] [CrossRef] [PubMed]
- Vincent, T.L. Of mice and men: Converging on a common molecular understanding of osteoarthritis. Lancet Rheumatol. 2020, 2, e633–e645. [Google Scholar] [CrossRef]
- Chou, C.-H.; Jain, V.; Gibson, J.; Attarian, D.E.; Haraden, C.A.; Yohn, C.B.; Laberge, R.-M.; Gregory, S.; Kraus, V.B. Synovial cell cross-talk with cartilage plays a major role in the pathogenesis of osteoarthritis. Sci. Rep. 2020, 10, 10868. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Luo, J.; Guo, J.; Yao, X.; Jing, X.; Guo, F. The PI3K/AKT/mTOR signaling pathway in osteoarthritis: A narrative review. Osteoarthr. Cartil. 2020, 28, 400–409. [Google Scholar] [CrossRef]
- Shin, T.-Y.; Lee, J.-K. Effect of Phlomis umbrosa Root on Mast Cell-Dependent Immediate-Type Allergic Reactions by Anal Therapy. Immunopharmacol. Immunotoxicol. 2003, 25, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Lee, S.H.; Lee, D.; Kim, H. Astragalus Extract Mixture HT042 Improves Bone Growth, Mass, and Microarchitecture in Prepubertal Female Rats: A Microcomputed Tomographic Study. Evid. Based Complement. Altern. Med. 2017, 2017, 5219418. [Google Scholar] [CrossRef]
- Kim, J.; Yang, S.; Choi, C.-Y. The Evaluation of the Effect of Herbal Extract on Osteoarthritis: In Vitro and In Vivo Study. Prev. Nutr. Food Sci. 2016, 21, 310–316. [Google Scholar] [CrossRef]
- Lee, J.E.; Lee, H.; Kim, M.H.; Yang, W.M. Osteogenic effects of Phlomis umbrosa via up-regulation of Runx2 in osteoporosis. Biomed. Rep. 2018, 10, 17–22. [Google Scholar] [CrossRef]
- Cao, T.; Fei, J.; Zu, G.; Han, G.; Lai, Z.; Ren, N.; Zhang, Q. Phylogenetic analysis and characterization of the complete chloroplast genome of Dipsacus asperoides, the endemic medicinal herb in China. Mitochondrial DNA Part B 2019, 4, 2557–2559. [Google Scholar] [CrossRef]
- Wang, C.; Gong, X.; Bo, A.; Zhang, L.; Zhang, M.; Zang, E.; Zhang, C.; Li, M. Iridoids: Research Advances in Their Phytochemistry, Biological Activities, and Pharmacokinetics. Molecules 2020, 25, 287. [Google Scholar] [CrossRef]
- Renaud, J.; Martinoli, M.-G. Considerations for the Use of Polyphenols as Therapies in Neurodegenerative Diseases. Int. J. Mol. Sci. 2019, 20, 1883. [Google Scholar] [CrossRef]
- Mohanan, P.; Subramaniyam, S.; Mathiyalagan, R.; Yang, D.-C. Molecular signaling of ginsenosides Rb1, Rg1, and Rg3 and their mode of actions. J. Ginseng Res. 2018, 42, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Han, J.; Guo, H.; Lv, D.; Wang, Y. Pathway and network analysis of genes related to osteoporosis. Mol. Med. Rep. 2019, 20, 985–994. [Google Scholar] [CrossRef] [PubMed]
- Dor, Y.; Cedar, H. Principles of DNA methylation and their implications for biology and medicine. Lancet 2018, 392, 777–786. [Google Scholar] [CrossRef]
- Chen, D.; Shen, J.; Zhao, W.; Wang, T.; Han, L.; Hamilton, J.L.; Im, H.-J. Osteoarthritis: Toward a comprehensive understanding of pathological mechanism. Bone Res. 2017, 5, 16044. [Google Scholar] [CrossRef]
- Zeggini, E.; Panoutsopoulou, K.; Southam, L.; Rayner, N.W.; Day-Williams, A.G.; Lopes, M.C.; Boraska, V.; Esko, T.; Evangelou, E.; Hoffman, A.; et al. Identification of new susceptibility loci for osteoarthritis (arcOGEN): A genome-wide association study. Lancet 2012, 380, 815–823. [Google Scholar] [CrossRef]
- Ata, A.; Kalhari, K.S.; Samarasekera, R. Chemical constituents of Barleria prionitis and their enzyme inhibitory and free radical scavenging activities. Phytochem. Lett. 2009, 2, 37–40. [Google Scholar] [CrossRef]
- Siddiqi, M.H.; Siddiqi, M.Z.; Ahn, S.; Kang, S.; Kim, Y.-J.; Natarajan, S.; Yang, N.-U.; Yang, D.-C. Ginseng saponins and the treatment of osteoporosis: Mini literature review. J. Ginseng Res. 2013, 37, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shen, J.; Yang, X.; Jin, Y.; Yang, Z.; Wang, R.; Zhang, F.; Linhardt, R.J. Akebia saponin D reverses corticosterone hypersecretion in an Alzheimer’s disease rat model. Biomed. Pharmacother. 2018, 107, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.-M.; Wu, L.-G.; Cai, J.-W.; Wu, L.-T.; Liang, M. Dexamethasone suppresses osteogenesis of osteoblast via the PI3K/Akt signaling pathway in vitro and in vivo. J. Recept. Signal Transduct. 2019, 39, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.; Park, M.K. Glucocorticoid-Induced Diabetes Mellitus: An Important but Overlooked Problem. Endocrinol. Metab. 2017, 32, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhao, J.; Zhang, X. Inhibition of uncoupling protein 2 by genipin reduces insulin-stimulated glucose uptake in 3T3-L1 adipocytes. Arch. Biochem. Biophys. 2009, 486, 88–93. [Google Scholar] [CrossRef]
- Xiao, Y.-P.; Tian, F.-M.; Dai, M.-W.; Wang, W.-Y.; Shao, L.-T.; Zhang, L. Are estrogen-related drugs new alternatives for the management of osteoarthritis? Arthritis Res. 2016, 18, 1–9. [Google Scholar] [CrossRef]
- Mathiyalagan, R.; Wang, C.; Kim, Y.J.; Castro-Aceituno, V.; Ahn, S.; Subramaniyam, S.; Simu, S.Y.; Jiménez-Pérez, Z.E.; Yang, D.C.; Jung, S.-K. Preparation of Polyethylene Glycol-Ginsenoside Rh1 and Rh2 Conjugates and Their Efficacy against Lung Cancer and Inflammation. Molecules 2019, 24, 4367. [Google Scholar] [CrossRef]
- Bosch, M.H.V.D. Osteoarthritis year in review 2020: Biology. Osteoarthr. Cartil. 2021, 29, 143–150. [Google Scholar] [CrossRef]
- Zhang, H.; Cai, D.; Bai, X. Macrophages regulate the progression of osteoarthritis. Osteoarthr. Cartil. 2020, 28, 555–561. [Google Scholar] [CrossRef]
- Alford, A.I.; Kozloff, K.M.; Hankenson, K.D. Extracellular matrix networks in bone remodeling. Int. J. Biochem. Cell Biol. 2015, 65, 20–31. [Google Scholar] [CrossRef]
- Hastings, J.F.; Skhinas, J.N.; Fey, D.; Croucher, D.R.; Cox, T.R. The extracellular matrix as a key regulator of intracellular signalling networks. Br. J. Pharmacol. 2019, 176, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Hu, X.; Cheng, J.; Zhang, X.; Zhao, F.; Shi, W.; Ren, B.; Yu, H.; Yang, P.; Li, Z.; et al. A small molecule promotes cartilage extracellular matrix generation and inhibits osteoarthritis development. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Englert, N.A.; Spink, B.C.; Spink, D.C. Persistent and non-persistent changes in gene expression result from long-term estrogen exposure of MCF-7 breast cancer cells. J. Steroid Biochem. Mol. Biol. 2011, 123, 140–150. [Google Scholar] [CrossRef]
- Das, A.; Mantena, S.R.; Kannan, A.; Evans, D.B.; Bagchi, M.K.; Bagchi, I.C. De novo synthesis of estrogen in pregnant uterus is critical for stromal decidualization and angiogenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 12542–12547. [Google Scholar] [CrossRef] [PubMed]
- Dip, R.; Lenz, S.; Gmuender, H.; Naegeli, H. Pleiotropic combinatorial transcriptomes of human breast cancer cells exposed to mixtures of dietary phytoestrogens. Food Chem. Toxicol. 2009, 47, 787–795. [Google Scholar] [CrossRef]
- Fillmore, C.M.; Gupta, P.B.; Rudnick, J.A.; Caballero, S.; Keller, P.J.; Lander, E.S.; Kuperwasser, C. Estrogen expands breast cancer stem-like cells through paracrine FGF/Tbx3 signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 21737–21742. [Google Scholar] [CrossRef]
- Krueger, F.; Andrews, S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011, 27, 1571–1572. [Google Scholar] [CrossRef] [PubMed]
- Akalin, A.; Kormaksson, M.; Li, S.; Garrett-Bakelman, F.E.; Figueroa, M.E.; Melnick, A.; Mason, C.E. methylKit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012, 13, R87. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- The Alliance of Genome Resources Consortium; Agapite, J.; Albou, L.-P.; Aleksander, S.; Argasinska, J.; Arnaboldi, V.; Attrill, H.; Bello, S.M.; A Blake, J.; Blodgett, O.; et al. Alliance of Genome Resources Portal: Unified model organism research platform. Nucleic Acids Res. 2020, 48, D650–D658. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Laulederkind, S.J.F.; Hayman, G.T.; Wang, S.-J.; Hoffman, M.J.; Smith, J.R.; Bolton, E.R.; De Pons, J.; Tutaj, M.A.; Tutaj, M.; Thota, J.; et al. Rat Genome Databases, Repositories, and Tools. In Advanced Structural Safety Studies; Springer: Clifton, NJ, USA, 2019; Volume 2018, pp. 71–96. [Google Scholar]



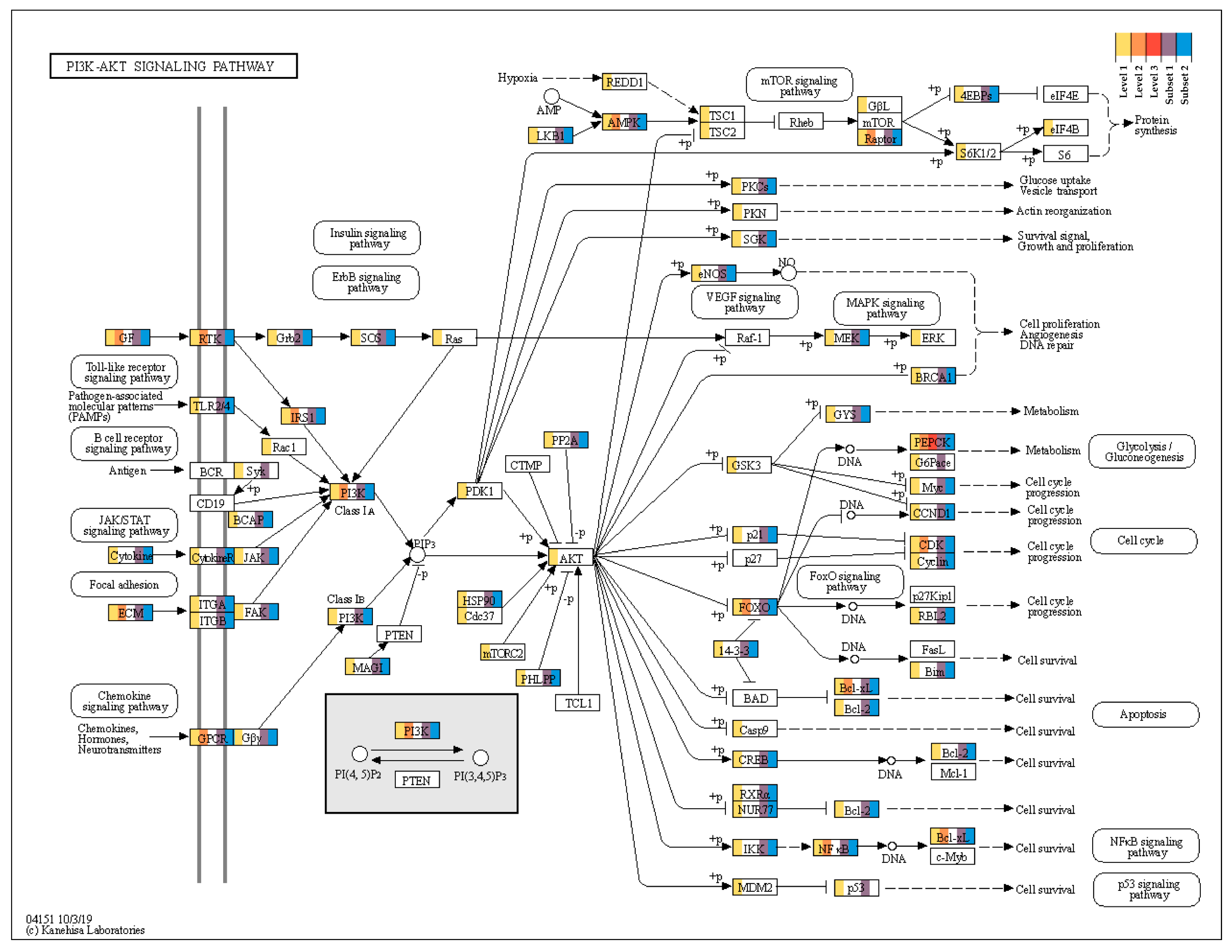
| Term | Count | % | p-Value | Genes | Fold Enrichment | FDR |
|---|---|---|---|---|---|---|
| rno04151:PI3K/AKT signaling pathway | 29 | 3.70 | 0.00 | Vegfa, Col3a1, Angpt4, Lamc1, Nfkb1, Col2a1, Prkaa1, Col11a2, Kitlg, Col4a3, Irs1, Col1a1, Rptor, Foxo3, Egfr, Bcl2l1, Fgfr3, Fgf2, Igf1r, Lama4, Cdk6, Efna5, Chad, Fgfr2, Pck1, Epha2, Lpar3, Pik3r2, Jak1 | 2.08 | 0.08 |
| rno05200:Pathways in cancer | 28 | 3.58 | 0.01 | Vegfa, Lamc1, Nfkb1, Kitlg, Col4a3, Rxrb, Egfr, Mecom, Bcl2l1, Fgfr3, Fgf2, Igf1r, Wnt4, Ptch1, Lama4, Axin2, Fzd9, Cdk6, Ctnnb1, Prkcg, Ppard, Gna13, Fgfr2, Hif1a, Lpar3, Pik3r2, Jak1, Brca2 | 1.72 | 0.12 |
| rno04144:Endocytosis | 22 | 2.81 | 0.00 | Pdcd6ip, Smurf1, Pard3, Chmp4c, Mvb12b, RT1-M6-2, Git2, Chmp1b, Dnm1, Smurf2, Rab11fip4, Acap1, Fgfr2, Egfr, Nedd4l, Rab31, Dnajc6, Zfyve27, Fgfr3, RT1-T24-3, Igf1r, Wipf1 | 1.99 | 0.12 |
| rno04014:Ras signaling pathway | 21 | 2.68 | 0.00 | Rasgrf2, Vegfa, Angpt4, Rgl1, Nfkb1, Shc3, Prkcg, Gab1, Efna5, Kitlg, Fgfr2, Egfr, Bcl2l1, Shc4, Pla2g12a, Fgfr3, Fgf2, Epha2, Rasa3, Pik3r2, Igf1r | 2.22 | 0.12 |
| rno05205:Proteoglycans in cancer | 18 | 2.30 | 0.00 | Vegfa, Fzd9, Ank3, Hcls1, Ctnnb1, Prkcg, Gab1, Itpr2, Hbegf, Hif1a, Egfr, Fgf2, Pik3r2, Igf1r, Ptch1, Wnt4, Gpc3, Cd44 | 2.18 | 0.12 |
| rno04510:Focal adhesion | 18 | 2.30 | 0.01 | Vegfa, Col3a1, Lamc1, Ctnnb1, Shc3, Col2a1, Prkcg, Col11a2, Parva, Col4a3, Chad, Col1a1, Dock1, Egfr, Shc4, Pik3r2, Igf1r, Lama4 | 2.09 | 0.12 |
| rno04015:Rap1 signaling pathway | 17 | 2.17 | 0.02 | Pard3, Vegfa, Angpt4, Ctnnb1, Prkcg, Efna5, Kitlg, Adora2b, Fgfr2, Egfr, Fgfr3, Gnao1, Fgf2, Epha2, Lpar3, Pik3r2, Igf1r | 1.92 | 0.23 |
| rno04020:Calcium signaling pathway | 16 | 2.04 | 0.01 | Sphk2, Prkcg, Adra1b, Itpr2, Ppp3ca, Nos1, Adora2b, Orai1, Itpkb, Egfr, Cacna1a, Grin2c, Atp2b1, Vdac3, Gnal, Ptk2b | 2.11 | 0.15 |
| rno04910:Insulin signaling pathway | 14 | 1.79 | 0.00 | Prkab1, Shc3, Prkaa1, Ptprf, Irs1, Ppp1r3c, Rptor, Hk2, Prkab2, Pck1, Shc4, Rhoq, Pik3r2, Acacb | 2.44 | 0.12 |
| rno05206:MicroRNAs in cancer | 14 | 1.79 | 0.01 | Vegfa, Cdk6, Nfkb1, Prkcg, Irs1, Slc7a1, Prkce, Rptor, Egfr, Shc4, Reck, Fgfr3, Trim71, Cd44 | 2.39 | 0.12 |
| rno04152:AMPK signaling pathway | 13 | 1.66 | 0.01 | Scd2, Prkab1, Prkaa1, Lepr, Pfkfb3, Irs1, Rptor, Foxo3, Prkab2, Pck1, Pik3r2, Acacb, Igf1r | 2.47 | 0.12 |
| rno04932:Non-alcoholic fatty liver disease (NAFLD) | 13 | 1.66 | 0.03 | Ndufa4l2, Cox4i2, Prkab1, Nfkb1, Prkaa1, Lepr, Ndufs7, Irs1, Prkab2, Casp7, Ndufb8, Ndufs8, Pik3r2 | 1.96 | 0.41 |
| rno04931:Insulin resistance | 12 | 1.53 | 0.01 | Irs1, Prkce, Ppp1r3c, Prkab2, Prkab1, Nfkb1, Pck1, Prkaa1, Ptprf, Rps6ka2, Pik3r2, Acacb | 2.66 | 0.12 |
| rno04512:ECM-receptor interaction | 11 | 1.40 | 0.00 | Col3a1, Col1a1, Chad, Lamc1, Col2a1, Col11a2, Gp9, Col4a3, Cd47, Lama4, Cd44 | 3.01 | 0.12 |
| rno04974:Protein digestion and absorption | 11 | 1.40 | 0.00 | Col3a1, Col1a1, Kcnk5, Col2a1, Col11a2, Atp1a4, Col4a3, Slc7a8, Kcnq1, Col17a1, Eln | 3.01 | 0.12 |
| rno04066:HIF-1 signaling pathway | 11 | 1.40 | 0.01 | Vegfa, Serpine1, Hk2, Hif1a, Angpt4, Egfr, Nfkb1, Prkcg, Pfkfb3, Pik3r2, Igf1r | 2.63 | 0.15 |
| rno05146:Amoebiasis | 11 | 1.40 | 0.02 | Col3a1, Col1a1, Lamc1, Nfkb1, Col2a1, Prkcg, Col11a2, Col4a3, Pik3r2, Gnal, Lama4 | 2.39 | 0.23 |
| rno04920:Adipocytokine signaling pathway | 10 | 1.28 | 0.00 | Irs1, Rxrb, Prkab2, Prkab1, Nfkb1, Pck1, Prkaa1, Lepr, Acacb, Nfkbib | 3.25 | 0.12 |
| rno05100:Bacterial invasion of epithelial cells | 9 | 1.15 | 0.02 | Dock1, Hcls1, Ctnnb1, Shc3, Septin8, Shc4, Gab1, Dnm1, Pik3r2 | 2.71 | 0.24 |
| rno04915:Estrogen signaling pathway | 9 | 1.15 | 0.04 | Hbegf, Fkbp5, Egfr, Shc3, Shc4, Gabbr1, Gnao1, Itpr2, Pik3r2 | 2.28 | 0.48 |
| rno05212:Pancreatic cancer | 8 | 1.02 | 0.02 | Vegfa, Egfr, Cdk6, Bcl2l1, Nfkb1, Jak1, Pik3r2, Brca2 | 3.05 | 0.23 |
| rno04520:Adherens junction | 8 | 1.02 | 0.03 | Pard3, Ssx2ip, Lmo7, Egfr, Ctnnb1, Ptprf, Ptprm, Igf1r | 2.67 | 0.38 |
| rno04730:Long-term depression | 7 | 0.89 | 0.04 | Gna13, Cacna1a, Prkcg, Gnao1, Itpr2, Nos1, Igf1r | 2.75 | 0.48 |
| rno05230:Central carbon metabolism in cancer | 7 | 0.89 | 0.05 | Fgfr2, Hk2, Hif1a, Slc7a5, Egfr, Fgfr3, Pik3r2 | 2.67 | 0.48 |
| rno05214:Glioma | 7 | 0.89 | 0.05 | Egfr, Cdk6, Shc3, Shc4, Prkcg, Pik3r2, Igf1r | 2.62 | 0.49 |
| rno00220:Arginine biosynthesis | 4 | 0.51 | 0.05 | Got2, Acy1, Gpt2, Nos1 | 4.87 | 0.48 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, Y.; Subramaniyam, S.; Chun, J.-M.; Jeon, J.-H.; Hong, J.-M.; Jung, H.; Seong, B.; Kim, C. Genome-Wide Differential Methylation Profiles from Two Terpene-Rich Medicinal Plant Extracts Administered in Osteoarthritis Rats. Plants 2021, 10, 1132. https://doi.org/10.3390/plants10061132
Shin Y, Subramaniyam S, Chun J-M, Jeon J-H, Hong J-M, Jung H, Seong B, Kim C. Genome-Wide Differential Methylation Profiles from Two Terpene-Rich Medicinal Plant Extracts Administered in Osteoarthritis Rats. Plants. 2021; 10(6):1132. https://doi.org/10.3390/plants10061132
Chicago/Turabian StyleShin, Younhee, Sathiyamoorthy Subramaniyam, Jin-Mi Chun, Ji-Hyeon Jeon, Ji-Man Hong, Hojin Jung, Boseok Seong, and Chul Kim. 2021. "Genome-Wide Differential Methylation Profiles from Two Terpene-Rich Medicinal Plant Extracts Administered in Osteoarthritis Rats" Plants 10, no. 6: 1132. https://doi.org/10.3390/plants10061132
APA StyleShin, Y., Subramaniyam, S., Chun, J.-M., Jeon, J.-H., Hong, J.-M., Jung, H., Seong, B., & Kim, C. (2021). Genome-Wide Differential Methylation Profiles from Two Terpene-Rich Medicinal Plant Extracts Administered in Osteoarthritis Rats. Plants, 10(6), 1132. https://doi.org/10.3390/plants10061132








