Resistance Mechanism to Metsulfuron-Methyl in Polypogon fugax
Abstract
:1. Introduction
2. Results
2.1. Whole-Plant Dose Response to ALS Inhibitors
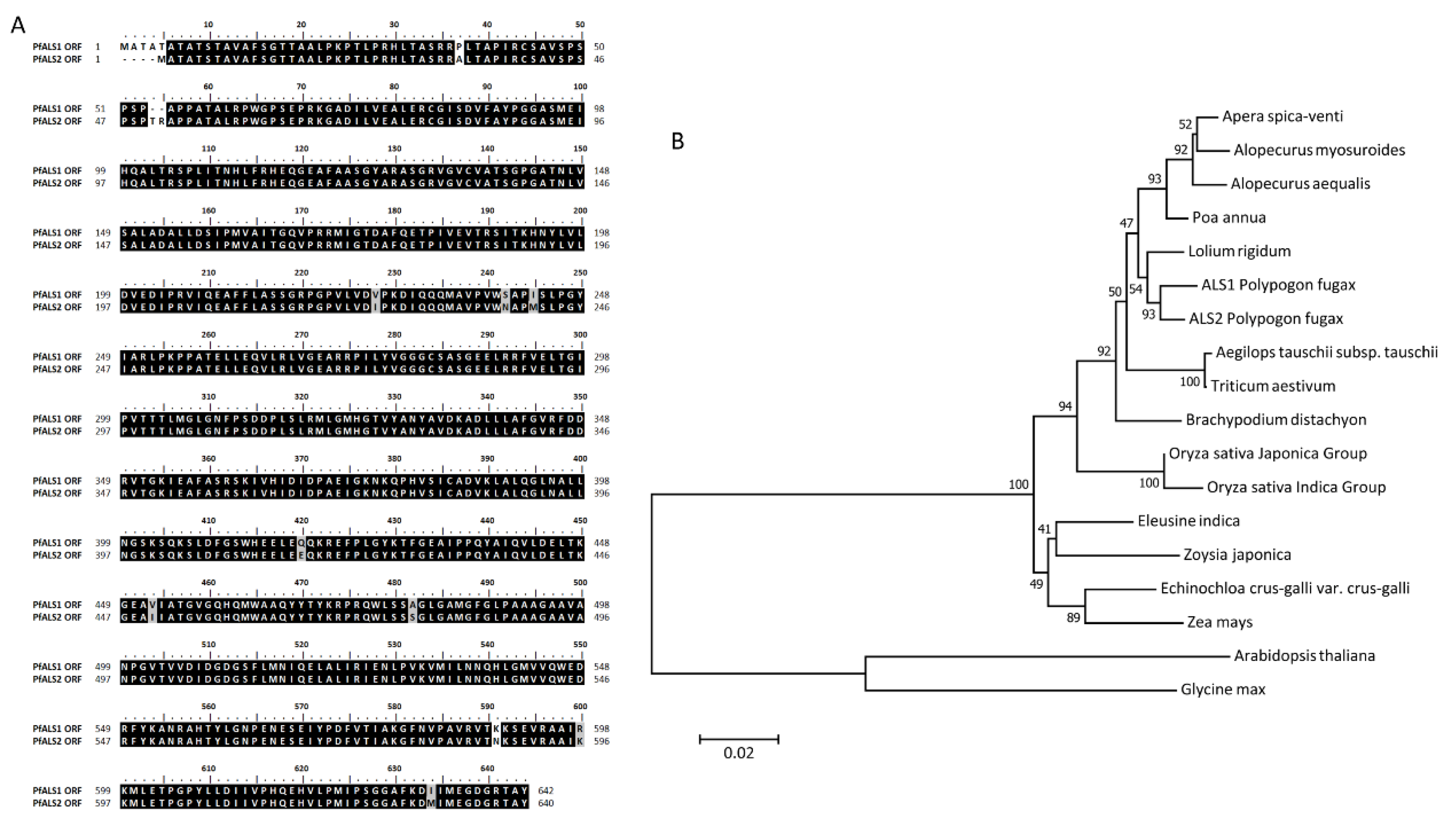
2.2. The Analysis of ALS Gene Sequence in S and R Biotypes
2.3. ALS Enzyme Activity Assay
2.4. GST Activity Assay
2.5. Effect of Malathion on Metsulfuron-Methyl Resistance
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Condition
4.2. Whole-Plant Bioassays for Resistance Confirmation
4.3. Isolation and Sequencing of ALS Genes from P. fugax
4.4. ALS Enzyme Activity Assay
4.5. GST Activity Assay
4.6. Effect of Malathion on Metsulfuron-Methyl Resistance
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Li, Y.H. Weed Flora of China, 1st ed.; China Agriculture Press: Beijing, China, 1998; pp. 1313–1315. (In Chinese) [Google Scholar]
- Xu, Z.H.; Qi, H.Y.; Lu, Y.L.; Yang, W.D.; Xie, G.X. Weed Identification and Management, 1st ed.; Zhejiang University: Hangzhou, China, 2014; 372p. (In Chinese) [Google Scholar]
- Chen, Q.H.; Zhou, X.G.; Zheng, S.J.; Luo, C.H.; Tang, Y.Z.; Zhang, J.J. Weed Occurrence and Control by Different Tillage Methods in Wheat Fields in Sichuan Province. Weed Sci. 2013, 31, 12–15. (In Chinese) [Google Scholar]
- He, C.; Wei-Jun, Z.; Yan, J. Development, Status of Damage and Control Strategy of Cornfield Weed in Shanghai. J. Shanghai Jiaotong Univ. 2004, 4, 393–399. (In Chinese) [Google Scholar]
- Wang, K.; Qiang, Q. Quantitative analysis of distribution of weed communities in wheat fields in the south of Jiangsu province. J. Biomathemat. 2005, 1, 107–114. (In Chinese) [Google Scholar] [CrossRef]
- Tang, W.; Zhou, F.; Chen, J.; Zhou, X. Resistance to ACCase-inhibiting Herbicides in an Asia Minor Bluegrass (Polypogon fugax) Population in China. Pestic. Biochem. Physiol. 2014, 108, 16–20. [Google Scholar] [CrossRef] [PubMed]
- De Prado, R.; Osuna, M.; Fischer, A. Resistance to ACCase inhibitor herbicides in a green foxtail (Setaria viridis) biotype in Europe. Weed Sci. 2004, 52, 506–512. [Google Scholar] [CrossRef]
- Maneechote, C.; Samanwong, S.; Zhang, X.Q.; Powles, S.B. Resistance ro ACCase-inhibiting herbicides in sprangletop (Leptochloa chinensis). Weed Sci. 2005, 53, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Powles, S.B.; Qin, Y. Evolution in Action: Plants Resistant to Herbicides. Annu. Rev. Plant. Biol. 2010, 61, 317–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Délye, C.; Pernin, F.; Michel, S. Universal PCR assays detecting mutations in acetyl-coenzyme A carboxylase or acetolactate synthase that endow herbicide resistance in grass weeds. Weed Res. 2001, 51, 353–362. [Google Scholar] [CrossRef]
- Délye, C.; Zhang, X.Q.; Séverine, M.; Annick, M.; Powles, S.B. Molecular bases for sensitivity to acetyl-coenzyme A carboxylase inhibitors in black-grass. Plant Physiol. 2005, 137, 794–806. [Google Scholar] [CrossRef] [Green Version]
- Yu, Q.; Abdallah, I.; Han, H.; Owen, M.; Powles, S.B. Distinct non-target site mechanisms endow resistance to glyphosate, ACCase and ALS-inhibiting herbicides in multiple herbicide-resistant Lolium rigidum. Planta 2009, 230, 713–723. [Google Scholar] [CrossRef]
- Li, L.; Bi, Y.; Liu, W.; Yuan, G.; Wang, J. Molecular basis for resistance to fenoxaprop-p-ethyl in American sloughgrass (Beckmannia syzigachne Steud.). Pestic. Biochem. Phys. 2013, 105, 118–121. [Google Scholar] [CrossRef]
- Beckie, H.J. Herbicide-Resistant Weeds: Management Tactics and Practices. Weed. Technol. 2006, 20, 793–814. [Google Scholar] [CrossRef]
- Norsworthy, J.K.; Ward, S.M.; Shaw, D.R.; Llewellyn, R.S.; Nichols, R.L.; Webster, T.M.; Bradley, K.W.; Frisvold, G.; Powles, S.B.; Burgos, N.R. Reducing the Risks of Herbicide Resistance: Best Management Practices and Recommendations. Weed Sci. 2012, 60, 31–62. [Google Scholar] [CrossRef] [Green Version]
- Iwakami, S.; Hashimoto, M.; Matsushima, K.I.; Watanabe, H.; Hamamura, K.; Uchino, A. Multiple-herbicide resistance in Echinochloa crus-galli var. formosensis, an allohexaploid weed species, in dry-seeded rice. Pestic. Biochem. Physiol. 2015, 119, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Santaella, J.; Heredia, A.; Prado, R.D. Basis of selectivity of cyhalofop-butyl in Oryza sativa L. Planta 2006, 223, 191–199. [Google Scholar] [CrossRef]
- Shaner, D. Role of Translocation as A Mechanism of Resistance to Glyphosate. Weed Sci. 2009, 57, 118–123. [Google Scholar] [CrossRef]
- Lopez-Martinez, N.; Gonzalez-Gutierrez, J.; De Prado, R. Propanil activity, uptake and metabolism in resistant Echinochloa spp. biotypes. Weed Res. 2001, 41, 187–196. [Google Scholar] [CrossRef]
- Julio, M.; Fernando, B.; Rafael, D.P. Resistance to chlortoluron in a downy brome (Bromus tectorum) biotype. Weed Sci. 2006, 54, 237–245. [Google Scholar]
- Scarabel, L.; Locascio, A.; Furini, A.; Sattin, M.; Varotto, S. Characterisation of ALS genes in the polyploid species Schoenoplectus mucronatus and implications for resistance management. Pest Manag. Sci. 2010, 66, 337–344. [Google Scholar] [CrossRef]
- Hanson, B.D.; Park, K.W.; Mallory-Smith, C.A.; Thill, D.C. Resistance of Camelina microcarpa to acetolactate synthase inhibiting herbicides. Weed Res. 2010, 44, 187–194. [Google Scholar] [CrossRef]
- Xu, X.; Liu, G.; Chen, S.; Li, B.; Liu, X.; Wang, X.; Fan, C.; Wang, G.; Ni, H. Mutation at residue 376 of ALS confers tribenuron-methyl resistance in flixweed (Descurainia sophia) populations from Hebei Province, China. Pestic Biochem. Physiol. 2015, 125, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Tranel, P.J.; Wright, T.R.; Heap, I.M. Mutations in Herbicide-resistant Weeds to ALS Inhibitors. Available online: http://www.weedscience.com (accessed on 31 May 2021).
- Preston, C.; Powles, S.B. Evolution of herbicide resistance in weeds: Initial frequency of target site-based resistance to acetolactate synthase-inhibiting herbicides in Lolium rigidum. Heredity 2002, 88, 8–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Zhu, X.; Wang, H.; Li, J.; Dong, L. Mechanism of resistance to fenoxaprop in Japanese foxtail (Alopecurus japonicus) from China. Pestic. Biochem. Physiol. 2013, 107, 25–31. [Google Scholar] [CrossRef]
- Yuan, J.S.; Tranel, P.J.; Stewart, C.N. Non-target-site herbicide resistance: A family business. Trends Plant Sci. 2007, 12, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Reade, J.; Milner, L.; Cobb, A. A role for glutathione S-transferases in resistance to herbicides in grasses. Weed Sci. 2004, 52, 468–474. [Google Scholar] [CrossRef]
- Burns, E.E.; Keith, B.K.; Refai, M.Y.; Bothner, B.; Dyer, W.E. Proteomic and biochemical assays of glutathione-related proteins in susceptible and multiple herbicide resistant Avena fatua L. Pestic. Biochem. Physiol. 2017, 140, 69–78. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Wu, C.; Guo, W.; Du, L.; Yuan, G.; Wang, J. Resistance Mechanisms to an Acetolactate Synthase (ALS) Inhibitor in Water Starwort (Myosoton aquaticum) Populations from China. Weed Sci. 2015, 63, 770–780. [Google Scholar] [CrossRef]
- Wright, A.A.; Rodriguez-Carres, M.; Sasidharan, R.; Koski, L.; Peterson, D.G.; Nandula, V.K.; Ray, J.D.; Bond, J.A.; Shaw, D.R. Multiple Herbicide–Resistant Junglerice (Echinochloa colona): Identification of Genes Potentially Involved in Resistance through Differential Gene Expression Analysis. Weed Sci. 2018, 66, 347–354. [Google Scholar] [CrossRef]
- Kumar, V.; Jha, P. First report of Ser653 Asn mutation endowing high-level resistance to imazamox in downy brome (Bromus tectorum L.). Pest Manag. Sci. 2017, 73, 2585–2591. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.G.; Chen, Y. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 2007, 43, 649–650. [Google Scholar] [CrossRef]
- Feng, Y.; Yuan, G.; Zhang, Y.; Dong, L.; Li, J. Mechanisms of Resistance to Pyroxsulam and ACCase Inhibitors in Japanese Foxtail (Alopecurus japonicus). Weed Sci. 2016, 64, 695–704. [Google Scholar] [CrossRef]
- Kruger, N.J. The Bradford Method for Protein Quantitation; Methods in Molecular Biology: Clifton, NJ, USA, 1994; Volume 32, pp. 9–15. [Google Scholar]




| Herbicide | Manufacturer/Suppliers | Application Rate (g a.i. ha−1) |
|---|---|---|
| Metsulfuron-methyl | Jiangsu Tianrong Group Co., Ltd., China | 0.19, 0.38, 0.75, 1.50, 3.00, 6.00, 12.0, 24.0, 48.0, 96.0, 192.0, 384.0 |
| Chlorsulfuron | Zhejiang Research Institute of Chemical Industry, China | 0.59, 1.17, 2.34, 4.69, 9.38, 18.75, 37.50, 75.00, 150.00, 300.00 |
| Pyribambenz isopropyl | Zhejiang Research Institute of Chemical Industry, China | 2.3, 4.7, 9.4, 18.8, 37.5, 75.0, 150.0, 300.0 |
| Monosulfuron | Zhejiang Research Institute of Chemical Industry, China | 1.41, 2.81, 5.63, 11.25, 22.50, 45, 90, 180, 360, 720, 1440 |
| Primer | Sequence (5′-3′) |
|---|---|
| ALS569-1246-F | TCACCAAGCACAACTAC |
| ALS569-1246-R | CTCATGCCACGAACTAA |
| LAD-1 | ACGATGGACTCCAGAGCGGCCGCVNVNNNGGAA |
| LAD-2 | ACGATGGACTCCAGAGCGGCCGCBNBNNNGGTT |
| LAD-3 | ACGATGGACTCCAGAGCGGCCGCVVNVNNNCCAA |
| LAD-4 | ACGATGGACTCCAGAGCGGCCGCBBNBNNNCGGT |
| AC1 | ACGATGGACTCCAGAG |
| ALS569LB-0a | GATGCAGAGCAGCCACCGCCAACATAAA |
| ALS569LB-1a | ACGATGGACTCCAGTCCGGCCGTGGCTTGGGCAGGCGGGCAATGTAC |
| ALS569LB-2a | TGCTGGATGTCTTTGGGGAT |
| ALS110LB-0a | GGATCCCAGTCAGCTCAACAAATC |
| ALS110LB-1a | ACGATGGACTCCAGTCCGGCCGAGGAAGAAGGCTTCCTGAATGAC |
| ALS110LB-2a | GACACCGCGGAGCACCTG |
| ALS1246RB-0a | TTATGTTGGCGGTGGCTGCTCTGCATC |
| ALS1246RB-1a | ACGATGGACTCCAGTCCGGCCTTGCATTTGGTGTGCGGTTTGATGAT |
| ALS1246RB-2a | CAAGAACAAGCAGCCGCATG |
| PfALS-F | CTCACCCAAACCCTCG |
| PfALS-R | GCACTTGTCGGTCATGTAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, X.; Wu, H.; Zhang, J.; Yang, Y.; Tang, W.; Lu, Y. Resistance Mechanism to Metsulfuron-Methyl in Polypogon fugax. Plants 2021, 10, 1309. https://doi.org/10.3390/plants10071309
Yu X, Wu H, Zhang J, Yang Y, Tang W, Lu Y. Resistance Mechanism to Metsulfuron-Methyl in Polypogon fugax. Plants. 2021; 10(7):1309. https://doi.org/10.3390/plants10071309
Chicago/Turabian StyleYu, Xiaoyue, Hanwen Wu, Jianping Zhang, Yongjie Yang, Wei Tang, and Yongliang Lu. 2021. "Resistance Mechanism to Metsulfuron-Methyl in Polypogon fugax" Plants 10, no. 7: 1309. https://doi.org/10.3390/plants10071309
APA StyleYu, X., Wu, H., Zhang, J., Yang, Y., Tang, W., & Lu, Y. (2021). Resistance Mechanism to Metsulfuron-Methyl in Polypogon fugax. Plants, 10(7), 1309. https://doi.org/10.3390/plants10071309







