Finding Needles in a Haystack: Using Geo-References to Enhance the Selection and Utilization of Landraces in Breeding for Climate-Resilient Cultivars of Upland Cotton (Gossypium hirsutum L.)
Abstract
:1. Introduction
2. Cotton Landraces: Diversity, Traits and Challenges in Breeding Applications
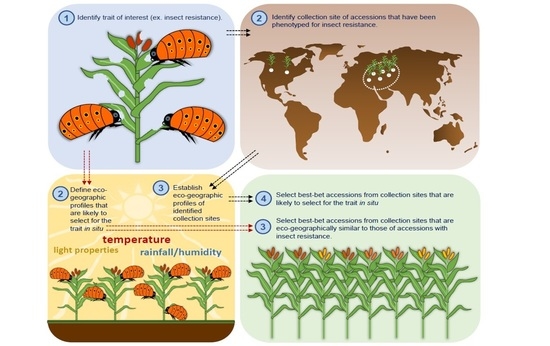
3. Focused Identification of Germplasm Strategy
4. Environmental Association Analysis
5. Perspectives on the Application of FIGS and Landscape Genomics in the Focused Selection of Cotton Landraces with Target Traits of Interest
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Suzuki, N.; Rivero, R.M.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and biotic stress combinations. New Phytol. 2014, 203, 32–43. [Google Scholar] [CrossRef]
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef]
- Available online: http://www.fao.org/docrep/010/a1075e/a1075e00.htm (accessed on 26 October 2020).
- Bebber, D.P.; Ramotowski, M.A.T.; Gurr, S.J. Crop pests and pathogens move polewards in a warming world. Nat. Clim. Chang. 2013, 3, 985–988. [Google Scholar] [CrossRef]
- Oosterhuis, D.M. Growth and development of the cotton plant. In Nitrogen Nutrition in Cotton: Practical Issues, Proceedings of the Southern Branch Workshop for Practicing Agronomists; Miley, W.N., Oosterhuis, D.M., Eds.; Publications of the American Society of Agronomy: Madison, WI, USA, 1990; pp. 1–24. [Google Scholar]
- Ray, R.L.; Fares, A.; Risch, E. Effects of drought ion crop production and cropping areas in Texas. Agri. Environ. Lett. 2018, 3, 170037. [Google Scholar] [CrossRef] [Green Version]
- Guerrero, B. The impact of agricultural drought losses on the Texas Economy; AgriLife Extension, Texas A&M University: College Station, TX, USA, 2012. [Google Scholar]
- Ullah, A.; Sun, H.; Yang, X.; Zhang, X. Drought coping strategies in cotton: Increased crop per drop. Plant Biotechnol. J. 2017, 15, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Salman, M.; Zia, Z.U.; Rana, I.A.; Maqsood, R.H.; Ahmad, S.; Bakhsh, A.; Azhar, M.T. Genetic effects conferring heat tolerance in upland cotton (Gossypium hirsutum L.). J. Cotton Res. 2019, 2, 9. [Google Scholar] [CrossRef]
- Wu, H.; Wang, X.; Xu, M.; Zhang, J. The effect of water deficit and waterlogging on the yield components of cotton. Crop Sci. 2018, 4, 1751–1761. [Google Scholar] [CrossRef]
- Qian, L.; Chen, X.; Wang, X.; Huang, S.; Luo, Y. The effects of flood, drought, and flood followed by drought on yield in cotton. Agronomy 2020, 10, 555. [Google Scholar] [CrossRef] [Green Version]
- Shim, J.; Mangat, P.K.; Angeles-Shim, R.B. Natural variation in wild Gossypium species as a tool to broaden the genetic base of cultivated cotton. J. Plant Sci. Curr. Res. 2018, 2, 005. [Google Scholar]
- Spindel, J.; McCouch, S. Ensuring and exploiting genetic diversity in rice. In Achieving Sustainable Cultivation of Rice; Breeding for higher yield and quality; Taylor & Francis Group: London, UK, 2017; Volume 1. [Google Scholar]
- Mercer, K.L.; Perales, H.R. Evolutionary response of landraces to climatic change in centers of diversity. Evol. Appl. 2010, 3, 480–493. [Google Scholar] [CrossRef]
- Frankel, O.H.; Brown, A.H.D.; Burdon, J.J. The Conservation of Plant Biodiversity, 2nd ed.; Cambridge University Press: Cambridge, UK, 1998; pp. 56–78. [Google Scholar]
- Redden, R. New approaches for crop genetic adaptation to the abiotic stresses predicted with climate change. Agronomy 2013, 3, 419–432. [Google Scholar] [CrossRef]
- Septiningsih, E.M.; Pamplona, A.M.; Sanchez, D.L.; Neeraja, C.N.; Vergara, G.V.; Heuer, S.; Ismail, A.M.; Mackill, D.J. Development of submergence-tolerant rice cultivars: The Sub1 locus and beyond. Ann. Bot. 2009, 103, 151–160. [Google Scholar] [CrossRef] [Green Version]
- Krishnamurthy, S.L.; Pundir, P.; Warraich, A.S.; Rathor, S.; Lokeshkumar, B.M.; Singh, N.K.; Sharma, P.C. Introgressed Saltol QTL lines improves the salinity tolerance in rice at seedling stage. Front. Plant Sci. 2020, 11, 833. [Google Scholar] [CrossRef] [PubMed]
- Angeles-Shim, R.B.; Reyes, V.P.; Del Valle, M.M.; Sunohara, H.; Shim, J.; Lapis, R.S.; Jena, K.K.; Ashikari, M.; Doi, K. Marker-assisted introgression of the quantitative resistance gene pi21 confers broad-spectrum resistance of rice to blast. Rice Sci. 2020, 27, 113–123. [Google Scholar] [CrossRef]
- Gano, B.; Dembele, J.S.B.; Tovignan, T.K.; Sine, B.; Vadez, V.; Diouf, D.; Audebert, A. Adaptation responses to early drought stress of West Africa sorghum varieties. Agronomy 2014, 44, 443. [Google Scholar]
- Khazaei, H.; Street, K.; Bari, A.; Mackay, M.; Stoddard, F.L. The FIGS (Focused Identification of Germplasm Strategy) approach identifies traits related to drought adaptation in Vicia faba genetic resources. PLoS ONE 2013, 8, e63107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubiales, D.; Fondevilla, S.; Fernandez-Aparicio, M. Development of pea breeding lines with resistance to Orobanche crenata derived from pea landraces and wild Pisum spp. Agronomy 2020, 11, 36. [Google Scholar] [CrossRef]
- Zhang, K.; Kuraparthy, V.; Fang, H.; Zhu, L.; Sood, S.; Jones, D.C. High-density linkage map construction and QTL analyses for fiber quality, yield and morphological traits using CottonSNP63K array in upland cotton (Gossypium hirsutum L.). BMC Genom. 2019, 20, 889. [Google Scholar] [CrossRef] [Green Version]
- Available online: http://www.icac.org (accessed on 18 January 2021).
- Campbell, B.T.; Saha, S.; Percy, R.; Frelichowski, J.; Jenkins, J.N.; Park, W.; Mayee, C.D.; Gotmare, V.; Dessauw, D.; Giband, M.; et al. Status of the global cotton germplasm resource. Crop Sci. 2010, 50, 1161–1179. [Google Scholar] [CrossRef] [Green Version]
- Abdurakhmonov, I.Y. World Cotton Germplasm Resources; Intech: Rijeka, Croatia, 2014; p. 320. [Google Scholar]
- Wambugu, P.W.; Ndjiondjop, M.N.; Henry, R.J. Role of genomics in promoting the utilization of plant genetic resources in genebanks. Brief. Funct. Genom. 2018, 17, 198–206. [Google Scholar] [CrossRef]
- Hinze, L.L.; Fang, D.D.; Gore, M.A.; Scheffler, B.E.; Yu, J.Z.; Frelichowski, J.; Percy, R.G. Molecular characterization of the Gossypium Diversity Reference Set of the US National Cotton Germplasm Collection. Theor. Appl. Genet. 2015, 128, 313–327. [Google Scholar] [CrossRef]
- Campbell, B.T.; Hugie, K.; Wu, J.; Jones, D.C. Assessing the breeding potential of thirteen day-neutral landrace accessions in an upland cotton breeding program. Crop Sci. 2019, 59, 1469–1478. [Google Scholar] [CrossRef]
- Hinze, L.L.; Hulse-Kemp, A.M.; Wilson, I.W.; Zhu, Q.H.; Llewellyn, D.J.; Taylor, J.M.; Spriggs, A.; Fang, D.D.; Ulloa, M.; Burke, J.J.; et al. Diversity analysis of cotton (Gossypium hirsutum L.) germplasm using the CottonSNP63K Array. BMC Plant Biol. 2017, 17, 37. [Google Scholar] [CrossRef] [Green Version]
- Zhu, G.; Li, W.; Wang, G.; Si, Q.; Cai, C.; Guo, W. Genetic basis of fiber improvement and decreased stress tolerance in cultivated versus semi-domesticated upland cotton. Front. Plant Sci. 2019, 10, 1572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shim, J.; Gannaban, R.B.; de los Reyes, B.G.; Angeles-Shim, R.B. Identification of novel sources of genetic variation for the improvement of cold germination ability in upland cotton (Gossypium hirsutum). Euphytica 2019, 215, 190. [Google Scholar] [CrossRef] [Green Version]
- Oosterhuis, D.M.; Wullschleger, S.D.; Stewart, J.M. Osmotic adjustment in commercial cultivars and wild types of cotton. Agron. Abstr. 1987, 97. [Google Scholar]
- Hou, S.; Zhu, G.; Li, Y.; Fu, J.; Niu, E.; Li, L.; Zhange, D.; Guo, W. Genome-wide association studies revel genetic variation and candidate genes of drought stress related traits in cotton (Gossypium hirsutum L.). Front. Plant Sci. 2018, 9, 1276. [Google Scholar] [CrossRef] [PubMed]
- Cushman, K.R.; Pabuayon, I.C.M.; Hinze, L.; Sweeney, M.E.; de los Reyes, B.G. Networks of physiological adjustments and defenses, and their synergy with sodium (Na+) homeostasis explain the hidden variation for salinity tolerance across the cultivated Gossypium hirsutum germplasm. Front. Plant Sci. 2020, 11, 588854. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.F.; Richmond, T.R. The genetics of flowering response in cotton. I. Fruiting behavior of Gossypium hirsutum var. Marie-Galante in a cross with a variety of cultivated American upland cotton. Genetics 1957, 42, 499–509. [Google Scholar] [CrossRef]
- McCarty, J.C.; Jenkins, J.N.; Parrot, W.L.; Creech, R.G. The conversion of photoperiodic primitive race stocks of cotton to day-neutral stocks. Bull. 4 Mississipi Agric. For. Exp. Strat. Miss. 1979, 4, 1–4. [Google Scholar]
- McCarty, J.C.; Jenkins, J.N. Registration of 14 primitive derived cotton germplasm lines with improved fiber strength. Crop Sci. 2005, 45, 2668–2669. [Google Scholar] [CrossRef]
- McCarty, J.C.; Jenkins, J.N. Registration of 21 day length-neutral flowering primitive cotton germplasm lines. Crop Sci. 2005, 45, 2134. [Google Scholar] [CrossRef]
- McCarty, J.C.; Jenkins, J.N. Registration of 16 day length-neutral flowering primitive cotton germplasm lines. Crop Sci. 2002, 42, 1755–1756. [Google Scholar] [CrossRef]
- McCarty, J.C.; Jenkins, J.N. Registration of 79 day-neutral primitive cotton germplasm lines. Crop Sci. 1993, 33, 351. [Google Scholar] [CrossRef]
- Campbell, B.T.; Greene, J.K.; Wu, J.; Jones, D.C. Assessing the breeding potential of day-neutral converted racestock germplasm in the Pee Dee cotton germplasm enhancement program. Euphytica 2014, 195, 453–465. [Google Scholar] [CrossRef]
- McCarty, J.C.; Wu, J.; Jenkins, J.N. Genetic diversity for agronomic and fiber traits in day-neutral accessions derived from primitive cotton germplasm. Euphytica 2006, 148, 283–293. [Google Scholar] [CrossRef]
- Sanders, R.; Bari, A.; Street, K.; Devlin, M. A New Approach to Mining Agricultural Gene Banks—To Speed the Pace of Research Innovation for Food Security; International Center for Agricultural Research in the Dry Areas (ICARDA): Beirut, Lebanon, 2013. [Google Scholar]
- Anglin, N.L.; Amri, A.; Kehel, Z.; Ellis, D. A case of need: Linking traits to genebank accessions. Biopreserv. Biobank. 2018, 15, 337–349. [Google Scholar] [CrossRef] [Green Version]
- Mackay, M.; Street, K. Focused identification of germplasm strategy—FIGS. In Proceedings of the 54th Australian Cereal Chemistry Conference and the 11th Wheat Breeders’ Assembly, editors. Cereal Chemistry Division, Royal Australian Chemical Institute (RACI), Melbourne, VIC, Australia, 21–24 September 2004; pp. 138–141. [Google Scholar]
- Bhullar, N.K.; Street, K.; Mackay, M.; Yahiaoui, N.; Keller, B. Unlocking wheat genetic resources for the molecular identification of previously undescribed functional alleles at the Pm3 resistance locus. Proc. Natl. Acad. Sci. USA 2009, 106, 9519–9524. [Google Scholar] [CrossRef] [Green Version]
- El Bouhssini, M.; Street, K.; Joubi, A.; Ibrahim, Z.; Rihawi, F. Sources of wheat resistance to Sunn pest, Eurygaster integriceps Puton, in Syria. Genet. Resour. Crop Evol. 2009, 56, 1065–1069. [Google Scholar] [CrossRef]
- Vikas, V.K.; Kumar, S.; Archak, S.; Tyagi, R.K.; Kumar, J.; Jacob, S.; Sivasamy, M.; Jayaprakash, P.; Saharan, M.S.; Basandrai, A.K.; et al. Screening of 19,460 genotypes of wheat species for resistance to powdery mildew and identification of potential candidates using focused identification of germplasm strategy (FIGS). Crop Sci. 2020, 60, 2857–2866. [Google Scholar] [CrossRef]
- Bari, A.; Street, K.; Mackay, M.; Endresen, D.T.F.; De Pauw, E.; Amri, A. Focused identification of germplasm strategy (FIGS) detects wheat stem rust resistance linked to environmental variables. Genet. Resour. Crop Evol. 2012, 59, 1465–1481. [Google Scholar] [CrossRef]
- El Bouhssini, M.; Street, K.; Amri, A.; Mackay, M.; Ogbonnaya, F.C.; Omran, A.; Abdalla, O.; Baum, M.; Dabbous, A.; Rihawi, F. Sources of resistance in bread wheat to Russian wheat aphid (Diuraphis noxia) in Syria identified using the Focused Identification of Germplasm Strategy (FIGS). Plant Breed. 2011, 130, 96–97. [Google Scholar] [CrossRef]
- Endresen, D.T.F.; Street, K.; Bari, A.; De Pauw, E. Predictive association between biotic stress traits and ecogeographic data for wheat and barley landraces. Crop Sci. 2011, 51, 2036–2055. [Google Scholar] [CrossRef]
- Egan, P.A.; Muola, A.; Stenberg, J.A. Capturing genetic variation in crop wild relatives: An evolutionary approach. Evol. Appl. 2018, 11, 1293–1304. [Google Scholar] [CrossRef] [Green Version]
- Stenberg, J.A.; Ortiz, R. Focused identification of germplasm strategy (FIGS): Polishing a rough diamond. Curr. Opin. Insect Sci. 2021, 45, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Bragg, J.G.; Supple1, M.A.; Andrew, R.L.; Borevitz, J.O. Genomic variation across landscapes: Insights and applications. New Phytol. 2015, 207, 953–967. [Google Scholar] [CrossRef]
- Rellstab, C.; Gugerli, F.; Eckert, A.; Hancock, A.; Holderegger, R. A practical guide to environmental association analysis in landscape genomics. Mol. Ecol. 2015, 24, 4348–4370. [Google Scholar] [CrossRef] [Green Version]
- Navarro, J.A.R.; Willcox, M.; Burgueño, J.; Romay, C.; Swarts, K.; Traschel, S.; Preciado, E.; Terron, A.; Delgado, H.V.; Vidal, V.; et al. A study of allelic diversity underlying flowering-time adaptation in maize landraces. Nat. Genet. 2017, 49, 476–480. [Google Scholar] [CrossRef]
- Lasky, J.R.; Upadhyaya, H.D.; Ramu, P.; Deshpande, S.; Hash, C.T.; Bonnette, J.; Juenger, T.E.; Hyma, K.; Acharya, C.; Mitchell, S.E.; et al. Genome-environment associations in sorghum landraces predict adaptive traits. Sci. Adv. 2015, 1, e1400218. [Google Scholar] [CrossRef] [Green Version]
- Turesson, G. The genotypical response of the plant species to the habitat. Hereditas 1922, 3, 211–350. [Google Scholar] [CrossRef]
- Huxley, J. Clines: An auxiliary taxonomic principle. Nature 1938, 142, 219–220. [Google Scholar] [CrossRef]
- Mitton, J.B.; Linhart, Y.B.; Hamrick, J.L.; Beckman, J.S. Observations on genetic structure and mating system of Ponderosa pine in Colorado front range. Theor. Appl. Genet. 1977, 51, 5–13. [Google Scholar] [CrossRef]
- Mourtzinis, S.; Specht, J.E.; Lindsey, L.E.; Wiebold, W.J.; Ross, J.; Nafziger, E.D.; Kandel, H.J.; Mueller, N.; Devillez, P.L.; Arriaga, F.J.; et al. Climate-induced reduction in US-wide soybean yields underpinned by region- and in-season specific responses. Nat. Plants 2015, 1, 14026. [Google Scholar] [CrossRef]
- Bandillo, N.B.; Anderson, J.E.; Kantar, B.; Stupar, R.M.; Specht, J.E.; Graef, G.L.; Lorenz, A.J. Dissecting the genetic basis of local adaptation in soybean. Sci. Rep. 2017, 7, 17195. [Google Scholar] [CrossRef]
- Available online: http://www.worldclim.org. (accessed on 18 May 2021).
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high-resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Available online: http://www.isric.org (accessed on 18 May 2021).
- Hengl, T.; de Jesus, J.M.; MacMillan, R.A.; Batjes, N.H.; Heuvelink, G.B.; Ribeiro, E.; Samuel-Rosa, A.; Kempen, B.; Leenaars, J.G.B.; Walsh, M.G.; et al. SoilGrids1km–global soil information based on automated mapping. PLoS ONE 2014, 9, e105992. [Google Scholar] [CrossRef] [Green Version]
- Cortes, L.T.; Zhang, Z.; Yu, J. Status and prospects of genome-wide association studies in plants. Plant Genome 2021, 14, e20077. [Google Scholar]
- Russell, J.; Mascher, M.; Dawson, I.K.; Kyriakidis, S.; Calixto, C.; Freund, F.; Bayer, M.; Milne, I.; Marshall-Griffiths, T.M.; Heinen, S.; et al. Exome sequencing of geographically diverse barley landraces and wild relatives gives insights into environmental adaptation. Nat. Genet. 2016, 48, 1024–1030. [Google Scholar] [CrossRef] [Green Version]
- Salvi, S.; Sponza, G.; Morgante, M.; Tomes, D.; Niu, X.; Fengler, K.A.; Meeley, R.; Ananiev, E.V.; Svitashev, S.; Bruggemann, E.; et al. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc. Natl. Acad. Sci. USA 2007, 104, 11376–11381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castelletti, S.; Tuberosa, R.; Pindo, M.; Salvi, S. A MITE transposon insertion is associated with differential methylation at the maize flowering time QTL Vgt1. G3 (Bethesda, Md) 2014, 4, 805–812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.; Muszynski, M.G.; Danilevskaya, O.N. The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell 2011, 23, 942–960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danilevskaya, O.N.; Meng, X.; Hou, Z.; Ananiev, E.V.; Simmons, C.R. A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol. 2008, 146, 250–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hung, H.Y.; Shannon, L.M.; Tian, F.; Bradbury, P.J.; Chen, C.; Flint-Garcia, S.A.; McMullen, M.D.; Ware, D.; Buckler, E.S.; Doebley, J.F.; et al. ZmCCT and the genetic basis of day-length adaptation underlying the post-domestication spread of maize. Proc. Natl. Acad. Sci. USA 2012, 109, E1913–E1921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, F.; Sakuma, Y.; Tran, L.S.P.; Maruyama, K.; Kidokoro, S.; Fujita, Y.; Fujita, M.; Umezawa, T.; Sawano, Y.; Miyazono, K.; et al. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 2008, 20, 1693–1707. [Google Scholar] [CrossRef] [Green Version]
- de Silva, K.; Laska, B.; Brown, C.; Sederoff, H.W.; Khodakovskaya, M. Arabidopsis thaliana calcium-dependent lipid-binding protein (AtCLB): A novel repressor of abiotic stress response. J. Exp. Bot. 2011, 62, 2679–2689. [Google Scholar] [CrossRef] [Green Version]
- Pandey, N.; Ranjan, A.; Pant, P.; Tripathi, R.K.; Ateek, F.; Pandey, H.P.; Patre, U.V.; Sawant, S.V. CAMTA 1 regulates drought responses in Arabidopsis thaliana. BMC Genom. 2013, 14, 216. [Google Scholar] [CrossRef] [Green Version]
- Redillas, M.C.; Jeong, J.S.; Kim, Y.S.; Jung, H.; Bang, S.W.; Choi, Y.D.; Ha, S.; Reuzeau, C.; Kim, J. The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol. J. 2012, 10, 792–805. [Google Scholar] [CrossRef] [PubMed]
- Distelfeld, A.; Avni, R.; Fischer, A.M. Senescence, nutrient remobilization, and yield in wheat and barley. J. Exp. Bot. 2014, 65, 3783–3798. [Google Scholar] [CrossRef] [Green Version]
- Yuen, C.Y.; Sedbrook, J.C.; Perrin, R.M.; Carroll, K.L.; Masson, P.H. Loss-of-function mutations of ROOT HAIR DEFECTIVE3 suppress root waving, skewing, and epidermal cell file rotation in Arabidopsis. Plant Physiol. 2005, 138, 701–714. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.F.; Wan, J.X.; Sun, Y.; Shi, Y.Z.; Braam, J.; Li, G.X.; Zheng, S.J. Xyloglucan endotransglucosylasehydrolase17 interacts with xyloglucan endotransglucosylase-hydrolase31 to confer xyloglucan endotransglucosylase action and affect aluminum sensitivity in Arabidopsis. Plant Physiol. 2014, 165, 1566–1574. [Google Scholar] [CrossRef]
- Yang, X.Y.; Zeng, Z.H.; Yan, J.Y.; Fan, W.; Bian, H.W.; Zhu, M.Y.; Yang, J.L.; Zheng, S.J. Association of specific pectin methylesterases with Al-induced root elongation inhibition in rice. Physiol. Plant. 2013, 148, 502–511. [Google Scholar] [CrossRef]
- Sanchez, J.; Mangat, P.K.; Angeles-Shim, R.B. Weathering the cold: Modifying membrane and storage fatty acid composition of seeds to improve cold germination ability in upland cotton (Gossypium hirsutum L.). Agronomy 2020, 9, 684. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, N.J.; Epstein, D.J.; Wang, D.; Vail, L.; Srinivasan, R.; Arnold, J.G. Possible impacts of global warming on the hydrology of the Ogallala aquifer region. Clim. Chang. 1999, 42, 677–692. [Google Scholar] [CrossRef]
- Terrell, B.L.; Johnson, P.N.; Segarra, E. Ogallala aquifer depletion: Economic impact on the Texas high plains. Water Policy 2002, 4, 33–46. [Google Scholar] [CrossRef]
- Ulloa, M.; Hutmacher, R.B.; Schramm, T.; Ellis, M.L.; Nichols, R.; Roberts, P.A.; Wright, S.D. Sources, selection and breeding of Fusarium wilt (Fusarium oxysporum f. sp. vasinfectum) race 4 (FOV4) resistance in upland (Gossypium hirsutum L.) cotton. Euphytica 2020, 216, 109. [Google Scholar] [CrossRef]
- Available online: https://npgsweb.ars-grin.gov/gringlobal/search.aspx (accessed on 18 May 2021).
- Hulse-Kemp, A.M.; Lemm, J.; Plieske, J.; Ashrafi, H.; Buyyarapu, R.; Fang, D.D.; Frelichowski, J.; Giband, M.; Hague, S.; Hinze, L.L.; et al. Development of a 63K SNP array for cotton and high-density mapping of intraspecific and interspecific populations of Gossypium spp. G3 (Bethesda) 2015, 5, 1187–1209. [Google Scholar] [CrossRef] [Green Version]
- Cai, C.; Zhu, G.; Zhang, T.; Guo, W. High-density 80K SNP array is a powerful tool for genotyping G. hirsutum accessions and genome analysis. BMC Genom. 2017, 18, 654. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shim, J.; Bandillo, N.B.; Angeles-Shim, R.B. Finding Needles in a Haystack: Using Geo-References to Enhance the Selection and Utilization of Landraces in Breeding for Climate-Resilient Cultivars of Upland Cotton (Gossypium hirsutum L.). Plants 2021, 10, 1300. https://doi.org/10.3390/plants10071300
Shim J, Bandillo NB, Angeles-Shim RB. Finding Needles in a Haystack: Using Geo-References to Enhance the Selection and Utilization of Landraces in Breeding for Climate-Resilient Cultivars of Upland Cotton (Gossypium hirsutum L.). Plants. 2021; 10(7):1300. https://doi.org/10.3390/plants10071300
Chicago/Turabian StyleShim, Junghyun, Nonoy B. Bandillo, and Rosalyn B. Angeles-Shim. 2021. "Finding Needles in a Haystack: Using Geo-References to Enhance the Selection and Utilization of Landraces in Breeding for Climate-Resilient Cultivars of Upland Cotton (Gossypium hirsutum L.)" Plants 10, no. 7: 1300. https://doi.org/10.3390/plants10071300
APA StyleShim, J., Bandillo, N. B., & Angeles-Shim, R. B. (2021). Finding Needles in a Haystack: Using Geo-References to Enhance the Selection and Utilization of Landraces in Breeding for Climate-Resilient Cultivars of Upland Cotton (Gossypium hirsutum L.). Plants, 10(7), 1300. https://doi.org/10.3390/plants10071300