Three β-Glucuronosyltransferase Genes Involved in Arabinogalactan Biosynthesis Function in Arabidopsis Growth and Development
Abstract
1. Introduction
2. Results
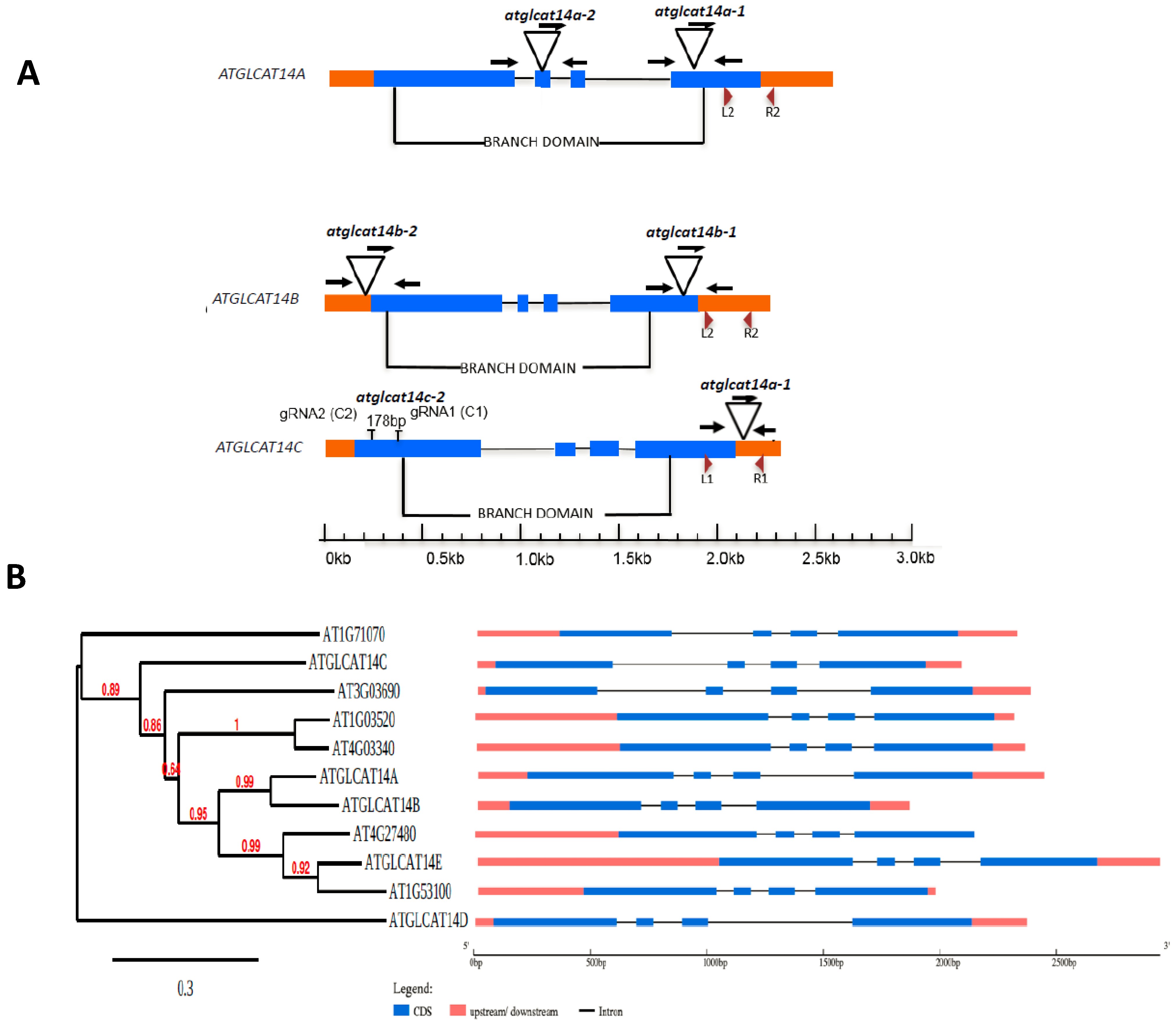
2.1. The CAZy GT14 Family
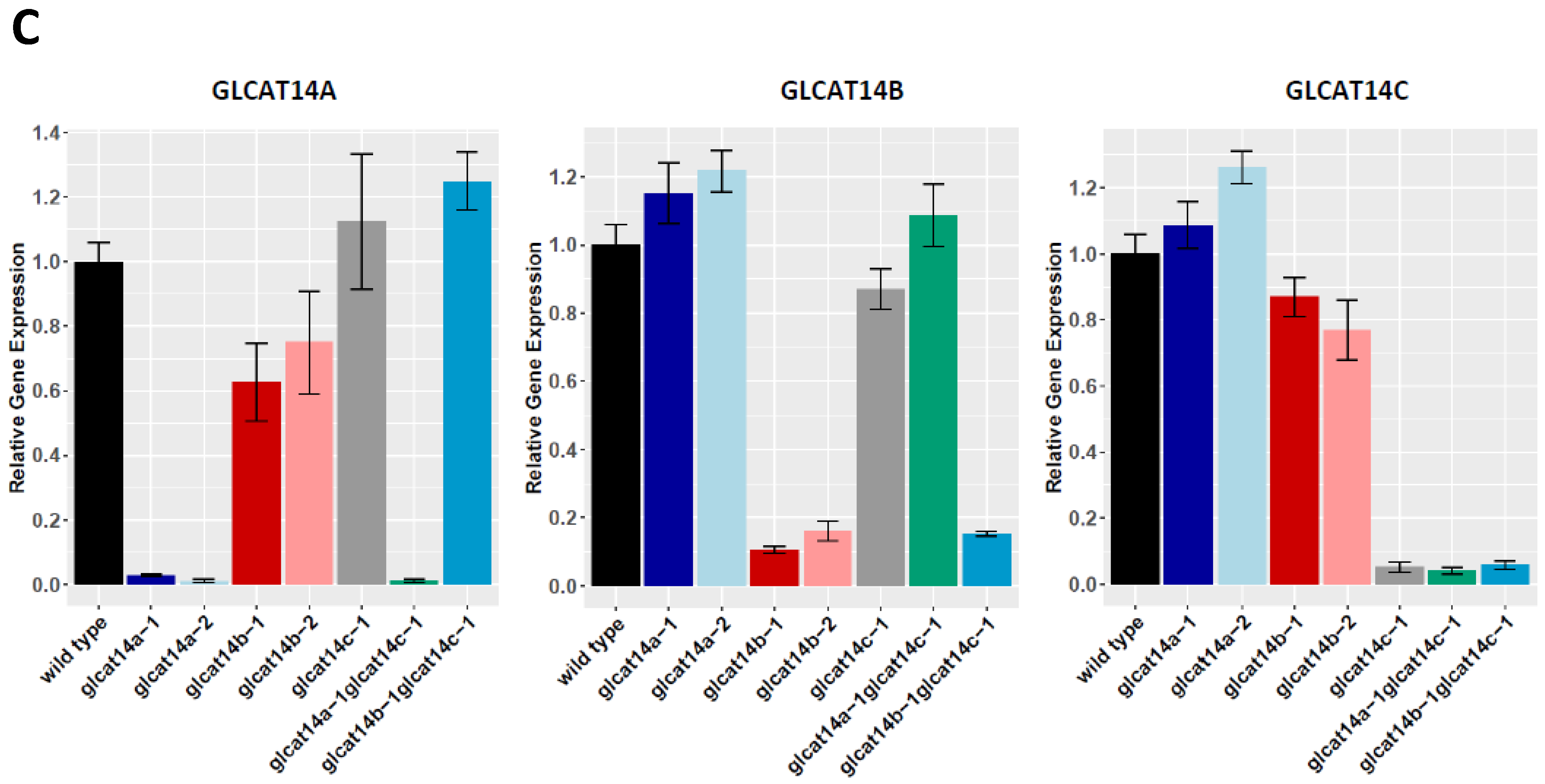
2.2. glcat14 Mutant Generation and Characterization
2.3. Subcellular Localization of GLCAT14A, GLCAT14B and GLCAT14C
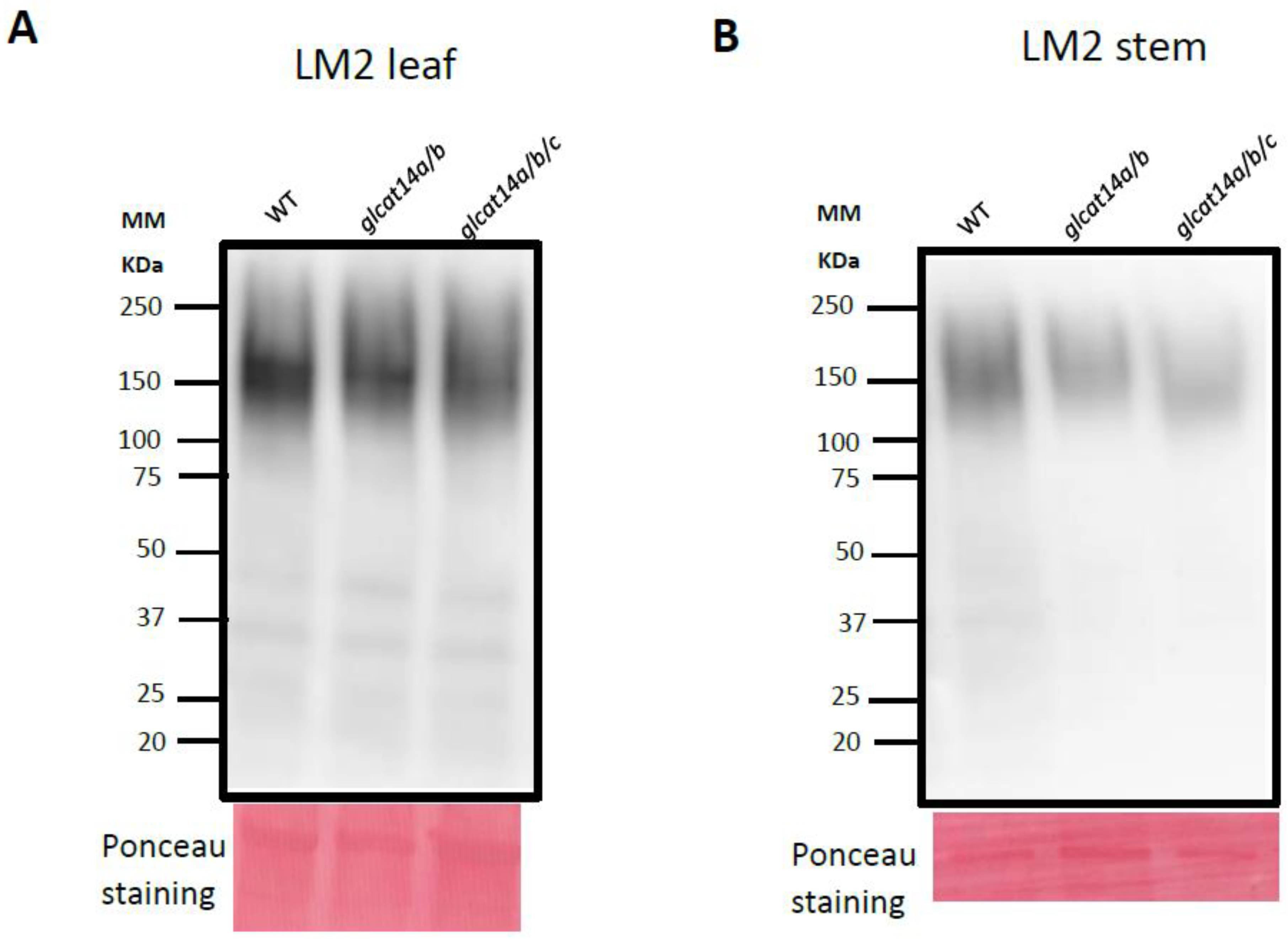
2.4. glcat14a/b and glcat14a/b/c Had Reduced Immunolabelling of LM2 Bound AGPs in Stem and Leaf
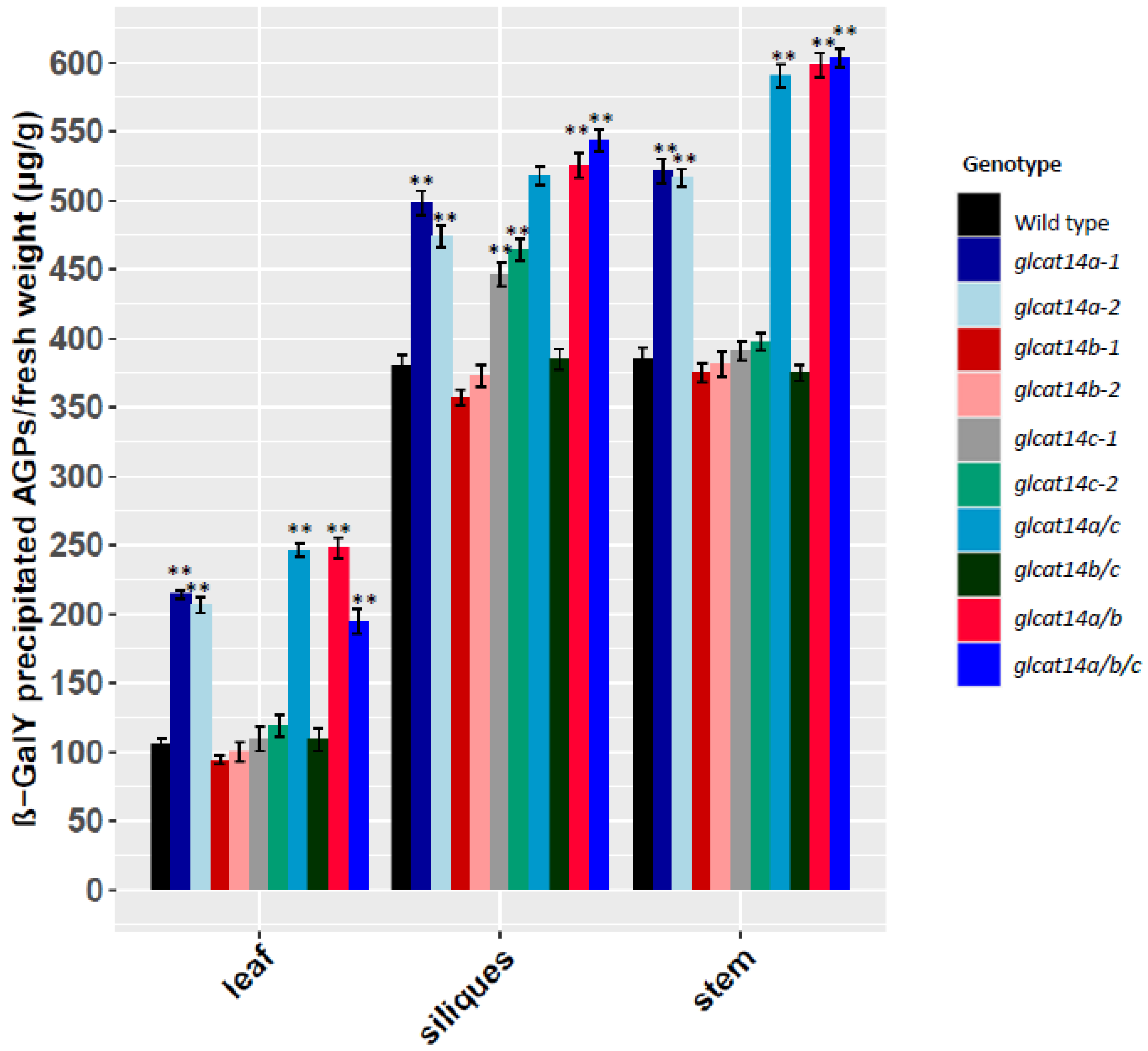
2.5. Quantification of β-D-Gal-Yariv Precipitated AGPs in glcat14 Mutants
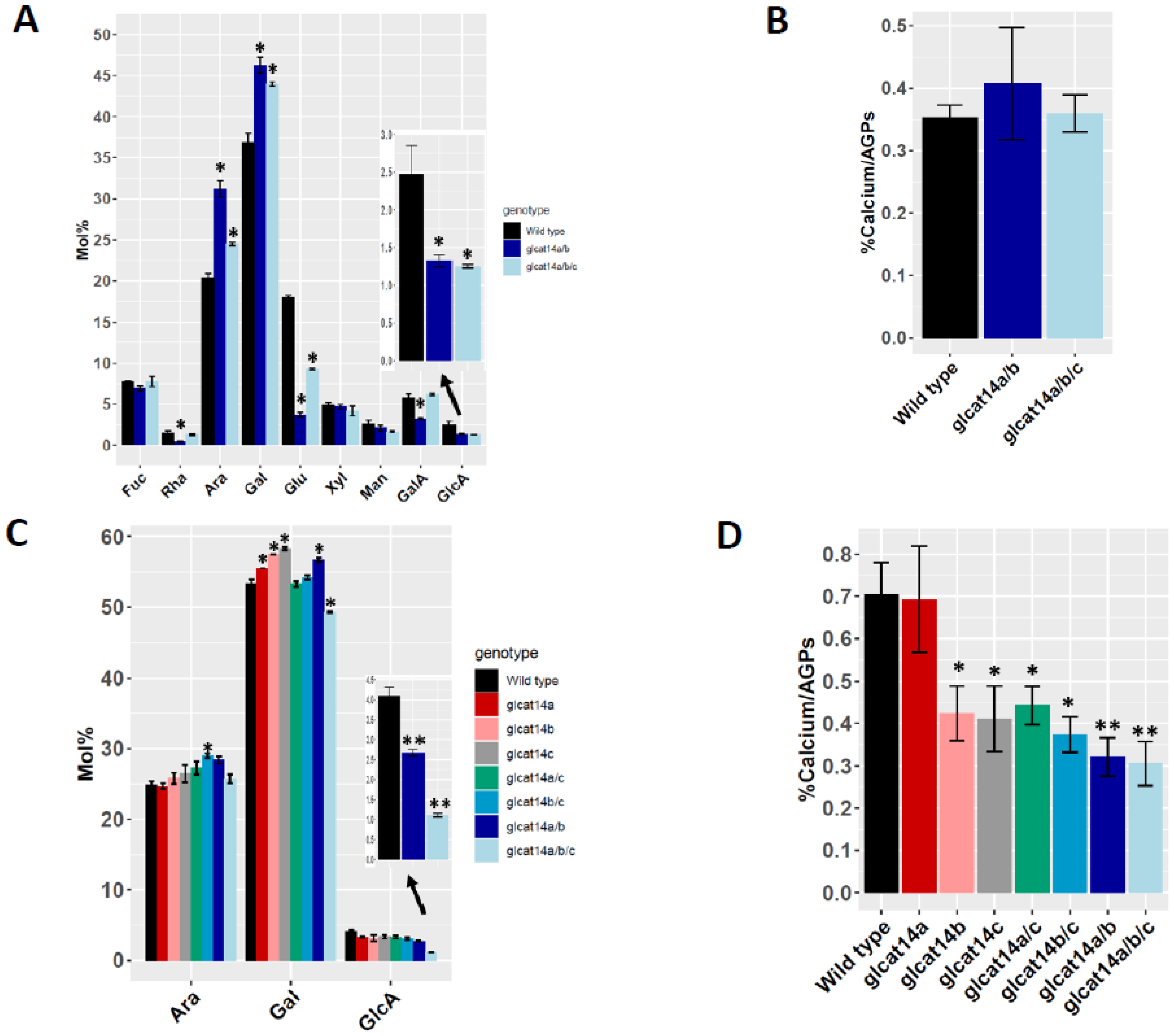
2.6. Monosaccharide Composition Analyses and Calcium Content
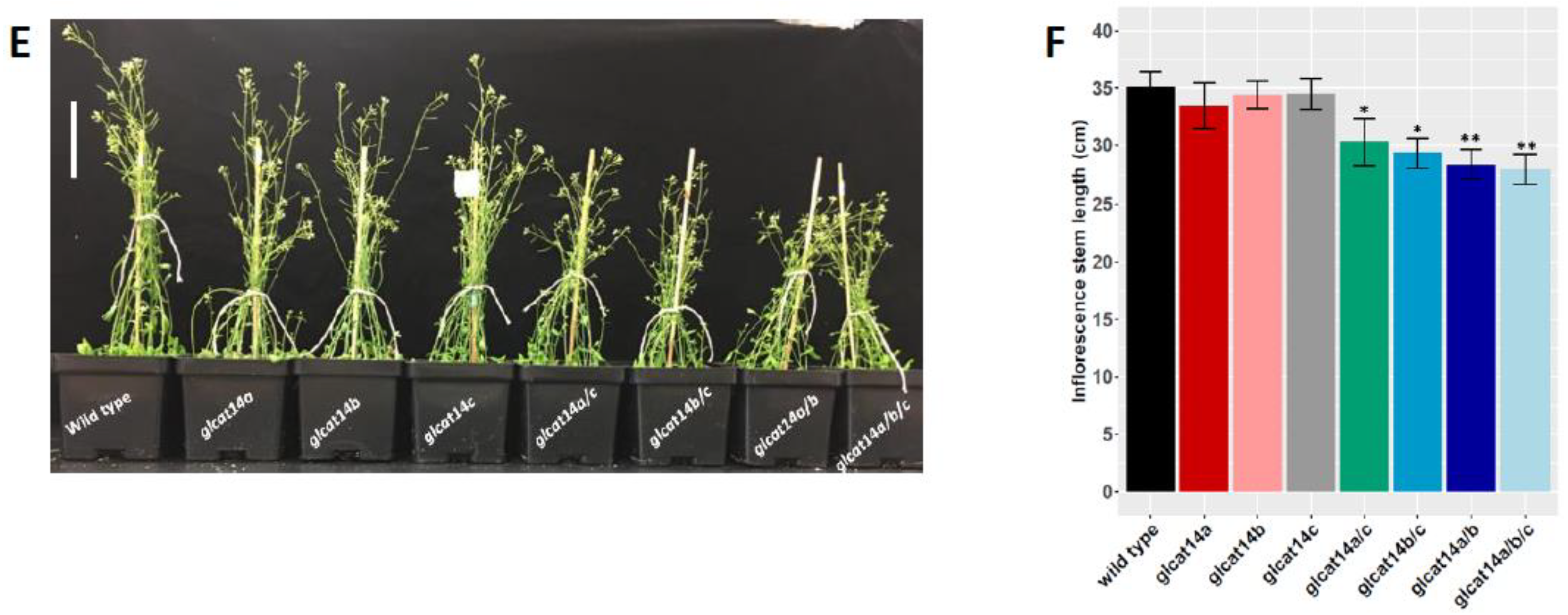
2.7. Both Single and Double glcat14 Mutant Display Pleiotropic Growth Defects
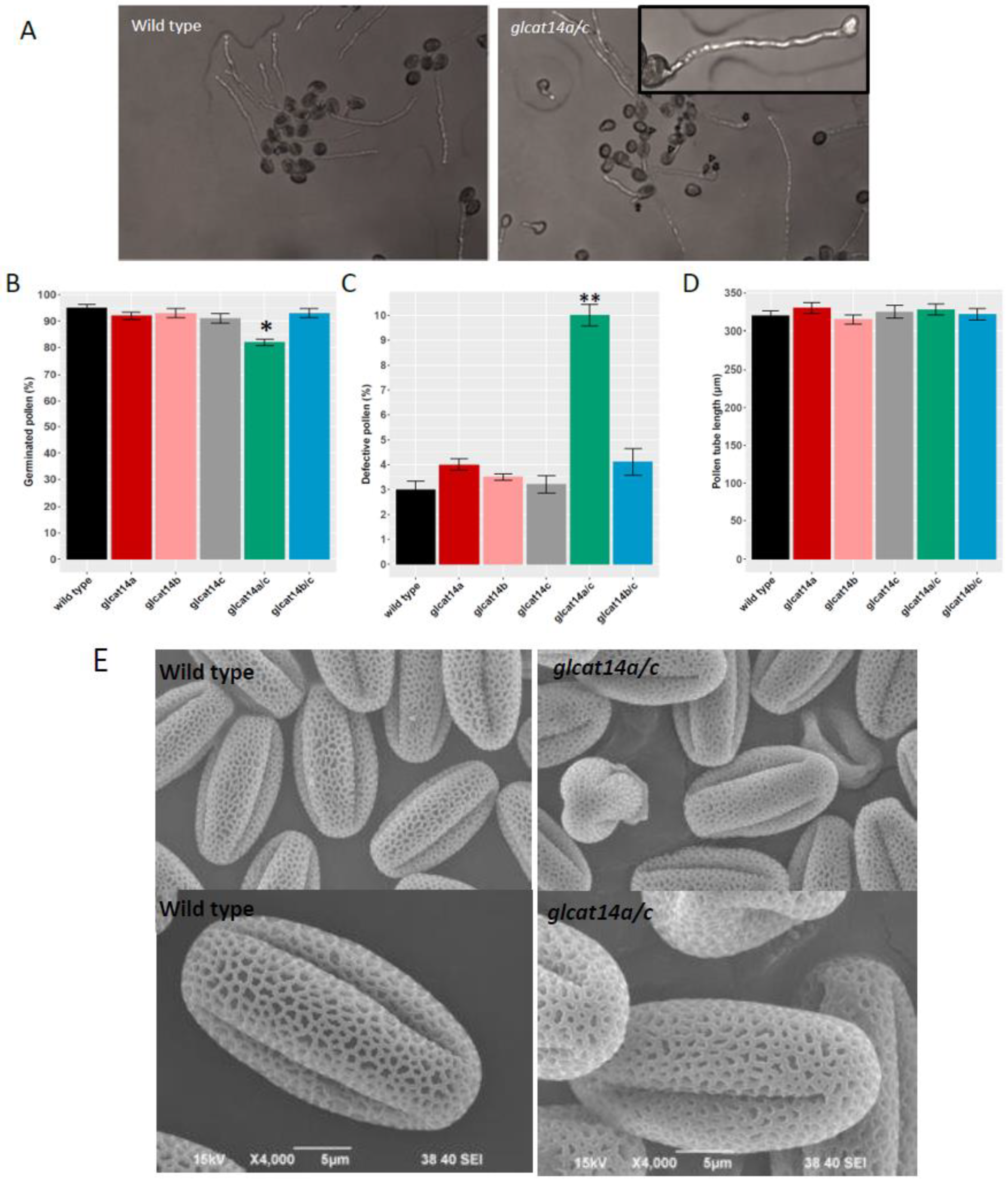
2.8. glcat14a/c Mutants Had Reduced Pollen Germination and Misshaped Pollen
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Quantitative RT-PCR
4.3. Bioinformatics
4.4. AGPs Quantification Using β-D-Gal-Yariv Reagent
4.5. Monosaccharide Composition Analysis by High Performance Anion Exchange Chromatography with Pulsed Amperometric Detection (HPAE-PAD)
4.6. Calcium Binding Assay
4.7. Seed Germination
4.8. Trichome Morphology
4.9. In Vitro Pollen Germination
4.10. Western Blotting Analysis
4.11. Cloning Procedures and Subcellular Localizations of GLCAT14A, GLCAT14B and GLCAT14C
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gaspar, Y.; Johnson, K.L.; McKenna, J.A.; Bacic, A.; Schultz, C.J. The complex structures of arabinogalactan-proteins and the journey towards understanding function. Plant Mol. Biol. 2001, 47, 161–176. [Google Scholar] [CrossRef]
- Showalter, A.M.; Keppler, B.; Lichtenberg, J.; Gu, D.; Welch, L.R. A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 2010, 153, 485–513. [Google Scholar] [CrossRef] [PubMed]
- Kieliszewski, M.J. The latest hype on Hyp-O-glycosylation codes. Phytochemistry 2001, 57, 319–323. [Google Scholar] [CrossRef]
- Ellis, M.; Egelund, J.; Schultz, C.J.; Bacic, A. Arabinogalactan-proteins: Key regulators at the cell surface? Plant Physiol. 2010, 153, 403–419. [Google Scholar] [CrossRef]
- Seifert, G.J.; Roberts, K. The biology of arabinogalactan proteins. Annu. Rev. Plant Biol. 2007, 58, 137–161. [Google Scholar] [CrossRef]
- Showalter, A.M. Arabinogalactan-proteins: Structure, expression and function. Cell Mol. Life Sci. 2001, 58, 1399–1417. [Google Scholar] [CrossRef] [PubMed]
- Knoch, E.; Dilokpimol, A.; Tryfona, T.; Poulsen, C.P.; Xiong, G.; Harholt, J. A β–glucuronosyltransferase from Arabidopsis thaliana involved in biosynthesis of type II arabinogalactan has a role in cell elongation during seedling growth. Plant J. 2013, 76, 1016–1029. [Google Scholar] [CrossRef]
- Tan, L.; Varnai, P.; Lamport, D.T.A.; Yuan, C.H.; Xu, J.F.; Qiu, F.; Kieliszewski, M.J. Plant O–hydroxyproline arabinogalactans are composed of repeating tri galactosyl subunits with short, bifurcated side chains. J. Biol. Chem. 2010, 285, 24575–24583. [Google Scholar] [CrossRef]
- Tryfona, T.; Liang, H.C.; Kotake, T. Carbohydrate structural analysis of wheat flour arabinogalactan protein. Carbohydr. Res. 2010, 345, 2648–2656. [Google Scholar] [CrossRef] [PubMed]
- Tryfona, T.; Liang, H.C.; Kotake, T.; Tsumuraya, Y.; Stephens, E.; Dupree, P. Structural characterization of Arabidopsis leaf arabino-galactan polysaccharides. Plant Physiol. 2012, 160, 653–666. [Google Scholar] [CrossRef]
- Dilokpimol, A.; Geshi, N. Arabidopsis thaliana glucuronosyltransferase in family GT14. Plant Signal. Behav. 2014, 9, 28891. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Hernandez, F.; Tryfona, T.; Rizza, A.; Yu, X.; Harris, M.O.; Webb, A.R.R.; Kotake, T.; Dupree, P. Calcium Binding by Arabinogalactan Polysaccharide Is Important for Normal Plant Development. Plant Cell. 2020, 32, 3346–3369. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Held, M.A.; Showalter, A.M. Elucidating the roles of three β-glucuronosyltransferases (GLCATs) acting on arabinogalactan-proteins using a CRISPR-Cas9 multiplexing approach in Arabidopsis. BMC Plant Biol. 2020, 20, 221. [Google Scholar]
- Bascom, C.S.; Hepler, P.K.; Bezanilla, M. Interplay between ions, the cytoskeleton, and cell wall properties during tip growth. Plant Physiol. 2018, 176, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Thor, K. Calcium—Nutrient and Messenger. Front. Plant Sci. 2019, 10, 440. [Google Scholar] [CrossRef]
- Lamport, D.T.A.; Várnai, P. Periplasmic arabinogalactan glycoproteins act as a calcium capacitor that regulates plant growth and development. New Phytol. 2013, 197, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Ajayi, O.O.; Showalter, A.M. Systems identification and characterization of β-glucuronosyltransferase genes involved in arabinogalactan-protein biosynthesis in plant genomes. Sci. Rep. 2020, 10, 20562. [Google Scholar] [CrossRef]
- Kitazawa, K.; Tryfona, T.; Yoshimi, Y.; Hayashi, Y.; Kawauchi, S.; Antonov, L.; Tanaka, H.; Takahashi, T.; Kaneko, S.; Dupree, P.; et al. β-Galactosyl Yariv reagent binds to the β-1,3-galactan of arabinogalactan proteins. Plant Physiol. 2013, 161, 1117–1126. [Google Scholar] [CrossRef]
- Gigli-Bisceglia, N.; Engelsdorf, T.; Hamann, T. Plant cell wall integrity maintenance in model plants and crop species-relevant cell wall components and underlying guiding principles. Cell. Mol. Life Sci. 2020, 77, 2049–2077. [Google Scholar] [CrossRef]
- Hepler, P.K. Calcium: A central regulator of plant growth and development. Plant Cell. 2005, 17, 2142–2155. [Google Scholar] [CrossRef]
- Vanneste, S.; Friml, J. Calcium: The missing link in auxin action. Plants 2013, 2, 650–675. [Google Scholar] [CrossRef] [PubMed]
- Corns, C.M.; Ludman, C.J. Some observations on the nature of the calcium cresolphthalein complexone reaction and its relevance to the clinical laboratory. Ann. Clin. Biochem. 1987, 24, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Scimeca, M.; Bischetti, S.; Lamsira, H.K.; Bonfiglio, R.; Bonanno, E. Energy Dispersive X-ray (EDX) microanalysis: A powerful tool in biomedical research and diagnosis. Eur. J. Histochem. 2018, 62, 2841. [Google Scholar] [CrossRef]
- Thimmaiah, C.; Shetty, P.; Shetty, S.B.; Natarajan, S.; Thomas, N. A Comparative analysis of the remineralization potential of CPP–ACP with Fluoride, Tri-Calcium Phosphate and Nano Hydroxyapatite using SEM/EDX–An in vitro study. J. Clin. Exp. Dent. 2019, 11, 1120–1126. [Google Scholar] [CrossRef] [PubMed]
- Coimbra, S.; Costa, M.; Jones, B.; Mendes, M.A.; Pereira, L.G. Pollen grain development is compromised in Arabidopsis agp6 agp11 null mutants. J. Exp. Bot. 2009, 60, 3133–3142. [Google Scholar] [CrossRef]
- Mizukami, A.G.; Inatsugi, R.; Jiao, J.; Kotake, T.; Kuwata, K.; Ootani, K.; Okuda, S.; Sankaranarayanan, S.; Sato, Y.; Maruyama, D.; et al. The AMOR arabinogalactan sugar chain induces pollen-tube competency to respond to ovular guidance. Curr. Biol. 2016, 26, 1091–1097. [Google Scholar] [CrossRef]
- Shimoda, R.; Okabe, K.; Kotake, T.; Matsuoka, K.; Koyama, T.; Tryfona, T.; Liang, H.C.; Dupree, P.; Tsumuraya, Y. Enzymatic fragmentation of carbohydrate moieties of radish arabinogalactan-protein and elucidation of the structures. Biosci. Biotechnol. Biochem. 2014, 78, 818–831. [Google Scholar] [CrossRef][Green Version]
- Ye, C.Y.; Li, T.; Tuskan, G.A.; Tschaplinski, T.J.; Yang, X. Comparative analysis of GT14/GT14-like gene family in Arabidopsis, Oryza, Populus, Sorghum and Vitis. Plant Sci. 2011, 181, 688–695. [Google Scholar] [CrossRef]
- Vaahtera, L.; Schulz, J.; Hamann, T. Cell wall integrity maintenance during plant development and interaction with the environment. Nat. Plants. 2019, 5, 924–932. [Google Scholar] [CrossRef]
- Hamann, T.; Denness, L. Cell wall integrity maintenance in plants: Lessons to be learned from yeast? Plant Signal. Behav. 2011, 6, 1706–1709. [Google Scholar] [CrossRef]
- Hamann, T. The plant cell wall integrity maintenance mechanism–A case study of a cell wall plasma membrane signaling network. Phytochemistry 2015, 112, 100–109. [Google Scholar] [CrossRef]
- Basu, D.; Tian, L.; Wang, W.; Bobbs, S.; Herock, H.; Travers, A.; Showalter, A.M. A small multigene hydroxyproline-O-galactosyltransferase family functions in arabinogalactan-protein glycosylation, growth and development in Arabidopsis. BMC Plant Biol. 2015, 15, 295. [Google Scholar] [CrossRef]
- Tan, L.; Eberhard, S.; Pattathil, S.; Warder, C.; Glushka, J.; Yuan, C.; Hao, Z.; Zhu, X.; Avci, U.; Miller, J.S. An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell. 2013, 25, 270–287. [Google Scholar] [CrossRef]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2007, 2, 718. [Google Scholar] [CrossRef]
- Guo, Y.; Xiong, L.; Song, C.; Gong, D.; Halfter, U.; Jian, K.; Zhu, J. A Calcium Sensor and Its Interacting Protein Kinase Are Global Regulators of Abscisic Acid Signaling in Arabidopsis. Dev. Cell 2002, 3, 233–244. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Gampala, S.S.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14, 15–45. [Google Scholar] [CrossRef] [PubMed]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Lamport, D. Preparation of arabinogalactan glycoproteins from plant tissue. BIO-Protoc. 2013, 3, 1–3. [Google Scholar] [CrossRef]
- ØBro, J.; Harholt, J.; Scheller, H.V.; Orfila, C. Rhamnogalacturonan I in Solanum tuberosum tubers contains complex arabinogalactan structures. Phytochemistry 2004, 65, 1429–1438. [Google Scholar] [CrossRef]
- Talbot, M.J.; White, R.G. Methanol fixation of plant tissue for Scanning Electron Microscopy improves preservation of tissue morphology and dimensions. Plant Methods 2013, 9, 36. [Google Scholar] [CrossRef]
- Fragkostefanakis, S.; Dandachi, F.; Kalaitzis, P. Expression of arabinogalactan proteins during tomato fruit ripening and in response to mechanical wounding, hypoxia and anoxia. Plant Physiol. Biochem. 2012, 52, 112–118. [Google Scholar] [CrossRef]
- Bradford, M.M. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Citovsky, V.; Lee, L.Y.; Vyas, S.; Glick, E.; Chen, M.H.; Vainstein, A.; Gafni, Y.; Gelvin, S.B.; Tzfira, T. Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J. Mol. Biol. 2006, 362, 1120–1131. [Google Scholar] [CrossRef]
- Batoko, H.; Zheng, H.Q.; Hawes, C.; Moore, I. A rab1 GTPase is required for transport between the endoplasmic reticulum and golgi apparatus and for normal golgi movement in plants. Plant Cell. 2000, 11, 2201–2218. [Google Scholar] [CrossRef]

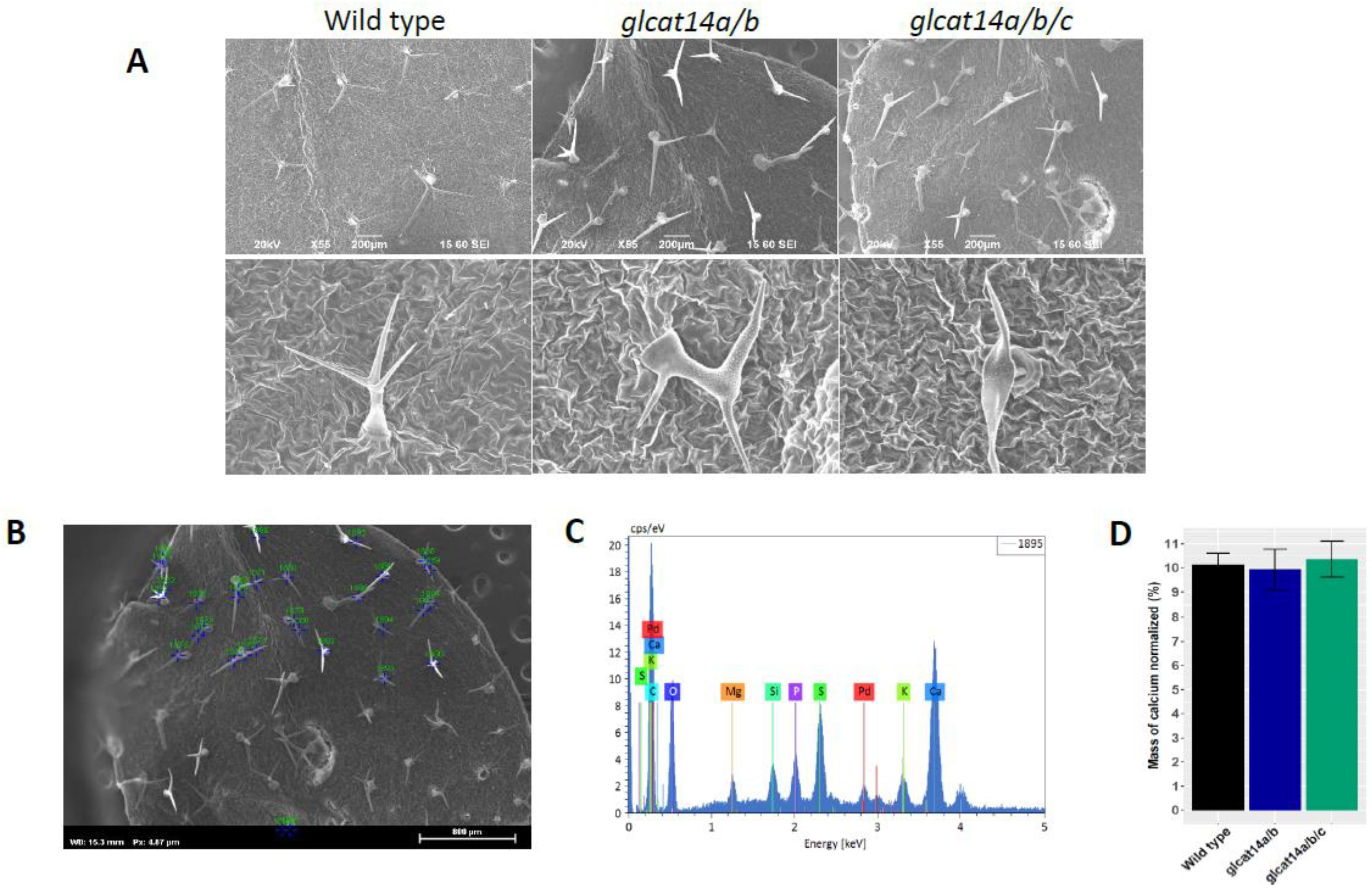
| Number of Trichome Branches | ||||
|---|---|---|---|---|
| Genotype | 1 | 2 | 3 | Total |
| Wild Type | 3.19 | 92.6 | 4.26 | 94 |
| glcat14a/b | 72.06 | 26.47 | 1.47 | 68 |
| glcat14a/b/c | 90.2 | 7.84 | 1.96 | 51 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ajayi, O.O.; Held, M.A.; Showalter, A.M. Three β-Glucuronosyltransferase Genes Involved in Arabinogalactan Biosynthesis Function in Arabidopsis Growth and Development. Plants 2021, 10, 1172. https://doi.org/10.3390/plants10061172
Ajayi OO, Held MA, Showalter AM. Three β-Glucuronosyltransferase Genes Involved in Arabinogalactan Biosynthesis Function in Arabidopsis Growth and Development. Plants. 2021; 10(6):1172. https://doi.org/10.3390/plants10061172
Chicago/Turabian StyleAjayi, Oyeyemi O., Michael A. Held, and Allan M. Showalter. 2021. "Three β-Glucuronosyltransferase Genes Involved in Arabinogalactan Biosynthesis Function in Arabidopsis Growth and Development" Plants 10, no. 6: 1172. https://doi.org/10.3390/plants10061172
APA StyleAjayi, O. O., Held, M. A., & Showalter, A. M. (2021). Three β-Glucuronosyltransferase Genes Involved in Arabinogalactan Biosynthesis Function in Arabidopsis Growth and Development. Plants, 10(6), 1172. https://doi.org/10.3390/plants10061172






