Exploration of Epigenetics for Improvement of Drought and Other Stress Resistance in Crops: A Review
Abstract
:1. Introduction
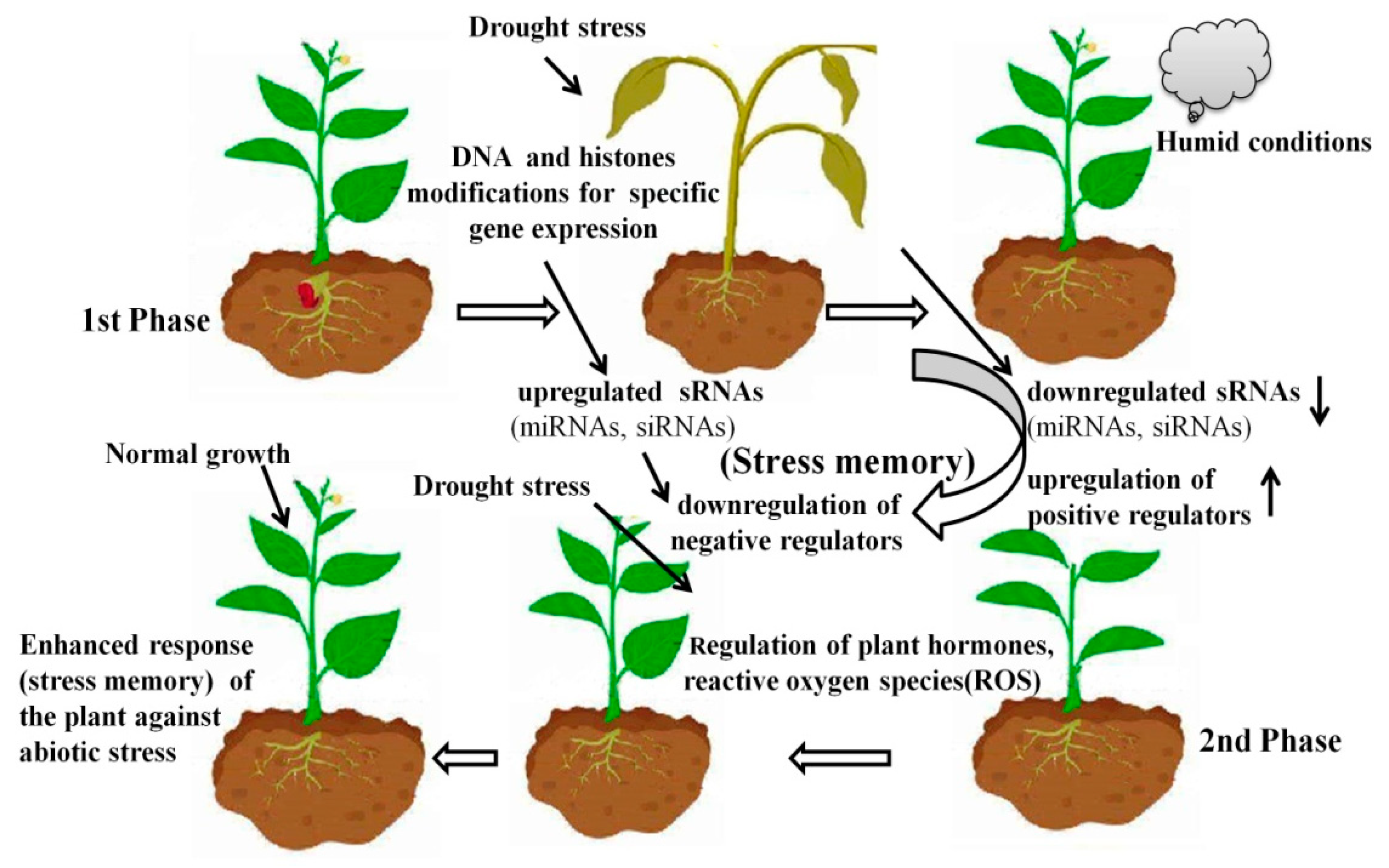
2. Stress Memory Enhances Abiotic Stress Resistance of Plants and Their Offspring
3. Memory Induced by One Type of Stress Can Increase Cross-Resistance and Transgenerational Memory
4. Stress Memory Is Mainly Regulated by Epigenetic Pathways
5. Application of Novel Plant Breeding Tools to Promote Abiotic Stress Resistance in Crops
6. Future Perspective for the Potential Application of Stress Memory for Genetic Improvement of Crops
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Pereira, A. Plant Abiotic Stress Challenges from the Changing Environment. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Porter, S.S.; Bantay, R.; Friel, C.A.; Garoutte, A.; Gdanetz, K.; Ibarreta, K.; Moore, B.M.; Shetty, P.; Siler, E.; Friesen, M.L. Beneficial microbes ameliorate abiotic and biotic sources of stress on plants. Funct. Ecol. 2020, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Lv, Y.; Jahan, N.; Chen, G.; Ren, D.; Guo, L. Sensing of Abiotic Stress and Ionic Stress Responses in Plants. Int. J. Mol. Sci. 2018, 19, 3298. [Google Scholar] [CrossRef] [Green Version]
- Zheng, X.; Chen, L.; Xia, H.; Wei, H.; Lou, Q.; Li, M.; Li, T.; Luo, L. Transgenerational epimutations induced by multi-generation drought imposition mediate rice plant’s adaptation to drought condition. Sci. Rep. 2017, 7, 39843. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.J.; Li, J.R.; Qi, S.L.; Lin, Q.F.; Jin, J.B.; Hua, X.J. Light affects salt stress-induced transcriptional memory of P5CS1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, E8335. [Google Scholar] [CrossRef] [Green Version]
- Latzel, V.; Rendina González, A.P.; Rosenthal, J. Epigenetic Memory as a Basis for Intelligent Behavior in Clonal Plants. Front. Plant Sci. 2016, 7, 1354. [Google Scholar] [CrossRef] [Green Version]
- Crisp, P.A.; Ganguly, D.; Eichten, S.R.; Borevitz, J.O.; Pogson, B.J. Reconsidering plant memory: Intersections between stress recovery, RNA turnover, and epigenetics. Sci. Adv. 2016, 2, e1501340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Y.; Fromm, M.; Avramova, Z. Multiple exposures to drought ’train’ transcriptional responses in Arabidopsis. Nat. Commun. 2012, 3, 740. [Google Scholar] [CrossRef]
- Molinier, J.; Ries, G.; Zipfel, C.; Hohn, B. Transgeneration memory of stress in plants. Nature 2006, 442, 1046–1049. [Google Scholar] [CrossRef] [PubMed]
- Thiebaut, F.; Hemerly, A.S.; Ferreira, P.C.G. A Role for Epigenetic Regulation in the Adaptation and Stress Responses of Non-model Plants. Front. Plant Sci. 2019, 10, 246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wojtyla, Ł.; Paluch-Lubawa, E.; Sobieszczuk-Nowicka, E.; Garnczarska, M. Drought Stress Memory and Subsequent Drought Stress Tolerance in Plants. In Priming-Mediated Stress and Cross-Stress Tolerance in Crop Plants; Elsevier: Amsterdam, The Netherlands, 2020; pp. 115–131. [Google Scholar] [CrossRef]
- Liew, Y.J.; Howells, E.J.; Wang, X.; Michell, C.T.; Burt, J.A.; Idaghdour, Y.; Aranda, M. Intergenerational epigenetic inheritance in reef-building corals. Nat. Clim. Chang. 2020, 10, 254–259. [Google Scholar] [CrossRef] [Green Version]
- Latutrie, M.; Gourcilleau, D.; Pujol, B. Epigenetic variation for agronomic improvement: An opportunity for vegetatively propagated crops. Am. J. Bot. 2019, 106, 1281–1284. [Google Scholar] [CrossRef]
- Kapazoglou, A.; Ganopoulos, I.; Tani, E.; Tsaftaris, A. Chapter Nine—Epigenetics, Epigenomics and Crop Improvement. In Advances in Botanical Research; Kuntz, M., Ed.; Academic Press: New York, NY, USA, 2018; Volume 86, pp. 287–324. [Google Scholar] [CrossRef]
- Kinoshita, T.; Seki, M. Epigenetic memory for stress response and adaptation in plants. Plant Cell Physiol. 2014, 55, 1859–1863. [Google Scholar] [CrossRef]
- Santos, A.P.; Serra, T.; Figueiredo, D.D.; Barros, P.; Lourenço, T.; Chander, S.; Oliveira, M.M.; Saibo, N.J.M. Transcription Regulation of Abiotic Stress Responses in Rice: A Combined Action of Transcription Factors and Epigenetic Mechanisms. OMICS J. Integr. Biol. 2011, 15, 839–857. [Google Scholar] [CrossRef] [Green Version]
- Bruce, T.J.A.; Matthes, M.C.; Napier, J.A.; Pickett, J.A. Stressful “memories” of plants: Evidence and possible mechanisms. Plant Sci. 2007, 173, 603–608. [Google Scholar] [CrossRef]
- Munne-Bosch, S.; Alegre, L. Cross-stress tolerance and stress “memory” in plants. Environ. Exp. Bot. 2013, 94, 1–88. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.-K. Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 2009, 12, 133–139. [Google Scholar] [CrossRef] [Green Version]
- Tombesi, S.; Frioni, T.; Poni, S.; Palliotti, A. Effect of water stress “memory” on plant behavior during subsequent drought stress. Environ. Exp. Bot. 2018, 150, 106–114. [Google Scholar] [CrossRef]
- Marcos, F.C.C.; Silveira, N.M.; Marchiori, P.E.R.; Machado, E.C.; Souza, G.M.; Landell, M.G.A.; Ribeiro, R.V. Drought tolerance of sugarcane propagules is improved when origin material faces water deficit. PLoS ONE 2018, 13, e0206716. [Google Scholar] [CrossRef] [Green Version]
- Walter, J.; Nagy, L.; Hein, R.; Rascher, U.; Beierkuhnlein, C.; Willner, E.; Jentsch, A. Do plants remember drought? Hints towards a drought-memory in grasses. Environ. Exp. Bot. 2011, 71, 34–40. [Google Scholar] [CrossRef]
- Janmohammadi, M.; Zolla, L.; Rinalducci, S. Low temperature tolerance in plants: Changes at the protein level. Phytochemistry 2015, 117, 76–89. [Google Scholar] [CrossRef]
- Ali, A.; Khan, M.; Sharif, R.; Mujtaba, M.; Gao, S.-J. Sugarcane Omics: An Update on the Current Status of Research and Crop Improvement. Plants 2019, 8, 344. [Google Scholar] [CrossRef] [Green Version]
- Marcos, F.C.C.; Silveira, N.M.; Mokochinski, J.B.; Sawaya, A.C.H.F.; Marchiori, P.E.R.; Machado, E.C.; Souza, G.M.; Landell, M.G.A.; Ribeiro, R.V. Drought tolerance of sugarcane is improved by previous exposure to water deficit. J. Plant Physiol. 2018, 223, 9–18. [Google Scholar] [CrossRef]
- Verhoeven, K.J.F.; Jansen, J.J.; van Dijk, P.J.; Biere, A. Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol. 2010, 185, 1108–1118. [Google Scholar] [CrossRef]
- Bilichak, A.; Kovalchuk, I. Transgenerational response to stress in plants and its application for breeding. J. Exp. Bot. 2016, 67, 2081–2092. [Google Scholar] [CrossRef] [PubMed]
- Vialou, V.; Feng, J.; Robison, A.J.; Nestler, E.J. Epigenetic mechanisms of depression and antidepressant action. Annu. Rev. Pharm. Toxicol. 2013, 53, 59–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, H.; Zhang, X.; Ma, W.; Song, J.; Rahman, S.U.; Wang, J.; Zhang, Y. Morphological and physiological responses to cyclic drought in two contrasting genotypes of Catalpa bungei. Environ. Exp. Bot. 2017, 138, 77–87. [Google Scholar] [CrossRef]
- Hu, T.; Liu, S.Q.; Amombo, E.; Fu, J.M. Stress memory induced rearrangements of HSP transcription, photosystem II photochemistry and metabolism of tall fescue (Festuca arundinacea Schreb.) in response to high-temperature stress. Front. Plant Sci. 2015, 6, 403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuartero, J.; Bolarin, M.C.; Asins, M.J.; Moreno, V. Increasing salt tolerance in the tomato. J. Exp. Bot. 2006, 57, 1045–1058. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Jin, Y.; Li, H.; Amombo, E.; Fu, J. Stress memory induced transcriptional and metabolic changes of perennial ryegrass (Lolium perenne) in response to salt stress. Physiol. Plant. 2016, 156, 54–69. [Google Scholar] [CrossRef]
- Wang, X.; Cai, J.; Liu, F.; Dai, T.; Cao, W.; Wollenweber, B.; Jiang, D. Multiple heat priming enhances thermo-tolerance to a later high temperature stress via improving subcellular antioxidant activities in wheat seedlings. Plant Physiol. Biochem. 2014, 74, 185–192. [Google Scholar] [CrossRef]
- Yactayo, W.; Ramírez, D.A.; Gutiérrez, R.; Mares, V.; Posadas, A.; Quiroz, R. Effect of partial root-zone drying irrigation timing on potato tuber yield and water use efficiency. Agric. Water Manag. 2013, 123, 65–70. [Google Scholar] [CrossRef]
- Watkinson, J.I.; Hendricks, L.; Sioson, A.A.; Vasquez-Robinet, C.; Stromberg, V.; Heath, L.S.; Schuler, M.; Bohnert, H.J.; Bonierbale, M.; Grene, R. Accessions of Solanum tuberosum ssp. andigena show differences in photosynthetic recovery after drought stress as reflected in gene expression profiles. Plant Sci. 2006, 171, 745–758. [Google Scholar] [CrossRef]
- Ramírez, D.A.; Rolando, J.L.; Yactayo, W.; Monneveux, P.; Mares, V.; Quiroz, R. Improving potato drought tolerance through the induction of long-term water stress memory. Plant Sci. 2015, 238, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Elkoca, E.; Haliloglu, K.; Esitken, A.; Ercisli, S. Hydro- and osmopriming improve chickpea germination. Acta Agric. Scand. Sect. B Soil Plant Sci. 2007, 57, 193–200. [Google Scholar] [CrossRef]
- Kaur, S.; Gupta, A.K.; Kaur, N. Effect of osmo- and hydropriming of chickpea seeds on seedling growth and carbohydrate metabolism under water deficit stress. Plant Growth Regul. 2002, 37, 17–22. [Google Scholar] [CrossRef]
- Farhoudi, R.; Sharifzadeh, F.; Poustini, K.; Makkizadeh, M.T.; Kochak Por, M. The effects of NaCl priming on salt tolerance in canola (Brassica napus) seedlings grown under saline conditions. Seed Sci. Technol. 2007, 35, 754–759. [Google Scholar] [CrossRef] [Green Version]
- Verkest, A.; Byzova, M.; Martens, C.; Willems, P.; Verwulgen, T.; Slabbinck, B.; Rombaut, D.; Van de Velde, J.; Vandepoele, K.; Standaert, E.; et al. Selection for Improved Energy Use Efficiency and Drought Tolerance in Canola Results in Distinct Transcriptome and Epigenome Changes. Plant Physiol. 2015, 168, 1338–1350. [Google Scholar] [CrossRef]
- Liu, Y.; Han, J.; Chen, Z.; Wu, H.; Dong, H.; Nie, G. Engineering cell signaling using tunable CRISPR–Cpf1-based transcription factors. Nat. Commun. 2017, 8, 2095. [Google Scholar] [CrossRef] [Green Version]
- Pandita, V.K.; Anand, A.; Nagarajan, S.; Seth, R.; Sinha, S.N. Solid matrix priming improves seed emergence and crop performance in okra. Seed Sci. Technol. 2010, 38, 665–674. [Google Scholar] [CrossRef]
- Patade, V.Y.; Bhargava, S.; Suprasanna, P. Halopriming mediated salt and iso-osmotic PEG stress tolerance and, gene expression profiling in sugarcane (Saccharum officinarum L.). Mol. Biol. Rep. 2012, 39, 9563–9572. [Google Scholar] [CrossRef]
- Fleta-Soriano, E.; Munné-Bosch, S. Stress Memory and the Inevitable Effects of Drought: A Physiological Perspective. Front. Plant Sci. 2016, 7, 143. [Google Scholar] [CrossRef] [Green Version]
- Khalil, F.; Naiyan, X.; Tayyab, M.; Pinghua, C. Screening of EMS-Induced Drought-Tolerant Sugarcane Mutants Employing Physiological, Molecular and Enzymatic Approaches. Agronomy 2018, 8, 226. [Google Scholar] [CrossRef] [Green Version]
- Jisha, K.C.; Puthur, J.T. Halopriming of seeds imparts tolerance to NaCl and PEG induced stress in Vigna radiata (L.) Wilczek varieties. Physiol. Mol. Biol. Plants 2014, 20, 303–312. [Google Scholar] [CrossRef] [Green Version]
- Tajdoost, S.; Farboodnia, T.; Heidari, R. Salt pretreatment enhance salt tolerance in Zea mays L. seedlings. Pak. J. Biol. Sci. 2007, 10, 2086–2090. [Google Scholar] [CrossRef] [Green Version]
- Tan, M.-P. Analysis of DNA methylation of maize in response to osmotic and salt stress based on methylation-sensitive amplified polymorphism. Plant Physiol. Biochem. 2010, 48, 21–26. [Google Scholar] [CrossRef]
- Chen, K.; Fessehaie, A.; Arora, R. Selection of Reference Genes for Normalizing Gene Expression during Seed Priming and Germination Using qPCR in Zea mays and Spinacia oleracea. Plant Mol. Biol. Rep. 2012, 30, 478–487. [Google Scholar] [CrossRef]
- Shan, X.; Wang, X.; Yang, G.; Wu, Y.; Su, S.; Li, S.; Liu, H.; Yuan, Y. Analysis of the DNA methylation of maize (Zea mays L.) in response to cold stress based on methylation-sensitive amplified polymorphisms. J. Plant Biol. 2013, 56, 32–38. [Google Scholar] [CrossRef]
- Rehman, H.u.; Iqbal, H.; Basra, S.M.A.; Afzal, I.; Farooq, M.; Wakeel, A.; Wang, N. Seed priming improves early seedling vigor, growth and productivity of spring maize. J. Integr. Agric. 2015, 14, 1745–1754. [Google Scholar] [CrossRef]
- Yan, K.; Xu, H.; Cao, W.; Chen, X. Salt priming improved salt tolerance in sweet sorghum by enhancing osmotic resistance and reducing root Na+ uptake. Acta Physiol. Plant. 2015, 37, 203. [Google Scholar] [CrossRef] [Green Version]
- Cheng, C.; Yun, K.-Y.; Ressom, H.W.; Mohanty, B.; Bajic, V.B.; Jia, Y.; Yun, S.J.; de los Reyes, B.G. An early response regulatory cluster induced by low temperature and hydrogen peroxide in seedlings of chilling-tolerant japonica rice. BMC Genom. 2007, 8, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gayacharan; Joel, A.J. Epigenetic responses to drought stress in rice (Oryza sativa L.). Physiol. Mol. Biol. Plants 2013, 19, 379–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goswami, A.; Banerjee, R.; Raha, S. Drought resistance in rice seedlings conferred by seed priming. Protoplasma 2013, 250, 1115–1129. [Google Scholar] [CrossRef]
- Mostofa, M.G.; Yoshida, N.; Fujita, M. Spermidine pretreatment enhances heat tolerance in rice seedlings through modulating antioxidative and glyoxalase systems. Plant Growth Regul. 2014, 73, 31–44. [Google Scholar] [CrossRef]
- Wang, W.; Huang, F.; Qin, Q.; Zhao, X.; Li, Z.; Fu, B. Comparative analysis of DNA methylation changes in two rice genotypes under salt stress and subsequent recovery. Biochem. Biophys. Res. Commun. 2015, 465, 790–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Xu, F.; Wu, Y.; Hu, H.-h.; Dai, X.-f. Progress of potato staple food research and industry development in China. J. Integr. Agric. 2017, 16, 2924–2932. [Google Scholar] [CrossRef]
- Garg, R.; Narayana Chevala, V.; Shankar, R.; Jain, M. Divergent DNA methylation patterns associated with gene expression in rice cultivars with contrasting drought and salinity stress response. Sci. Rep. 2015, 5, 14922. [Google Scholar] [CrossRef] [Green Version]
- Mouradi, M.; Bouizgaren, A.; Farissi, M.; Latrach, L.; Qaddoury, A.; Ghoulam, C. Seed osmopriming improves plant growth, nodulation, chlorophyll fluorescence and nutrient uptake in alfalfa (Medicago sativa L.)—Rhizobia symbiosis under drought stress. Sci. Hortic. 2016, 213, 232–242. [Google Scholar] [CrossRef]
- Du, Y.L.; Wang, Z.Y.; Fan, J.W.; Turner, N.C.; He, J.; Wang, T.; Li, F.M. Exogenous abscisic acid reduces water loss and improves antioxidant defence, desiccation tolerance and transpiration efficiency in two spring wheat cultivars subjected to a soil water deficit. Funct. Plant Biol. 2013, 40, 494–506. [Google Scholar] [CrossRef]
- Li, X.; Cai, J.; Liu, F.; Dai, T.; Cao, W.; Jiang, D. Exogenous Abscisic Acid Application during Grain Filling in Winter Wheat Improves Cold Tolerance of Offspring’s Seedlings. J. Agron. Crop Sci. 2014, 200, 467–478. [Google Scholar] [CrossRef]
- Lemmens, E.; Deleu, L.J.; De Brier, N.; De Man, W.L.; De Proft, M.; Prinsen, E.; Delcour, J.A. The Impact of Hydro-Priming and Osmo-Priming on Seedling Characteristics, Plant Hormone Concentrations, Activity of Selected Hydrolytic Enzymes, and Cell Wall and Phytate Hydrolysis in Sprouted Wheat (Triticum aestivum L.). ACS Omega 2019, 4, 22089–22100. [Google Scholar] [CrossRef] [Green Version]
- Abid, M.; Tian, Z.; Zahoor, R.; Ata-Ul-Karim, S.T.; Daryl, C.; Snider, J.L.; Dai, T. Chapter Two—Pre-Drought Priming: A Key Drought Tolerance Engine in Support of Grain Development in Wheat. In Advances in Agronomy; Sparks, D.L., Ed.; Academic Press: New York, NY, USA, 2018; Volume 152, pp. 51–85. [Google Scholar] [CrossRef]
- Fan, H.H.; Wei, J.; Li, T.C.; Li, Z.P.; Guo, N.; Cai, Y.P.; Lin, Y. DNA methylation alterations of upland cotton (Gossypium hirsutum) in response to cold stress. Acta Physiol. Plant. 2013, 35, 2445–2453. [Google Scholar] [CrossRef]
- Wang, M.; Wang, Q.; Zhang, B. Response of miRNAs and their targets to salt and drought stresses in cotton (Gossypium hirsutum L.). Gene 2013, 530, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, M.; Fu, R.; Qian, X.; Rong, P.; Zhang, Y.; Jiang, P.; Wang, J.; Lu, X.; Wang, D.; et al. Epigenetic mechanisms of salt tolerance and heterosis in Upland cotton (Gossypium hirsutum L.) revealed by methylation-sensitive amplified polymorphism analysis. Euphytica 2016, 208, 477–491. [Google Scholar] [CrossRef]
- Gao, S.; Yang, L.; Zeng, H.Q.; Zhou, Z.S.; Yang, Z.M.; Li, H.; Sun, D.; Xie, F.; Zhang, B. A cotton miRNA is involved in regulation of plant response to salt stress. Sci. Rep. 2016, 6, 19736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, X.; Chen, X.; Mu, M.; Wang, J.; Wang, X.; Wang, D.; Yin, Z.; Fan, W.; Wang, S.; Guo, L.; et al. Genome-Wide Analysis of Long Noncoding RNAs and Their Responses to Drought Stress in Cotton (Gossypium hirsutum L.). PLoS ONE 2016, 11, e0156723. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Wang, X.; Chen, X.; Shu, N.; Wang, J.; Wang, D.; Wang, S.; Fan, W.; Guo, L.; Guo, X.; et al. Single-base resolution methylomes of upland cotton (Gossypium hirsutum L.) reveal epigenome modifications in response to drought stress. BMC Genom. 2017, 18, 297. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Xu, H.; Mischke, S.; Meinhardt, L.W.; Zhang, D.; Zhu, X.; Li, X.; Fang, W. Exogenous abscisic acid significantly affects proteome in tea plant (Camellia sinensis) exposed to drought stress. Hortic. Res. 2014, 1, 14029. [Google Scholar] [CrossRef] [Green Version]
- Eskandari, H.; Kazemi, K. Effect of seed priming on germination properties and seedling establishment of cowpea (Vigna sinensis). Not. Sci. Biol. 2011, 3, 113–116. [Google Scholar] [CrossRef] [Green Version]
- Boucelha, L.; Djebbar, R. Influence of different pre-germination treatments of Vigna unguiculata (L.) Walp. seeds on germination performance and water stress tolerance. Biotechnol. Agron. Société Environ. 2015, 19, 160–172. [Google Scholar]
- Xin, C.; Hou, R.; Wu, F.; Zhao, Y.; Xiao, H.; Si, W.; Ali, M.E.; Cai, L.; Guo, J. Analysis of cytosine methylation status in potato by methylation-sensitive amplified polymorphisms under low-temperature stress. J. Plant Biol. 2015, 58, 383–390. [Google Scholar] [CrossRef]
- Banik, P.; Zeng, W.; Tai, H.; Bizimungu, B.; Tanino, K. Effects of drought acclimation on drought stress resistance in potato (Solanum tuberosum L.) genotypes. Environ. Exp. Bot. 2016, 126, 76–89. [Google Scholar] [CrossRef]
- Boguszewska-Mańkowska, D.; Pieczyński, M.; Wyrzykowska, A.; Kalaji, H.M.; Sieczko, L.; Szweykowska-Kulińska, Z.; Zagdańska, B. Divergent strategies displayed by potato (Solanum tuberosum L.) cultivars to cope with soil drought. J. Agron. Crop Sci. 2018, 204, 13–30. [Google Scholar] [CrossRef] [Green Version]
- Guedes, F.A.d.F.; Nobres, P.; Rodrigues Ferreira, D.C.; Menezes-Silva, P.E.; Ribeiro-Alves, M.; Correa, R.L.; DaMatta, F.M.; Alves-Ferreira, M. Transcriptional memory contributes to drought tolerance in coffee (Coffea canephora) plants. Environ. Exp. Bot. 2018, 147, 220–233. [Google Scholar] [CrossRef]
- Slaughter, A.; Daniel, X.; Flors, V.; Luna, E.; Hohn, B.; Mauch-Mani, B. Descendants of Primed Arabidopsis Plants Exhibit Resistance to Biotic Stress. Plant Physiol. 2012, 158, 835. [Google Scholar] [CrossRef] [Green Version]
- Sani, E.; Herzyk, P.; Perrella, G.; Colot, V.; Amtmann, A. Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol. 2013, 14, R59. [Google Scholar] [CrossRef] [Green Version]
- Amooaghaie, R.; Nikzad, K. The role of nitric oxide in priming-induced low-temperature tolerance in two genotypes of tomato. Seed Sci. Res. 2013, 23, 123–131. [Google Scholar] [CrossRef] [Green Version]
- Omidvar, V.; Fellner, M. DNA Methylation and Transcriptomic Changes in Response to Different Lights and Stresses in 7B-1 Male-Sterile Tomato. PLoS ONE 2015, 10, e0121864. [Google Scholar] [CrossRef] [Green Version]
- Umezawa, T.; Shimizu, K.; Kato, M.; Ueda, T. Enhancement of salt tolerance in soybean with NaCl pretreatment. Physilogia Plant. 2000, 110, 59–63. [Google Scholar] [CrossRef]
- Lecube, M.L.; Noriega, G.O.; Santa Cruz, D.M.; Tomaro, M.L.; Batlle, A.; Balestrasse, K.B. Indole acetic acid is responsible for protection against oxidative stress caused by drought in soybean plants: The role of heme oxygenase induction. Redox Rep. 2014, 19, 242–250. [Google Scholar] [CrossRef]
- Chen, R.; Li, M.; Zhang, H.; Duan, L.; Sun, X.; Jiang, Q.; Zhang, H.; Hu, Z. Continuous salt stress-induced long non-coding RNAs and DNA methylation patterns in soybean roots. BMC Genom. 2019, 20, 730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.-W.; Zeng, X.-Y.; Leng, Y.; Feng, L.; Kang, X.-H. Indole-3-butyric acid mediates antioxidative defense systems to promote adventitious rooting in mung bean seedlings under cadmium and drought stresses. Ecotoxicol. Environ. Saf. 2018, 161, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Christou, A.; Manganaris, G.A.; Papadopoulos, I.; Fotopoulos, V. Hydrogen sulfide induces systemic tolerance to salinity and non-ionic osmotic stress in strawberry plants through modification of reactive species biosynthesis and transcriptional regulation of multiple defence pathways. J. Exp. Bot. 2013, 64, 1953–1966. [Google Scholar] [CrossRef] [PubMed]
- Christou, A.; Filippou, P.; Manganaris, G.A.; Fotopoulos, V. Sodium hydrosulfide induces systemic thermotolerance to strawberry plants through transcriptional regulation of heat shock proteins and aquaporin. BMC Plant Biol. 2014, 14, 42. [Google Scholar] [CrossRef] [Green Version]
- Christou, A.; Manganaris, G.A.; Fotopoulos, V. Systemic mitigation of salt stress by hydrogen peroxide and sodium nitroprusside in strawberry plants via transcriptional regulation of enzymatic and non-enzymatic antioxidants. Environ. Exp. Bot. 2014, 107, 46–54. [Google Scholar] [CrossRef]
- Banerjee, S.; Sirohi, A.; Ansari, A.A.; Gill, S.S. Role of small RNAs in abiotic stress responses in plants. Plant Gene 2017, 11, 180–189. [Google Scholar] [CrossRef]
- Zhang, M.; An, P.; Li, H.; Wang, X.; Zhou, J.; Dong, P.; Zhao, Y.; Wang, Q.; Li, C. The miRNA-Mediated Post-Transcriptional Regulation of Maize in Response to High Temperature. Int. J. Mol. Sci. 2019, 20, 1754. [Google Scholar] [CrossRef] [Green Version]
- Frost, C.J.; Mescher, M.C.; Carlson, J.E.; De Moraes, C.M. Plant defense priming against herbivores: Getting ready for a different battle. Plant Physiol. 2008, 146, 818–824. [Google Scholar] [CrossRef] [Green Version]
- Shi, W.; Lawas, L.; Raju, B.; Jagadish, S. Acquired thermo-tolerance and trans-generational heat stress response at flowering in rice. J. Agron. Crop Sci. 2016, 202, 309–319. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.; Chen, J.; Wang, X.; Cai, J.; Zhou, Q.; Dai, T.; Cao, W.; Jiang, D. Parental Drought-Priming Enhances Tolerance to Post-anthesis Drought in Offspring of Wheat. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Achievements and Challenges in Understanding Plant Abiotic Stress Responses and Tolerance. Plant Cell Physiol. 2011, 52, 1569–1582. [Google Scholar] [CrossRef] [Green Version]
- Whittle, C.; Otto, S.; Johnston, M.O.; Krochko, J. Adaptive epigenetic memory of ancestral temperature regime in Arabidopsis thaliana. Botany 2009, 87, 650–657. [Google Scholar] [CrossRef] [Green Version]
- Ding, Y.; Avramova, Z.; Fromm, M. The Arabidopsis trithorax-like factor ATX1 functions in dehydration stress responses via ABA-dependent and ABA-independent pathways. Plant J. Cell Mol. Biol. 2011, 66, 735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Migicovsky, Z.; Yao, Y.; Kovalchuk, I. Transgenerational phenotypic and epigenetic changes in response to heat stress in Arabidopsis thaliana. Plant Signal Behav. 2014, 9, e27971. [Google Scholar] [CrossRef] [Green Version]
- Ganguly, D.R.; Crisp, P.A.; Eichten, S.R.; Pogson, B.J. The Arabidopsis DNA Methylome Is Stable under Transgenerational Drought Stress. Plant Physiol. 2017, 175, 1893–1912. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Wang, X.; Zhong, J.; Zhou, Q.; Wang, X.; Cai, J.; Dai, T.; Cao, W.; Jiang, D. Drought priming induces thermo-tolerance to post-anthesis high-temperature in offspring of winter wheat. Environ. Exp. Bot. 2016, 127, 26–36. [Google Scholar] [CrossRef]
- Tabassum, T.; Farooq, M.; Ahmad, R.; Zohaib, A.; Wahid, A. Seed priming and transgenerational drought memory improves tolerance against salt stress in bread wheat. Plant Physiol. Biochem. 2017, 118, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Li, Y.; Duan, W.; Huang, F.; Hou, X. Cold acclimation alters DNA methylation patterns and confers tolerance to heat and increases growth rate in Brassica rapa. J. Exp. Bot. 2017, 68, 1213–1224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatzig, S.V.; Nuppenau, J.-N.; Snowdon, R.J.; Schießl, S.V. Drought stress has transgenerational effects on seeds and seedlings in winter oilseed rape (Brassica napus L.). BMC Plant Biol. 2018, 18, 297. [Google Scholar] [CrossRef]
- Ou, X.; Zhang, Y.; Xu, C.; Lin, X.; Zang, Q.; Zhuang, T.; Jiang, L.; von Wettstein, D.; Liu, B. Transgenerational inheritance of modified DNA methylation patterns and enhanced tolerance induced by heavy metal stress in rice (Oryza sativa L.). PLoS ONE 2012, 7, e41143. [Google Scholar] [CrossRef] [Green Version]
- Forestan, C.; Aiese Cigliano, R.; Farinati, S.; Lunardon, A.; Sanseverino, W.; Varotto, S. Stress-induced and epigenetic-mediated maize transcriptome regulation study by means of transcriptome reannotation and differential expression analysis. Sci. Rep. 2016, 6, 30446. [Google Scholar] [CrossRef] [Green Version]
- İşeri, Ö.D.; Körpe, D.A.; Sahin, F.I.; Haberal, M. Hydrogen peroxide pretreatment of roots enhanced oxidative stress response of tomato under cold stress. Acta Physiol. Plant. 2013, 35, 1905–1913. [Google Scholar] [CrossRef]
- Zhang, X.; Shen, L.; Li, F.; Meng, D.; Sheng, J. Arginase induction by heat treatment contributes to amelioration of chilling injury and activation of antioxidant enzymes in tomato fruit. Postharvest Biol. Technol. 2013, 79, 1–8. [Google Scholar] [CrossRef]
- Bilichak, A.; Ilnytskyy, Y.; Wóycicki, R.; Kepeshchuk, N.; Fogen, D.; Kovalchuk, I. The elucidation of stress memory inheritance in Brassica rapa plants. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Kellenberger, R.T.; Desurmont, G.A.; Schlüter, P.M.; Schiestl, F.P. Trans-generational inheritance of herbivory-induced phenotypic changes in Brassica rapa. Sci. Rep. 2018, 8, 3536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Streb, P.; Aubert, S.; Gout, E.; Feierabend, J.; Bligny, R. Cross tolerance to heavy-metal and cold-induced photoinhibiton in leaves of Pisum sativum acclimated to low temperature. Physiol. Mol. Biol. Plants 2008, 14, 185–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foyer, C.H.; Rasool, B.; Davey, J.W.; Hancock, R.D. Cross-tolerance to biotic and abiotic stresses in plants: A focus on resistance to aphid infestation. J. Exp. Bot. 2016, 67, 2025–2037. [Google Scholar] [CrossRef]
- Lämke, J.; Bäurle, I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 2017, 18, 1–11. [Google Scholar] [CrossRef]
- Atkinson, N.J.; Urwin, P.E. The interaction of plant biotic and abiotic stresses: From genes to the field. J. Exp. Bot. 2012, 63, 3523–3543. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Vignjevic, M.; Jiang, D.; Jacobsen, S.; Wollenweber, B. Improved tolerance to drought stress after anthesis due to priming before anthesis in wheat (Triticum aestivum L.) var. Vinjett. J. Exp. Bot. 2014, 65, 6441–6456. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Xin, C.; Cai, J.; Zhou, Q.; Dai, T.; Cao, W.; Jiang, D. Heat Priming Induces Trans-generational Tolerance to High Temperature Stress in Wheat. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Topbjerg, H.B.; Jiang, D.; Liu, F. Drought priming at vegetative stage improves the antioxidant capacity and photosynthesis performance of wheat exposed to a short-term low temperature stress at jointing stage. Plant Soil 2015, 393, 307–318. [Google Scholar] [CrossRef]
- Li, X.; Liu, F. Drought Stress Memory and Drought Stress Tolerance in Plants: Biochemical and Molecular Basis. In Drought Stress Tolerance in Plants, Vol 1: Physiology and Biochemistry; Hossain, M.A., Wani, S.H., Bhattacharjee, S., Burritt, D.J., Tran, L.-S.P., Eds.; Springer International Publishing: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]
- Wang, X.; Vignjevic, M.; Liu, F.; Jacobsen, S.; Jiang, D.; Wollenweber, B. Drought priming at vegetative growth stages improves tolerance to drought and heat stresses occurring during grain filling in spring wheat. Plant Growth Regul. 2015, 75, 677–687. [Google Scholar] [CrossRef]
- Rajashekar, C.B.; Panda, M.J.S.H. Water stress is a component of cold acclimation process essential for inducing full freezing tolerance in strawberry. Sci. Hortic. 2014, 174, 54–59. [Google Scholar] [CrossRef] [Green Version]
- Kreyling, J.; Wiesenberg, G.L.; Thiel, D.; Wohlfart, C.; Huber, G.; Walter, J.; Jentsch, A.; Konnert, M.; Beierkuhnlein, C. Cold hardiness of Pinus nigra Arnold as influenced by geographic origin, warming, and extreme summer drought. Environ. Exp. Bot. 2012, 78, 99–108. [Google Scholar] [CrossRef]
- Blodner, C.; Skroppa, T.; Johnsen, O.; Polle, A. Freezing tolerance in two Norway spruce (Picea abies [L.] Karst.) progenies is physiologically correlated with drought tolerance. J. Plant Physiol. 2005, 162, 549–558. [Google Scholar] [CrossRef]
- Trewavas, A. What is plant behaviour? Plant Cell Environ. 2009, 32, 606–616. [Google Scholar] [CrossRef]
- Vu, W.T.; Chang, P.L.; Moriuchi, K.S.; Friesen, M.L. Genetic variation of transgenerational plasticity of offspring germination in response to salinity stress and the seed transcriptome of Medicago truncatula. BMC Evol. Biol. 2015, 15, 59. [Google Scholar] [CrossRef] [Green Version]
- Choi, C.S.; Sano, H. Abiotic-stress induces demethylation and transcriptional activation of a gene encoding a glycerophosphodiesterase-like protein in tobacco plants. Mol. Genet. Genom. 2007, 277, 589–600. [Google Scholar] [CrossRef]
- Mozgova, I.; Mikulski, P.; Pecinka, A.; Farrona, S. Epigenetic Mechanisms of Abiotic Stress Response and Memory in Plants. In Epigenetics in Plants of Agronomic Importance: Fundamentals and Applications; Alvarez-Venegas, R., De-la-Peña, C., Casas-Mollano, J., Eds.; Springer: Cham, Switzerland, 2019. [Google Scholar] [CrossRef]
- Gallusci, P.; Dai, Z.; Génard, M.; Gauffretau, A.; Leblanc-Fournier, N.; Richard-Molard, C.; Vile, D.; Brunel-Muguet, S. Epigenetics for Plant Improvement: Current Knowledge and Modeling Avenues. Trends Plant Sci. 2017, 22, 610–623. [Google Scholar] [CrossRef] [PubMed]
- Cortijo, S.; Wardenaar, R.; Colomé-Tatché, M.; Gilly, A.; Etcheverry, M.; Labadie, K.; Caillieux, E.; Hospital, F.; Aury, J.-M.; Wincker, P.; et al. Mapping the Epigenetic Basis of Complex Traits. Science 2014, 343, 1145. [Google Scholar] [CrossRef]
- Lang, Z.; Wang, Y.; Tang, K.; Tang, D.; Datsenka, T.; Cheng, J.; Zhang, Y.; Handa, A.K.; Zhu, J.-K. Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl. Acad. Sci. USA 2017, 114, E4511. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Lang, Z.; Zhu, J.-K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef]
- Klumpp, A.; Ansel, W.; Fomin, A.; Schnirring, S.; Pickl, C. Influence of climatic conditions on the mutations in pollen mother cells of Tradescantia clone 4430 and implications for the Trad-MCN bioassay protocol. Hereditas 2004, 141, 142–148. [Google Scholar] [CrossRef]
- Cruzan, M.B.; Streisfeld, M.A.; Schwoch, J.A. Phenotypic Effects of Somatic Mutations Accumulating during Vegetative Growth. bioRxiv 2018. [Google Scholar] [CrossRef]
- Alsdurf, J.; Anderson, C.; Siemens, D.H. Epigenetics of drought-induced trans-generational plasticity: Consequences for range limit development. AOB Plants 2015, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.-M.; Tao, X.; Wang, Y.; Ma, D.-W.; Li, D.; Yang, H.; Ma, X.-R. Genomics. Analysis of DNA methylation of perennial ryegrass under drought using the methylation-sensitive amplification polymorphism (MSAP) technique. Mol. Genet. 2014, 289, 1075–1084. [Google Scholar]
- Madlung, A.; Comai, L. The effect of stress on genome regulation and structure. Ann. Bot. 2004, 94, 481–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habu, Y.; Kakutani, T.; Paszkowski, J. Epigenetic developmental mechanisms in plants: Molecules and targets of plant epigenetic regulation. Curr. Opin. Genet. Dev. 2001, 11, 215–220. [Google Scholar] [CrossRef]
- Hirayama, T.; Shinozaki, K. Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 2010, 61, 1041–1052. [Google Scholar] [CrossRef] [PubMed]
- Springer, N.M.; Schmitz, R.J. Exploiting induced and natural epigenetic variation for crop improvement. Nat. Reviews. Genet. 2017, 18, 563–575. [Google Scholar] [CrossRef]
- Vanyushin, B.F.; Ashapkin, V.V. DNA methylation in higher plants: Past, present and future. Biochim. Et Biophys. Acta (Bba) Gene Regul. Mech. 2011, 1809, 360–368. [Google Scholar] [CrossRef]
- Finnegan, E.J.; Kovac, K.A. Plant DNA methyltransferases. Plant Mol. Biol. 2000, 43, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Matzke, M.; Kanno, T.; Huettel, B.; Daxinger, L.; Matzke, A.J.M. Targets of RNA-directed DNA methylation. Curr. Opin. Plant Biol. 2007, 10, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Tompa, R.; McCallum, C.M.; Delrow, J.; Henikoff, J.G.; van Steensel, B.; Henikoff, S. Genome-Wide Profiling of DNA Methylation Reveals Transposon Targets of CHROMOMETHYLASE3. Curr. Biol. 2002, 12, 65–68. [Google Scholar] [CrossRef] [Green Version]
- Lindroth, A.M.; Cao, X.; Jackson, J.P.; Zilberman, D.; McCallum, C.M.; Henikoff, S.; Jacobsen, S.E. Requirement of CHROMOMETHYLASE3 for Maintenance of CpXpG Methylation. Science 2001, 292, 2077. [Google Scholar] [CrossRef] [Green Version]
- Goodrich, J.; Tweedie, S. Remembrance of things past: Chromatin remodeling in plant development. Annu. Rev. Cell Dev. Biol. 2002, 18, 707–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marconi, G.; Pace, R.; Traini, A.; Raggi, L.; Lutts, S.; Chiusano, M.; Guiducci, M.; Falcinelli, M.; Benincasa, P.; Albertini, E. Use of MSAP markers to analyse the effects of salt stress on DNA methylation in rapeseed (Brassica napus var. oleifera). PLoS ONE 2013, 8, e75597. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.-l.; Yu, S.-x.; Ye, W.-w.; Wang, H.-m.; Wang, J.-j.; Fang, B.-x. Study on DNA Cytosine Methylation of Cotton (Gossypium hirsutum L.) Genome and Its Implication for Salt Tolerance. Agric. Sci. China 2010, 9, 783–791. [Google Scholar] [CrossRef]
- Steward, N.; Ito, M.; Yamaguchi, Y.; Koizumi, N.; Sano, H. Periodic DNA methylation in maize nucleosomes and demethylation by environmental stress. J. Biol. Chem. 2002, 277, 37741–37746. [Google Scholar] [CrossRef] [Green Version]
- Hao, Y.J.; You, C.X.; Deng, X.X. Analysis of ploidy and the patterns of amplified fragment length polymorphism and methylation sensitive amplified polymorphism in strawberry plants recovered from cryopreservation. Cryo Lett. 2002, 23, 37–46. [Google Scholar]
- Hashida, S.-n.; Kitamura, K.; Mikami, T.; Kishima, Y. Temperature shift coordinately changes the activity and the methylation state of transposon Tam3 in Antirrhinum majus. Plant Physiol. 2003, 132, 1207–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashida, S.-N.; Uchiyama, T.; Martin, C.; Kishima, Y.; Sano, Y.; Mikami, T. The temperature-dependent change in methylation of the Antirrhinum transposon Tam3 is controlled by the activity of its transposase. Plant Cell 2006, 18, 104–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kou, H.P.; Li, Y.; Song, X.X.; Ou, X.F.; Xing, S.C.; Ma, J.; Von Wettstein, D.; Liu, B. Heritable alteration in DNA methylation induced by nitrogen-deficiency stress accompanies enhanced tolerance by progenies to the stress in rice (Oryza sativa L.). J. Plant Physiol. 2011, 168, 1685–1693. [Google Scholar] [CrossRef]
- Fieldes, M.A.; Schaeffer, S.M.; Krech, M.J.; Brown, J.C. DNA hypomethylation in 5-azacytidine-induced early-flowering lines of flax. Tag. Theor. Appl. Genetics. Theor. Und Angew. Genet. 2005, 111, 136–149. [Google Scholar] [CrossRef]
- Wang, H.; Feng, Q.; Zhang, M.; Yang, C.; Sha, W.; Liu, B. Alteration of DNA methylation level and pattern in sorghum (Sorghum bicolor L.) pure-lines and inter-line F1 hybrids following low-dose laser irradiation. J. Photochem. Photobiol. B Biol. 2010, 99, 150–153. [Google Scholar] [CrossRef]
- Jiang, C.; Mithani, A.; Belfield, E.J.; Mott, R.; Hurst, L.D.; Harberd, N.P. Environmentally responsive genome-wide accumulation of de novo Arabidopsis thaliana mutations and epimutations. Genome Res. 2014, 24, 1821–1829. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.S.; Pan, Y.J.; Zhao, X.Q.; Dwivedi, D.; Zhu, L.H.; Ali, J.; Fu, B.Y.; Li, Z.K. Drought-induced site-specific DNA methylation and its association with drought tolerance in rice (Oryza sativa L.). J. Exp. Bot. 2011, 62, 1951–1960. [Google Scholar] [CrossRef] [PubMed]
- Paun, O.; Bateman, R.M.; Fay, M.F.; Hedren, M.; Civeyrel, L.; Chase, M.W. Stable epigenetic effects impact adaptation in allopolyploid orchids (Dactylorhiza: Orchidaceae). Mol. Biol. Evol. 2010, 27, 2465–2473. [Google Scholar] [CrossRef] [Green Version]
- Hofmeister, B.T.; Lee, K.; Rohr, N.A.; Hall, D.W.; Schmitz, R.J. Stable inheritance of DNA methylation allows creation of epigenotype maps and the study of epiallele inheritance patterns in the absence of genetic variation. Genome Biol. 2017, 18, 155. [Google Scholar] [CrossRef] [PubMed]
- Vasquez-Robinet, C.; Mane, S.P.; Ulanov, A.V.; Watkinson, J.I.; Stromberg, V.K.; De Koeyer, D.; Schafleitner, R.; Willmot, D.B.; Bonierbale, M.; Bohnert, H.J.; et al. Physiological and molecular adaptations to drought in Andean potato genotypes. J. Exp. Bot. 2008, 59, 2109–2123. [Google Scholar] [CrossRef] [Green Version]
- Lacal, I.; Ventura, R. Epigenetic Inheritance: Concepts, Mechanisms and Perspectives. Front Mol Neurosci. 2018, 11. [Google Scholar] [CrossRef] [Green Version]
- Norouzitallab, P.; Baruah, K.; Vanrompay, D.; Bossier, P. Can epigenetics translate environmental cues into phenotypes? Sci. Total Environ. 2019, 647, 1281–1293. [Google Scholar] [CrossRef]
- Reynolds, M.; Langridge, P. Physiological breeding. Curr. Opin. Plant Biol. 2016, 31, 162–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiaz, S.; Ahmad, S.; Noor, M.A.; Wang, X.; Younas, A.; Riaz, A.; Riaz, A.; Ali, F. Applications of the CRISPR/Cas9 System for Rice Grain Quality Improvement: Perspectives and Opportunities. Int. J. Mol. Sci. 2019, 20, 888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheng, Z.; Fiaz, S.; Li, Q.; Chen, W.; Wei, X.; Xie, L.; Jiao, G.; Shao, G.; Tang, S.; Wang, J.; et al. Molecular breeding of fragrant early-season hybrid rice using the BADH2 gene. Pak. J. Bot. 2019, 51, 2089–2095. [Google Scholar] [CrossRef]
- Barman, H.N.; Sheng, Z.; Fiaz, S.; Zhong, M.; Wu, Y.; Cai, Y.; Wang, W.; Jiao, G.; Tang, S.; Wei, X.; et al. Generation of a new thermo-sensitive genic male sterile rice line by targeted mutagenesis of TMS5 gene through CRISPR/Cas9 system. BMC Plant Biol. 2019, 19, 109. [Google Scholar] [CrossRef] [Green Version]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef] [Green Version]
- Piatek, A.; Ali, Z.; Baazim, H.; Li, L.; Abulfaraj, A.; Al-Shareef, S.; Aouida, M.; Mahfouz, M.M. RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol. J. 2015, 13, 578–589. [Google Scholar] [CrossRef]
- Duan, Y.-B.; Li, J.; Qin, R.-Y.; Xu, R.-F.; Li, H.; Yang, Y.-C.; Ma, H.; Li, L.; Wei, P.-C.; Yang, J.-B. Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant Mol. Biol. 2016, 90, 49–62. [Google Scholar] [CrossRef]
- Lou, D.; Wang, H.; Liang, G.; Yu, D. OsSAPK2 Confers Abscisic Acid Sensitivity and Tolerance to Drought Stress in Rice. Front. Plant Sci. 2017, 8, 993. [Google Scholar] [CrossRef] [Green Version]
- Fiaz, S.; Wang, X.; Younas, A.; Alharthi, B.; Riaz, A.; Ali, H. Apomixis and strategies to induce apomixis to preserve hybrid vigor for multiple generations. Gm Crop. Food 2020, 12, 57–70. [Google Scholar] [CrossRef]
- Nguyen, D.; Rieu, I.; Mariani, C.; van Dam, N.M. How plants handle multiple stresses: Hormonal interactions underlying responses to abiotic stress and insect herbivory. Plant Mol. Biol. 2016, 91, 727–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reinders, J.; Wulff, B.B.; Mirouze, M.; Mari-Ordonez, A.; Dapp, M.; Rozhon, W.; Bucher, E.; Theiler, G.; Paszkowski, J. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 2009, 23, 939–950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johannes, F.; Porcher, E.; Teixeira, F.K.; Saliba-Colombani, V.; Simon, M.; Agier, N.; Bulski, A.; Albuisson, J.; Heredia, F.; Audigier, P.; et al. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 2009, 5, e1000530. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Johzuka-Hisatomi, Y.; Terada, R.; Nakamura, I.; Iida, S. The MET1b gene encoding a maintenance DNA methyltransferase is indispensable for normal development in rice. Plant Mol. Biol. 2014, 85, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Hung, Y.-H.; Liu, F.; Zhang, X.-Q.; Xiao, W.; Hsieh, T.-F. Sexual and Non-sexual Reproduction: Inheritance and Stability of Epigenetic Variations and Consequences for Breeding Application. Adv. Bot. Res. 2018, 88, 117–163. [Google Scholar] [CrossRef]
- Danchin, E.; Pocheville, A.; Rey, O.; Pujol, B.; Blanchet, S. Epigenetically facilitated mutational assimilation: Epigenetics as a hub within the inclusive evolutionary synthesis. Biol. Rev. 2019, 94, 259–282. [Google Scholar] [CrossRef] [Green Version]
- Rendina González, A.P.; Preite, V.; Verhoeven, K.J.F.; Latzel, V. Transgenerational Effects and Epigenetic Memory in the Clonal Plant Trifolium repens. Front. Plant Sci. 2018, 9, 1677. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Tanino, K.K.; Robinson, S.J. Stable Epigenetic Variants Selected from an Induced Hypomethylated Fragaria vesca Population. Front. Plant Sci. 2016, 7, 1768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gourcilleau, D.; Mousset, M.; Latutrie, M.; Marin, S.; Delaunay, A.; Maury, S.; Pujol, B. Assessing Global DNA Methylation Changes Associated with Plasticity in Seven Highly Inbred Lines of Snapdragon Plants (Antirrhinum majus). Genes 2019, 10, 256. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Crop Species | Stress Resistance | Treatment/Pathway | References |
|---|---|---|---|
| Chickpea (Cicer arietinum) | Drought | Water and osmotic stress | [37,38] |
| Canola (Brassica napus) | Salt/drought | NaCl priming of seeds, halo-tolerant plant growth-promoting rhizobacteria (PGPR), increased energy use efficiency | [39,40,41] |
| Okra (Abelmoschus esculentus) | Low temperature | Solid matrix priming (SMP) | [42] |
| Sugarcane (Saccharum officinarum) | Drought/salinity | Seed (plants) priming with NaCl and PEG, drought stress memory | [21,25,43,44,45] |
| Mung bean (Vigna radiate) | Drought/salinity | Halopriming of seeds with NaCl and PEG | [46] |
| Maize (Zea Mays) | Low temperature/salt | Water, cold, NaCl, osmotic and hormonal stress | [47,48,49,50,51] |
| Sorghum (Sorghum bicolar) | Salinity | Salt priming of seedlings | [52] |
| Spinach (Spinacia oleracea) | Low temperature | Osmopriming | [49] |
| Rice (Oryza japonica/indica) | Low/high temperature/drought/salt | ABA and H2O2, salt, hydro/dehydro, osmotic, spermidine treatment of seedlings, DNA methylation, gene expression and smRNA and multi-generation drought imposition for abiotic stress tolerance | [53,54,55,56,57,58,59] |
| Alfalfa (Medicago sativa) | Drought | Seed osmotic treatment with PEG | [60] |
| Wheat (Triticum aestivum) | Drought/thermo | Water and osmotic seed priming, pre-drought/heat stress, exogenous ABA application | [33,61,62,63,64] |
| Cotton (Gossypium hirsutum) | Drought/salt/cold | NaCl and PEG treatment/miRNAs/lncRNAs expression, cold stress, DNA methylation | [65,66,67,68,69,70] |
| Tea plant (Camellia sinensis) | Drought | PEG and exogenous ABA-treated | [71] |
| Cowpea (Vigna unguiculata) | Drought | Water, osmotic, and hormonal seed stress | [72,73] |
| Potato (Solanum tuberosum) | Drought/low temperature | Long-term water stress memory, drought, and low temperature | [36,74,75,76] |
| Coffee (Coffea canephora) | Drought | Transcriptional memory | [77] |
| Arabidopsis (Arabidopsis thaliana) | Drought/salinity/biotic stress | β-amino-butyric acid, hyperosmotic priming of seedlings | [78,79] |
| Tomato (Lycopersicon esculentum) | Salinity/abiotic stresses | Priming of seeds with NaCl, ABA, and mannitol induced nitric oxide stresses | [80,81] |
| Soybean (Glycine max) | Drought/salt | Indole acetic acid, NaCl stress on seedlings induced long non-coding RNAs and DNA methylation | [82,83,84] |
| Mung bean (Vigna radiate) | Drought/heavy metals | Indole-3-butyric acid | [85] |
| Strawberry (Fragaria ananassa) | Salinity and non-ionic osmotic/thermotolerance | Hydrogen sulfide/sodium hydrosulfide/hydrogen peroxide and sodium nitroprusside priming of roots | [86,87,88] |
| Crop Species | Stress Resistance | Primary Exposure/Treatment | Transgenerational Physical Response | References |
|---|---|---|---|---|
| Arabidopsis (Arabidopsis thaliana) | Drought, extreme light adoption | β–aminobutyric acid (BABA), dehydration stress, salt and heat stress, short wavelength radiations | Descendants exhibit biotic and abiotic stress resistance, phenotypic changes for increased flexibility | [5,9,78,96,97,98] |
| Wheat (Triticum aestivum) | Drought/salt/heat | Terminal drought/water, osmotic and heat priming of first-generation plants | Drought memory improved resistance against salt stress and drought, and thermotolerance | [67,99,100] |
| Canola (Brassica napus) | Cold/heat/drought | Cold acclimation | Heat/drought resistance, increased growth and yield | [101,102] |
| Rice (Oryza sativa) | Abiotic | Heavy metals, sublethal heat exposure, drought | Enhanced tolerance through heritable changes in gene expression and DNA methylation | [4,92,103] |
| Maize (Zea mays) | Drought/salt | Osmotic stress | Through epigenetic mechanisms, better response to abiotic stresses | [104] |
| Tomato (Lycopersicon sculentum) | Cold | Hydrogen peroxide pretreatment of roots, arginase induction by heat treatment of fruit | Enhanced oxidative stress response, amelioration of chilling injury, and activation of antioxidant enzymes | [105,106] |
| Turnip/Field mustard (Brassica rapa/campestris) | Heat/cold shock/biotic | Heat/salinity/drought/biotic | Stress-induced transgenerational inheritance and cross-protection | [107,108] |
| Pea (Pisum sativum) | Heavy metals | Acclimated to low temperature | Cold-induced photo-inhibition | [109] |
| Alfalfa (Medicago sativa) | Drought | Drought stress/osmopriming | Enhanced growth under drought | [60] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, C.; Ali, K.; Yan, K.; Fiaz, S.; Dormatey, R.; Bi, Z.; Bai, J. Exploration of Epigenetics for Improvement of Drought and Other Stress Resistance in Crops: A Review. Plants 2021, 10, 1226. https://doi.org/10.3390/plants10061226
Sun C, Ali K, Yan K, Fiaz S, Dormatey R, Bi Z, Bai J. Exploration of Epigenetics for Improvement of Drought and Other Stress Resistance in Crops: A Review. Plants. 2021; 10(6):1226. https://doi.org/10.3390/plants10061226
Chicago/Turabian StyleSun, Chao, Kazim Ali, Kan Yan, Sajid Fiaz, Richard Dormatey, Zhenzhen Bi, and Jiangping Bai. 2021. "Exploration of Epigenetics for Improvement of Drought and Other Stress Resistance in Crops: A Review" Plants 10, no. 6: 1226. https://doi.org/10.3390/plants10061226








