Coffee Disease Visualization and Classification
Abstract
:1. Introduction
- We present a guided approach that achieved 98% accuracy in coffee disease classification. Further, in this study, we provide visualization of coffee disease, which exclusively highlights the region responsible for classification.
- In this study, we implement three visualization approaches: Grad-CAM, Grad-CAM++, and Score-CAM. We also provided a visual comparison of those approaches.
- In this study, we demonstrate the relevance of visualization in coffee disease classification. In support of our argument, we present two models and compare their accuracy and visualization results. By comparing the naïve approach and guided approach, this paper will provide new researchers with insights into the factors to consider when applying visualization and classification.
2. Materials and Methods
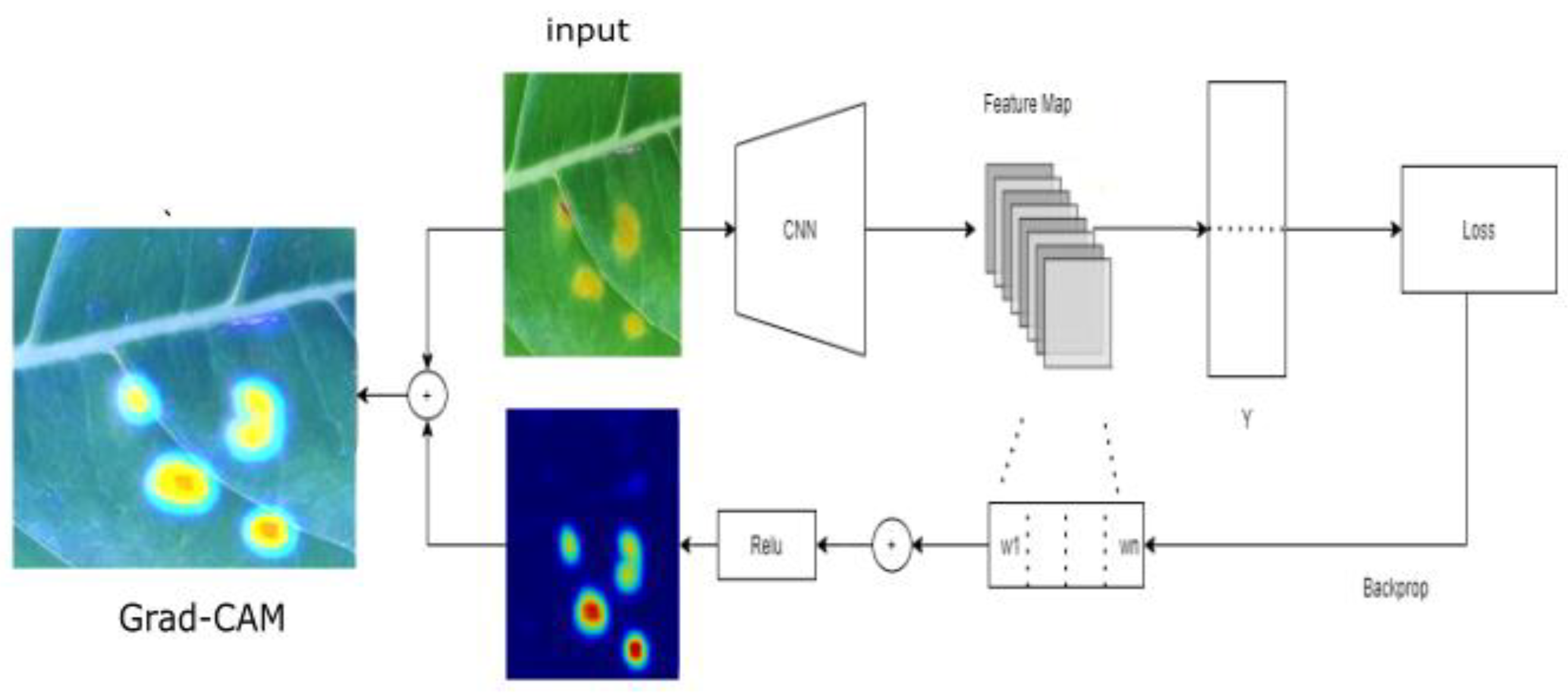
2.1. Visualization Method
2.1.1. Grad-CAM
2.1.2. Grad-CAM++
2.1.3. Score-CAM
2.2. Visualization of Coffee Disease
2.2.1. Coffee Leaf Images Dataset
2.2.2. Naïve Approach
2.2.3. Guided Approach
3. Results and Discussion
3.1. Naïve Approach
3.2. Guided Approach
3.3. Comparison of Visualization Methods
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Getachew, S. Status of Forest Coffee (Coffea arabica L.) Diseases in the Afromontane Rainforest Areas of Ethiopia: A review. Greener J. Agric. Sci. 2017, 7, 19–31. [Google Scholar] [CrossRef]
- Teferi, D. Status of Major Coffee Diseases of Coffea arabica L. in Afromontane Rainforests of Ethiopia. A Review. Food Sci. Qual. Manag. 2018, 76, 35–40. [Google Scholar]
- Degaga, J. Review on Coffee Production and Marketing in Ethiopia. J. Mark. Consum. Res. 2020, 67, 7–15. [Google Scholar] [CrossRef]
- Moat, J.; Williams, J.; Baena, S.; Wilkinson, T.; Gole, T.W.; Challa, Z.K.; Demissew, S.; Davis, A.P. Resilience potential of the Ethiopian coffee sector under climate change. Nat. Plants 2017, 3, 17081. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, X. Plant diseases and pests detection based on deep learning: A review. Plant Methods 2021, 17, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, T.F. Handbook of Approximation Algorithms and Metaheuristics; Chapman and Hall/CRC: New York, NY, USA, 2007; 1432p. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Oquab, M.; Bottou, L.; Laptev, I.; Sivic, J. Is object localization for free?—Weakly-supervised learning with convolutional neural networks. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Zhou, Z.-H. A brief introduction to weakly supervised learning. Natl. Sci. Rev. 2018, 5, 44–53. [Google Scholar] [CrossRef] [Green Version]
- Zhou, M.; Bai, Y.; Zhang, W.; Zhao, T.; Mei, T. Look-Into-Object: Self-Supervised Structure Modeling for Object Recognition. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 11771–11780. [Google Scholar]
- Yao, Q.; Gong, X. Saliency Guided Self-Attention Network for Weakly and Semi-Supervised Semantic Segmentation. IEEE Access 2020, 8, 14413–14423. [Google Scholar] [CrossRef]
- Sun, G.; Wang, W.; Dai, J.; Van Gool, L. Mining Cross-Image Semantics for Weakly Supervised Semantic Segmentation. In Computer Vision—ECCV 2020; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2020; pp. 347–365. [Google Scholar] [CrossRef]
- Fan, J.; Zhang, Z.; Song, C.; Tan, T. Learning Integral Objects With Intra-Class Discriminator for Weakly-Supervised Semantic Segmentation. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 4282–4291. [Google Scholar]
- Minhas, M.S.; Zelek, J. Semi-supervised anomaly detection using autoencoders. arXiv 2020, arXiv:2001.03674. [Google Scholar]
- Zeiler, M.D.; Fergus, R. Visualizing and understanding convolutional networks. In Computer Vision—ECCV 2014; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2014; pp. 818–833. [Google Scholar]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning Deep Features for Discriminative Localization. In Proceedings of the CVPR 2016, 2016 IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; IEEE: New York, NY, USA, 2016; pp. 2921–2929. [Google Scholar]
- Yoo, D.; Park, S.; Lee, J.-Y.; Kweon, I.S. Multi-scale pyramid pooling for deep convolutional representation. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Boston, MA, USA, 7–12 June 2015; pp. 71–80. [Google Scholar] [CrossRef]
- Jo, S.; Yu, I.-J. Puzzle-CAM: Improved localization via matching partial and full features. arXiv 2021, arXiv:2101.11253. [Google Scholar]
- Kumar, M.; Gupta, P.; Madhav, P. Sachin Disease Detection in Coffee Plants Using Convolutional Neural Network. In Proceedings of the 2020 5th International Conference on Communication and Electronics Systems (ICCES), Coimbatore, India, 10–12 June 2020; pp. 755–760. [Google Scholar]
- Manso, G.L.; Knidel, H.; Krohling, R.A.; Ventura, J.A. A smartphone application to detection and classification of coffee leaf miner and coffee leaf rust. arXiv 2019, arXiv:1904.00742. [Google Scholar]
- Zhang, S.; Wu, X.; You, Z.; Zhang, L. Leaf image based cucumber disease recognition using sparse representation classification. Comput. Electron. Agric. 2017, 134, 135–141. [Google Scholar] [CrossRef]
- Yeh, C.-H.; Lin, M.-H.; Chang, P.-C.; Kang, L.-W. Enhanced Visual Attention-Guided Deep Neural Networks for Image Classification. IEEE Access 2020, 8, 163447–163457. [Google Scholar] [CrossRef]
- Hsiao, J.-K.; Kang, L.-W.; Chang, C.-L.; Lin, C.-Y. Comparative study of leaf image recognition with a novel learning-based approach. In Proceedings of the 2014 Science and Information Conference, London, UK, 27–29 August 2014; pp. 389–393. [Google Scholar] [CrossRef]
- Xiong, J.; Yu, D.; Liu, S.; Shu, L.; Wang, X.; Liu, Z. A Review of Plant Phenotypic Image Recognition Technology Based on Deep Learning. Electronics 2021, 10, 81. [Google Scholar] [CrossRef]
- Singh, V.; Misra, A. Detection of plant leaf diseases using image segmentation and soft computing techniques. Inf. Process. Agric. 2017, 4, 41–49. [Google Scholar] [CrossRef] [Green Version]
- Kalvakolanu, A.T. Plant disease detection using deep learning. arXiv 2020, arXiv:2003.05379. [Google Scholar]
- Saleem, M.H.; Khanchi, S.; Potgieter, J.; Arif, K.M. Image-Based Plant Disease Identification by Deep Learning Meta-Architectures. Plants 2020, 9, 1451. [Google Scholar] [CrossRef] [PubMed]
- Tabernik, D.; Šela, S.; Skvarč, J.; Skočaj, D. Segmentation-based deep-learning approach for surface-defect detection. J. Intell. Manuf. 2020, 31, 759–776. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.; Taylor, C.J.; Cootes, T. Small Defect Detection Using Convolutional Neural Network Features and Random Forests. In Computer Vision—ECCV 2018 Workshops; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2018; pp. 398–412. [Google Scholar] [CrossRef]
- Chen, Y.-F.; Yang, F.-S.; Su, E.; Ho, C.-C. Automatic Defect Detection System Based on Deep Convolutional Neural Networks. In Proceedings of the 2019 International Conference on Engineering, Science, and Industrial Applications (ICESI), Tokyo, Japan, 22–24 August 2019. [Google Scholar] [CrossRef]
- Sorte, L.X.B.; Ferraz, C.T.; Fambrini, F.; Goulart, R.D.R.; Saito, J.H. Coffee Leaf Disease Recognition Based on Deep Learning and Texture Attributes. Procedia Comput. Sci. 2019, 159, 135–144. [Google Scholar] [CrossRef]
- Hong, H.; Lin, J.; Huang, F. Tomato disease detection and classification by deep learning. In Proceedings of the 2020 International Conference on Big Data, Artificial Intelligence and Internet of Things Engineering (ICBAIE), Fuzhou, China, 12–14 June 2020; pp. 25–29. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. Int. J. Comput. Vis. 2020, 128, 336–359. [Google Scholar] [CrossRef] [Green Version]
- Chattopadhay, A.; Sarkar, A.; Howlader, P.; Balasubramanian, V.N. Grad-CAM++: Generalized Gradient-Based Visual Explanations for Deep Convolutional Networks. In Proceedings of the 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), Lake Tahoe, NV, USA, 12–15 March 2018; pp. 839–847. [Google Scholar]
- Wang, H.; Wang, Z.; Du, M.; Yang, F.; Zhang, Z.; Ding, S.; Mardziel, P.; Hu, X. Score-CAM: Score-Weighted Visual Explanations for Convolutional Neural Networks. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Seattle, WA, USA, 14–19 June 2020; pp. 111–119. [Google Scholar] [CrossRef]
- Parraga-Alava, J.; Cusme, K.; Loor, A.; Santander, E. RoCoLe: A robusta coffee leaf images dataset. Mendeley Data V2 2019. [Google Scholar] [CrossRef]
- Hindorf, H.; Omondi, C.O. A review of three major fungal diseases of Coffea arabica L. in the rainforests of Ethiopia and progress in breeding for resistance in Kenya. J. Adv. Res. 2011, 2, 109–120. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Qin, X.; Zhang, Z.; Huang, C.; Dehghan, M.; Zaiane, O.R.; Jagersand, M. U2-Net: Going deeper with nested U-structure for salient object detection. Pattern Recognit. 2020, 106, 107404. [Google Scholar] [CrossRef]














| Epoch | Naïve Approach | Proposed Approach (Guided Approach) |
|---|---|---|
| 4 | 72% | 71% |
| 5 | 73% | 72% |
| 6 | 75% | 86% |
| 10 | 74% | 98% |
| 14 | 75% | 98% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yebasse, M.; Shimelis, B.; Warku, H.; Ko, J.; Cheoi, K.J. Coffee Disease Visualization and Classification. Plants 2021, 10, 1257. https://doi.org/10.3390/plants10061257
Yebasse M, Shimelis B, Warku H, Ko J, Cheoi KJ. Coffee Disease Visualization and Classification. Plants. 2021; 10(6):1257. https://doi.org/10.3390/plants10061257
Chicago/Turabian StyleYebasse, Milkisa, Birhanu Shimelis, Henok Warku, Jaepil Ko, and Kyung Joo Cheoi. 2021. "Coffee Disease Visualization and Classification" Plants 10, no. 6: 1257. https://doi.org/10.3390/plants10061257
APA StyleYebasse, M., Shimelis, B., Warku, H., Ko, J., & Cheoi, K. J. (2021). Coffee Disease Visualization and Classification. Plants, 10(6), 1257. https://doi.org/10.3390/plants10061257






