Abstract
Fruit trees provide essential nutrients to humans by contributing to major agricultural outputs and economic growth globally. However, major constraints to sustainable agricultural productivity are the uncontrolled proliferation of the population, and biotic and abiotic stresses. Tree mutation breeding has been substantially improved using different physical and chemical mutagens. Nonetheless, tree plant breeding has certain crucial bottlenecks including a long life cycle, ploidy level, occurrence of sequence polymorphisms, nature of parthenocarpic fruit development and linkage. Genetic engineering of trees has focused on boosting quality traits such as productivity, wood quality, and resistance to biotic and abiotic stresses. Recent technological advances in genome editing provide a unique opportunity for the genetic improvement of woody plants. This review examines application of the CRISPR-Cas system to reduce disease susceptibility, alter plant architecture, enhance fruit quality, and improve yields. Examples are discussed of the contemporary CRISPR-Cas system to engineer easily scorable PDS genes, modify lignin, and to alter the flowering onset, fertility, tree architecture and certain biotic stresses.
Keywords:
mutagenesis; TILLING; genome editing; targeted mutation; CRISPR-Cas; transgene-free; fruit trees 1. Introduction
Conventional breeding has been the sole source of genetic improvement in fruit crops for various traits. Classical approaches to introduce a promising trait in an elite cultivar require the introgression of related alleles through multiple generations of selection. For example, the introduction of a disease-resistance trait into a high yielding cultivar is commenced by crossing it with a disease-resistant cultivar, followed by recurrent backcrossing with the elite parent to sustain the genetic potential of the elite cultivar besides conserving the newly introduced resistance allele. Usually, the whole process encompasses several generations to stabilize the resistance allele in the elite background. Fruit crop breeding have certain limitations, which may include outcross reproduction, prolonged juvenility, and enormous genome landscapes [,]; therefore, it requires decades to improve such traits. The obligate outcrossing nature of fruit trees amalgamates classical breeding for genotypic and phenotypic traits. A relevant example to illustrate this dilemma is the development of resistance to apple scab. Hough et al. [] conducted a wide range of crosses between an elite apple cultivar and a genetically compatible wild-type cultivar as the source of resistance to apple scab. However, over several decades of continuous breeding, the resultant cultivars lost the fruit quality traits []. The application of marker-assisted selection, such as marker-assisted breeding (MAB), marker-assisted selection (MAS) and genome-wide association mapping (GWAS) for quantitative trait loci (QTLs), may contribute to shorten the selection process, but not bypass the generations of backcrossing []. For example, apples, cucumbers, mandarins, peaches, and strawberries have been substantially improved [,,]. Fast-track breeding approaches may possibly overcome extreme juvenility in fruit trees via the transgenic expression of the desired genes. The breeding time for fruit trees can be shortened to one-fifth of the conventional crossbreeding approaches []. For example, the apple cultivar ‘Pinova’ was transformed to impart early flowering by expressing a MADS-box gene from Betula pendula []. In another study, null segregants of fire blight and apple scab resistant apples were generated within seven years []. Similarly, Endo et al. [] successfully substituted the genetic background of mandarin through an integrated transgenic approach and MAS to transfer CTV resistance from a transgenic trifoliate orange. However, to obtain the null segregants, the transgene should be segregated out from the elite parental background through backcrossing with the recurrent parent. The detachability of the T-DNA transgene can be confirmed through comparative genomic hybridization (CGG) and next-generation sequencing (NGS) approaches []. In fast-track breeding, MAS plays a critical role to increase the selection efficiency in the backcrossed progenies.
Several new strategies were introduced in the middle of the 20th century to enrich the genetic diversity of fruit trees. Mutagenesis has been used to facilitate plant breeding since the 1920s with the discovery that mutations induced with physical (gamma irradiation) or chemical mutagen treatment can be inherited []. Importantly, with the discovery of X-rays, a subsequent series of induced mutations were conceptualized in plants and animals (Figure 1). In 1934, the first commercial mutant tobacco variety was produced []; since then, mutant crop cultivars have been continuously registered globally (Figure 2A,B). It was not until 1963 that the first mutated apple cultivar “Mori-hou-fu 3A” was developed in Japan through gamma rays. The following year, a sweet cherry (Prunus avium L.) cultivar “Compact Lambert” was developed in Canada (Table 1). The use of chemical mutagen “EMS” was successfully applied for mutating apples to develop another mutant cultivar “Belrene” in France in 1970. The application of physical mutagens was also successful in tree breeding. For example, grapefruit (Citrus × paradisi) cultivar “Rio Red” and clementia (Citrus celementina) cultivar “Nero” were developed using thermal neutrons and fast neutrons in 1970 and 2006 in USA and Spain, respectively. Various hybridization methods were also developed to produce hybrids between sexually incompatible species by disrupting the meiotic cell division to form polyploids, followed by the restoration of meiosis. Additionally, hybridization approaches also included chromosomal additions/subtractions or the fusion of protoplasts from sexually incompatible species []. The genetic background of the elite crop cultivars was further broadened through chemical or physical mutagenesis to increase the desirable alleles in the elite lines.
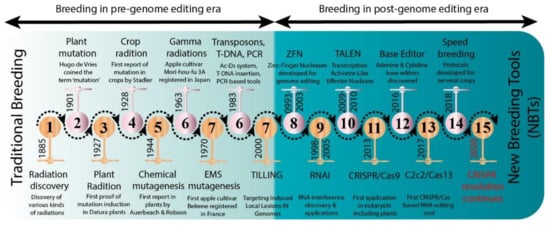
Figure 1.
Historic timeline for mutagenesis in plants.
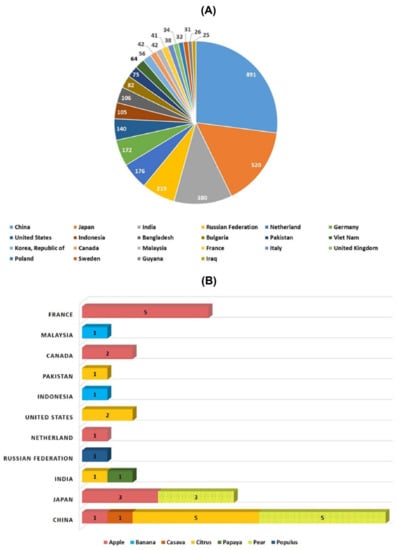
Figure 2.
Number of mutant varieties released in top 22 countries (A) and number of mutant tree varieties of assorted fruit species in selected countries (B).

Table 1.
Country-wise varietal approval developed by mutagen treatment.
The advent of plant transformation approaches including Agrobacterium-mediated, particle bombardment or electroporation-mediated and chemical transfections through protoplasts allowed the development of transgenic plants with the specific genetic constructs from any biological source. Although these methods involve in vitro culturing, many fruit tree species are recalcitrant to transformation and regeneration. However, fruit crops amenable to in vitro culturing were successfully transformed to directly introgress new genes without recurrent backcrossing []. However, the resulting transgenic plants faced regulatory complexities and gave rise to a dichotomy between the product and process regulatory framework []. These legalities have some practical implications because even if the gene transfer is intraspecific (involving plants from the same species), it still left some cargo of a transgene such as the remnants of the genetic markers or parts from the bacterial plasmid itself or the T-DNA of A. tumefaciens [].
Genome editing (GE) technology has revolutionized fruit crop breeding []. GE encompasses three types of specific site-directed nucleases (SNDs)—i.e., SDN-1, SDN-2, and SDN-3—to cause double-stranded breaks (DSBs) at pre-defined genome targets. The major advantage of SDNs is the targeted DNA cleavage and the subsequent use of cellular machinery to introduce the desired change during the repairing processes. These DSBs are imprecisely repaired by the cell DNA repair mechanism and cause insertion and deletion (indels) mutation over all dysfunctions of the gene of interest without introducing any foreign element into the genome. The entire process is well-regulated and, at the sequence level, the indel mutants cannot be distinguished from natural variations and/or irradiated or chemical mutants []. Four SDN-based GE techniques include meganucleases, zing-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the most recent clustered regularly interspaced short palindromic repeat/CRISPR-associated proteins (CRISPR/Cas), respectively. These GE techniques are based upon biological molecules with a DNA-binding domain and the cleavage activity (through nucleases). The mutants developed with chemical or physical mutagen treatment are usually hemizygous and their homozygosity is achieved through either filial segregations or recurrent backcrosses to fix the introduced mutations. Nevertheless, this is not a case with site-directed or targeted mutagenesis using GE, and GE shared another edge that is its multiplexing capabilities—i.e., simultaneous targeting of multiple genes or copies of a gene. This characteristic can be extremely useful to target homologous genes in polyploid fruit trees. The recent advancements in GE to substitute a single nucleotide allows individual base swapping in a DNA strand []. These developments in GE can help to overcome the GMO regulatory frameworks because it leaves no trace of a transgene or exogenous source in the targeted genome. Moreover, there is evidence that the remnants of Agrobacterium T-DNA have a role in the evolution of some plant families []. Thus, the boundary between natural and engineered crop species may become more blurred after such evidence and ultimately gain the attention of the scientific community to revise the regulatory framework, at least for GE crops.
2. Tree Breeding under Climate Change
The response of trees to any global climate change scenario is a pressing question for natural vegetation and man-made plantations []. Climate change is a major threat to tree plantations due to fluctuations in annual precipitation, drought, heat, salinity, and enhanced insect infestations []. Investigations to explore the ability and genetic basis of adaptation to global climate change in ecologically and industrially distinct tree species to cope with abiotic and biotic stresses are key research lines in plant science []. However, this knowledge has rarely been translated into conserving the genetic resources or bringing the genetic improvements to woody perennials.
The objective of most tree breeding programs is to gradually improve tree populations through recurrent selection cycles and verifications []. Traditionally, tree breeding mostly relies on phenotypically selecting superior candidates from the natural or planted stands. It constitutes the base population and further selection builds a pool of selected population with elite donors. Furthermore, these selected populations are then tested for progeny trials and the reselection of parents []. However, such selections may cause genetic erosions in the overall populations due to inbreeding depression. The production of hybrids and subsequent backcrossing may accelerate classical breeding with the aim of harnessing heterotic effects by virtue of dominance or over-dominance, tree adaptability and increased yield []. Among other potential applications, hybrid breeding has been widely applied to maximize the tree crown perimeter, tree height, conferring resistance to Fusarium spp. [], and to chestnut blight from wild donor tree plants into American chestnut populations [].
The most promising alternative approach towards tree breeding is molecular- marker-assisted selection (MAS) and molecular-assisted breeding (MAB) []. MAS and MAB tools can be effective in pyramiding simple Mendelian traits regulated by a few genes but have limited utility for selection against complex genetic traits in trees []. Moreover, MAS and MAB cannot be effective due to fluctuations in allelic frequencies over generations and therefore cannot explain genetic variations for complex traits []. To circumvent these limitations, the use of the genomic selection (GD) approach is suitable rather than phenotypic selection-based traditional breeding using MAS and MAB. Despite having a relatively short history, the technique has been successfully implemented in plant breeding. It can substantially reduce the long breeding cycles for tree breeding and positively enhance the genetic gain over time []. In the GS approach, a large number of molecular markers are used to analyze the cumulative effects of QTLs evenly distributed over the genome. Therefore, it makes GS much more efficient for tree breeding due to the possibility to assess the individual genomic estimated breeding value (GEBV) of a single plant. It involves four basic steps: (a) phenotyping and genotyping of the selected individuals from a breeding population, (b) generation of genomic prediction models, (c) model validation on the test population, and d) prediction of GEBV for non-phenotyped individuals and further selection []. Unlike MAS, there are no pre-requisites in GS for a prior information about marker linkage, or QTL localizations in the genome and their relative phenotypic effects [].
3. Mutagenesis as a Source of Genetic Variability in Tree Plants
Genetic improvement through conventional breeding necessarily requires recurrent selection cycles in fruit trees [] (Figure 3). However, a major limitation is the large number of crosses and the development of subsequent filial generations. This is even more challenging in fruit trees such that recurrent selection may take decades of continuous breeding efforts []. The lengthy breeding process can be accelerated in fruit frees with more advanced techniques such as MAS and GWAS for QTLs []. For example, many quality- and yield-related traits have been improved in apple, banana, mandarin, peach, and strawberry through conventional breeding coupled with mutagenesis, MAS, genetic engineering, MAB and others [,,]. The genetic improvements in fruit trees are, however, progressing at a slower pace, but the availability of pangenomes, broader understanding of genotypic and phenotypic interactions and fast-track breeding may hasten the development of fruit tree cultivars with better genetic makeup.
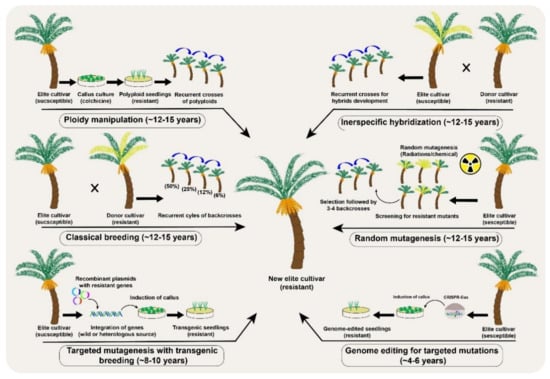
Figure 3.
A comparative analysis of different conventional and the new breeding tools (NBTs) to modify desirable genetic modifications in a date palm (Phoenix dactylifera L.) fruit crop.
Genetic improvement through conventional breeding is limited to sexually compatible crop plants []. Nevertheless, the genetic diversity of self-incompatible plants can be increased by mutagen treatment (physical or chemical) to induce new mutations in cultured cells, seeds, seedlings, or sometimes whole plants. Random mutations are preferred in seeds because the small number of cells in the developing embryo makes mutagenesis much easier and produces fewer chimeric plants []. During somatic mutations, a fewer number of cells or limited sectors in the apical meristem are affected, creating chimeric mutated plants. Such sectorial mutations involve genetic differences in either one or two layers of cells and is more frequent in vegetatively propagated fruit trees []. An effective way to dissociate chimerism in vegetatively propagated plant is through organogenesis or embryogenesis [,]. The mutation frequency and population structure of the mutants directly depend upon the type of mutagen and the time of exposure. Irrespective of the used mutagens, the ultimate induced mutations are random and therefore require a large screening population to identify the desired mutants []. Mutation breeding is advantageous over conventional breeding because it precludes segregation progenies while improving the genetic make-up during selection cycles.
High energy X-ray radiations were the earliest mutagens used to induce desired traits in fruit trees []. Currently, X-ray mutagenesis has been either replaced or supplemented with other more advanced physical mutagens such as fast neutrons, ionizing radiations and gamma rays. Besides bringing about beneficial mutations from single-nucleotide replacements to chromosomal aberrations, these mutagens may trigger DNA damage directly or indirectly in the form of oxygen radicals []. Physical mutagenesis has been successfully used to improve a number of traits in fruit trees—for example, improved heat tolerance in pineapple [], self-fertile in cherry fruits, fruit color in apple, bunch size in banana, short-statured papaya plants, disease resistant pear and growth earliness in grapevine [].
Among the chemical mutagens, ethylmethanesulfonate (EMS) is the most widely employed alkylating agent in fruit crops [], including banana and peach []. However, it is not suitable for vegetatively propagated fruit trees and perennial allogamous fruit trees because of their heterozygous genomes and prolonged life cycle. Although, chemical mutagens are extremely efficient in inducing desirable mutagenesis in whole plants or seeds, it is not recommended for tissue-cultured plants due to their extreme toxicity []. Chemical mutagens predominantly cause hemizygous point mutations and successive backcrosses are necessary to obtain a homozygous line and to stabilize the mutated gene of interest []. On the other hand, physical mutagenesis has a high risk of a collateral effect on non-targeted genes because the impact of physical mutagens produces multisite mutations of various sizes. For example, the use of fast neutron mutagenesis causes large deletions in the genome besides translocations and chromosomal loss [,]. Chemical mutagens are more affordable; however, these carry serious health and environmental risks. Moreover, chemically induced mutations are genetically less stable than physical mutations.
Polyploidy is another interesting natural phenomenon in plant evolution, adaptation, and speciation, which can also be induced using colchicine, for genetic improvements. Colchicine application induces autopolyploidy by blocking mitosis without interfering with DNA replication and ultimately doubles the chromosome numbers (Figure 3). The generation of triploid dessert apple and tetraploid grapevine cultivars are successful examples of autopolyploids in fruit crops []. Interspecific hybridizations have also been tested in some citrus cultivars, including the formation of natural hybrids []. However, as in conventional breeding, if the hybrids are fertile in perennial fruit trees, multiple backcrosses are still needed to remove the undesirable genetic background of the recessive parent. For example, scab resistance in apple took more than 40 years [], and the enhancement of sugar and antioxidants levels in elderberries took at least 10 years through the interspecific hybridization approach [].
Somaclonal variation is a natural phenomenon occurring during in vitro tissue culturing and can produce useful genetic variations in plants []. It includes DNA-related genetic or epigenetic variations, which induce phenotypic changes distinguishable from the original parent. Major causes include but are not limited to prolonged in vitro culturing, tissue culturing media composition, the presence of phytohormones and certain other mechanical factors during culturing []. Somaclones can be detected through morphological assessments of the off-type regenerants, biochemical response of explants, fingerprinting with protein or isozymes-based markers, and cytogenetic assessment [,]. In addition, more advanced DNA- or transposon-based molecular markers [] and the use of next-generation sequencing (NGS) screening have also been successfully applied to detect somaclonal variations in fruit tree breeding.
TILLING as a Powerful Tool in Mutation Breeding
Numerous significant genes from older mutant cultivars continue to serve as a foundation for modern gene pools in commercial cultivars. Nonetheless, the burden of unwanted genetic mutations and the development of new breeding tools (NBTs) have had an effect on the use of random mutation techniques []. Recent advancements in screening methods enable the detection of SNPs and complex traits at the molecular level, which are otherwise difficult to discern with conventional screening methods. The utilization of mutagenesis underwent a huge change with the development of TILLING (targeting induced local lesions in genomes) as a high-throughput mutant screening technique to identify point mutations at a specific locus in the mutated genome []. The TILLING technique redirected mutation breeding away from laborious forward genetics approaches to reverse genetics approaches, allowing plant breeders to detect mutations in known genes. Furthermore, TILLING has been accompanied with the more advanced next-generation sequencing (NGS) techniques to provide more practical solutions to bypass extensive mutant screening for the selected genes [].
The major mutation screening methods in TILLING include celery nuclease (CEL I) [], high-resolution melting (HRM) [] and NGS []. The mismatch-specific CEL I method is a popular TILLING technique, which is coupled with the LI-COR gel analyzer system. The HRM incorporates the PCR technique in which the monitoring of dsDNA product is monitored with a dsDNA-specific fluorescent dye followed by the formation of a high-resolution melting curve. The more advanced NGS technique has further facilitated the mutant screening in a TILLING population through whole-genome sequencing, de novo assembly and resequencing tools.
The basic procedure of TILLING includes mutation induction through chemical, physical or biological agents to produce an M1 population. These M1 plants are then allowed to self-pollinate and generate M2 plants. Total genomic DNA is isolated and subjected to eightfold DNA pooling followed by PCR amplification of the gene of interest. The recurrent heating and cooling steps form heteroduplexes, which are then incubated with CEL I endonuclease to cleave mismatches in these heteroduplexes. The cleaved DNA products are separated on a denaturing gel electrophoresis and the fluorescence is detected with a LI-COR DNA analyzer. The induced mutations are then verified by sequencing of the polymorphic individuals, respectively []. Although the CEL I -based TILLING platform has been widely used, the critical steps such as enzymatic digestions, cloning and gel electrophoresis make it time consuming. Moreover, insufficient genome sequence information of many plant species affects the efficacy of this TILLING platform []. Contrarily, the HRM-based TILLING offers more accurate, sensitive, and cost-effective mutant screening through PCR and analysis of the DNA melting curve. Nevertheless, detection of small insertions and deletions is difficult and limited to amplicons with a size <450 bp with HRM []. The NGS-based TILLING platform is comparatively a potential screening method with more accurate mutant screening. However, the high cost, the generation of a large sequencing dataset and the requirement of sophisticated bioinformatics tools still pose major challenges to its adoption in studying the genetics and genomics of mutagenic studies [].
4. Genomics and Genetic Engineering Perspectives of Trees
The genetic improvement of the tree plant genome can be accelerated through two distinct approaches: MAB through quantitative trait loci (QTL) mapping, and direct gene transfer through genetic engineering. The whole genomes of many tree plants have been completely sequenced; consequently, comprehensive genetic architecture of useful genetic traits are now available, which can be helpful for marker-assisted breeding, MAB [,]. The availability of such datasets can widely assist in genetic expression, and functional and comparative genomics. Moreover, recent developments in –omics and NGS technologies and, in parallel, more advanced bioinformatics tools, can expedite in-depth molecular studies in trees [,]. The transcriptomic, proteomic and metabolomics data sets of woody plants are dynamically bridging the gaps between tree genomes and genetic expression studies.
Genetic transformation can be improvised by inserting single or multiple genes directly into the elite background across the species or genus without long cycles of selections and screening [,], e.g., herbicide tolerance in populous []. The first application of genetic engineering in fruit trees was in papaya when papaya varieties ‘Sunset’ and ‘Kapoho’ were genetically modified by inserting the capsid protein (CP) gene of papaya ringspot virus (PRSV) to confer viral resistance. Consequently, the first transgenic papaya cultivar was developed in 1998 []. Recently, the USA approved a non-browning arctic apple cultivar [,]. Several other genetic traits for fruit quality, tree physiology and abiotic stress tolerance have been successfully engineered for transgenic apple, banana, papaya, and pineapple [,]. Current transgenic fruit trees approved in the USA include papaya against PRSV, plum against plum pox virus (PPV) [], apple with the non-browning trait [] and pineapple cultivar ‘Pinkglow’ []. Transgenic papaya plants have been successfully engineered to alter elite traits related to tree growth, nitrogen metabolism, lignin contents and abiotic stress tolerance [,]. Moreover, resistance in papaya was also conferred against phytophthora blight, papaya dieback disease (PDBD) and papaya ringspot virus (PRSV) in several studies []. Among non-transgenic approaches, dsRNA-mediated protection strategies have also been practiced in papaya against PRSV []. Similarly, eucalyptus species have also been genetically transformed to introduce genes from endogenous or heterologous sources to modify their salt tolerance status and secondary cell wall constituents []. Many pine softwood tree species have also been utilized for transgenic developments for various traits [].
5. Genome Editing in Precision Breeding
Precision breeding techniques encompass a broad range of technologies that enable the introduction of genetic variation into a plant genome. It combines and utilizes a variety of innovative technologies to engineer desired traits in plants in order to drive new agricultural advancements. In the recent years, the execution of contemporary genome-editing technologies has enabled researchers to easily, swiftly, and economically introduce site-specific modification at the desired DNA sequences in a wide range of cell types and organisms. The CRISPR-Cas based genome-editing technologies can be customized easily to target a desired locus and has brought an unparalleled revolution in agricultural sciences and precision plant breeding [,]. CRISPR-Cas tools can foster crop resilience and reduce chemical crop protection with a strong environmental and public health impact on crop production.
5.1. Principle and Types of CRISPR-Cas Systems
CRISPR-Cas systems are prokaryotic immune systems that protect cells by selectively and specifically cleaving the nucleic acids of the invaders, such as viruses and plasmids []. Since the discovery of the CRISPR system, several new versions of CRISPR-Cas have appeared []. Spurred by the interference ability of CRISPR-Cas systems, they are classified into two major classes—I and II—based on the structural variation and arrangement of Cas genes. Both classes are further sub-divided into six main types related to the type of nucleic acid they target []. Another benchmark difference between these two classes is the number of nucleases; class I has a single and class II has multiple effector nucleases. Makarova et al. [] appraised the evolutionary classification of the CRISPR-Cas systems, especially concerning class II and its variants. Among them, the most utilized Cas9 endonucleases are isolated from various microbes such as Francisella novicida, Staphylococcus aureus and Streptococcus pyogenes and belong to class II type II systems [,].
The Cas endonuclease function by accompanied CRISPR-RNAs (crRNAs) and trans-activating crRNAs which can recognize foreign nucleic acid sequences [,]. The working principle of CRISPR-Cas systems, as a prokaryotic immunity, is based on the integration of the invader’s nucleic acid fragments into the CRISPR locus during infection. Once integrated, the subsequent infections activate the transcription of the integrated fragments and are then recruited by the Cas to cleave the invader’s genome. Different types of Cas nucleases along with their variants, and their specificities, characteristics, and PAM recognitions sites have been summarized (Figure 4, Table 2). The applications of Cas9 have been increased immensely after the invention and customization of single-guide RNA (sgRNA) []. Cas9 proteins contain two unique nuclease domains: RuvC and HNH. The former domain cleaves the non-target DNA strand, while the latter domain cleaves the target DNA strand complementary to the sgRNA. The sgRNA is usually comprised of a unique 20 bp sequence and contains a short (usually 2–6 bp in length) essential DNA sequence at the 5′ end and designated as the “protospacer adjacent motif” (PAM) [,]. A sgRNA recognizes and binds to the target sequence, and then Cas nuclease induces double-stranded break (DSB) at the target DNA. After the induction of DSB, cell genome repair mechanisms become activated and repair the induced DSB either via non-homologous end joining (NHEJ) or high-fidelity homology-directed repair (HDR) []. During NHEJ repair, DSB is directly re-ligated without any homologous template, resulting in insertions, deletions (InDels) or substitutions. On the other hand, HDR is a precise repair pathway that can utilize either an endogenous or an exogenous DNA segment as a template to repair the DSB. HDR may introduce novel alleles, correct existing mutations, or insert a new sequence of interest [,].
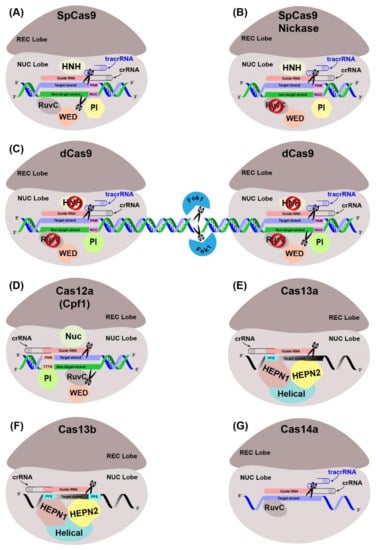
Figure 4.
Different Cas protein variants potentially useful for genome editing in perennial trees. (A) The CRISPR-Cas9-based system, generally comprised of Cas9, the crRNA, and tracrRNA. It entails hybridization and binding of this triad to the complementary sequence after recognizing a PAM sequence. The recognition lobe (REC) recognizes the crRNA:tracrRNA:target DNA complex, whereas the PAM sequence is intrinsically recognized by the PAM interaction domain (PI) of Cas protein. The HNH and RuvC domains of NUC lobe cleave target and non-target strands upstream of the PAM, respectively, by producing DSB. (B) The SpCas9 is engineered to reduce off-target activities by introducing a point mutation in RuvC domain, which results in only a single-stranded break (SSB). (C) RNA-guided Fok1 nuclease was fused with an inactive dead Cas9 (dCas9) to enhance on-target efficiency. (D) CRISPR-Cas 12a system uses a single crRNA. The Cas12a:crRNA duplex binds to the complementary target sequence with the help of 23–25 nt long gRNA. The Nuc domain cleaves the target strand at ~18 nt downstream to the PAM and the RuvC domain cleaves the non-target strand at ~23 nt downstream of the PAM. (E) CRISPR-Cas13a is engineered to target ssRNAs. Instead of a PAM sequence, the binding of Cas13a:crRNA duplex to the target site is mediated via a protospacer flanking sequence (PFS). The REC lobe recognizes the Cas13a:crRNA duplex and the ssRNA substrate is cleaved in a sequence-specific manner through HEPN1 and HEPN2 domains. (F) CRISPR-Cas13b has a distinct suppressor and enhancer Cas genes, which are expressed under two PFSs to target ssRNAs. (G) contemporary CRISPR-Cas 14a is a single effector system involving a Cas14a protein associated with crRNA and ~130 bp tracrRNA. The hybridization of crRNA and tracrRNA complex is accomplished independent to PAM and RuvC domain cleave the target ssDNA.
The class II single Cas protein systems are comprised of Cas9 (type II), Cas12 (type V), Cas13a–d (type VI) and Cas14a–c (type V-F) [,]. Among these Cas proteins, Cas9 and Cas12 nucleases target and cleave dsDNA, while different Cas13 (type VI) proteins such as Cas13a (C2c2), Cas13b (C2c6), Cas13c (C2c7) and Cas13d target and cleave the target RNA.
Type I CRISPR systems generally consist of different nucleases, such as Cas5, Cas6, Cas7, Cas8 and Cas11, in a different combination and target dsDNA. Type II consists of Cas5, Cas6, Cas7 and Csf1. Type III is the most abundant group of nucleases present in nearly a quarter of bacterial species and include Cas5, Cas6, Cas8 and Cas10 nucleases []. This type is further categorized into two subtypes based on Cas effectors; type III-A and type III-B and include Cas10-Csm and Cas10-Cmr. Another unique feature of type-III systems is their ability to target the nascent mRNAs and DNA sequence in transcriptionally active complexes without the requirement of PAM sequence.
5.2. CRISPR/Cas Based GE Strategy in Trees
The use of CRISPR-Cas systems in trees holds unmatched potential for resilience to biotic and abiotic stresses. Until now, these systems have been executed in different trees such as apple, banana, citrus, pear, and others (Table 3). Most of the CRISPR work in trees has been focused on the editing of the easily scorable Phytoene Desaturase (PDS) gene, which induces albino phenotypes due to reduced photosynthesis and carotenoid synthesis.
5.3. CRISPR-Mediated Genome Editing in Apples
Apple (Malus × domestica) ranks as one of the most produced temperate fruits in the world, with approximately 87 million tons of worldwide production in 2020 []. The first successful execution of the CRISPR-Cas system was achieved in the rootstock of the first generation of transgenic apple plants (cv. JM2) by editing the PDS gene with up to 31.8% editing efficiency []. The CRISPR-mediated GE of DIPM-1, DIPM-2, and DIPM-4 genes was achieved in the apple protoplast; however, the indel mutagenesis efficiencies were quite low and in the range of 0.5 to 6.9%, via DSBs []. Just a few years later, highly efficient, accurate and DNA-free GE procedures were evaluated in the apple protoplast to modify the PDS gene, based on CRISPR delivery either directly as CRISPR–Cas9 RNP complex or via plasmid. Nonetheless, direct delivery of CRISPR–Cas9 RNPs has a superiority over the plasmid-mediated delivery; direct delivery took about two to three weeks, whereas the plasmid-mediated delivery procedure required more than three months []. Initially, Chevreau et al. [] appraised an adventitious and highly efficient Agrobacterium-mediated CRISPR-Cas transformation method in ‘Gala’ apples and demonstrated that the presence of a surfactant (Silwet L-77, 0.002 per cent v/v) in bacterial suspension increased the average transformation efficiency (mean 5.8% and maximum 30%) []. Subsequently, the same group targeted two easily scorable genes—PDS and Terminal Flower 1 (TFL1)—via CRISPR-Cas9 and a characteristic albino phenotype was observed in 85% and early flowering in 93% of the apple transgenic lines. Sequencing of target zones revealed a variable frequency of mutation, insertions were more frequent than substitutions or deletions, and biallelic chimeras were more prevalent []. To deal with the high frequency of chimeras, an adventitious regeneration step from the leaves of T0 transgenic apples was opted. The results yielded 99% truncated alleles of RT0 plants with ~67% of plants having less heterogeneous editing profiles than the T0 []. In the same study, a CRISPR-Cas9-based cytidine base editor was successfully executed to modify PDS and acetolactate synthase (ALS) genes in apples.
Erwinia amylovora, a causative agent of fire blight disease, is a leading constraint to apple production []. It triggers its infection via a DspA/E effector that binds to the apple susceptibility factor, MdDIPM4. The knockdown of MdDIPM4 via a heat-inducible CRISPR/Cas9-FLP/FRT system yielded an editing efficiency of 75% in 57 transgenic lines. Seven GE lines challenged with the E. amylovora exhibited a substantial reduction in susceptibility, and almost all of the challenged lines revealed successful elimination of the transgene upon exposure to heat. Another major threat to apple is Botryosphaeria dothidea, which triggers the expression of MdCNGC2 and defense-related genes including MdPR1, MdPR2, MdPR4, MdPR5, MdPR8, and MdPR10a in apples. The CRISPR-Cas9-mediated knockdown of MdCNGC2 led to a reduction in lesions caused by B. dothidea [].
Apart from induced mutagenesis, a reliable and efficient method for the identification of prevalent viruses/viroids was developed based on a CRISPR-Cas12 platform that could detect most of the viruses in one hour and is highly reliable [].
5.4. CRISPR-Mediated Genome Editing in Banana
Bananas are the fourth largest food crop around the globe, one of the most important commercial fruits in the tropics and cultivated in 130 countries [,]. Traditional breeding is challenging in banana due to its complex triploid genome and parthenocarpic nature [], but modern techniques such as CRISPR-Cas have opened new horizons to tackle such problems. Recently, the CRISPR-Cas system was used to target the conserved domain of two RAS-PDS genes by a common sgRNA in the embryonic cell suspension culture of banana cv. Rasthali. The regenerated plantlets exhibited a range of albino to variegated phenotypes at a 59% mutation rate []. The multiplexed CRISPR-Cas approach using polycistronic tRNA-gRNA system was employed in banana cv. Cavendish to target exon1 of the PDS gene, which yielded albino phenotypes associated with triallelic deletions or insertions of 19 regenerated plants at a 100% mutation efficiency []. The results of these studies demonstrated the feasibility of GE in banana via CRISPR-based targeted genome mutation.

Table 2.
The characteristics, specificities, and nucleic acid targets of different Cas proteins and their variants.
Table 2.
The characteristics, specificities, and nucleic acid targets of different Cas proteins and their variants.
| Cas Type | Organism | Size (Amino Acids) | Class/ Type | PAM Site | Altered PAM | Types of End | Mutations | Plants | References |
|---|---|---|---|---|---|---|---|---|---|
| SpCas9 | Streptococcus pyogenes | 1368–1424 | 2/II | NGG | -- | Blunt/ds or ss | -- | Many plant species | [,,,] |
| SpCas9 VQR | S. pyogenes | 1372 | 2/II | NGA | Yes | Blunt/ds or ss | D1135V/R1335Q/T1337R | Rice | [] |
| SpCas9 EQR | S. pyogenes | 1372 | 2/II | NGAG | Yes | Blunt/ds or ss | D1135E/R1335Q/T1337R | - | [] |
| SpCas9 VRER | S. pyogenes | 1372 | 2/II | NGCG | Yes | Blunt/ds or ss | D1135V/G1218R/R1335E/T1337R | Rice | [] |
| SpCas9 D1135E | S. pyogenes | 1372 | 2/II | NAG/NGA | Yes | Blunt/ds or ss | D1135E | - | [] |
| SpCas9 QQR1 | S. pyogenes | 1372 | 2/II | NAAG | Yes | Blunt/ds or ss | G1218R/N1286Q/I1331F/D1332K/ R1333Q/R1335Q/T1337R | - | [] |
| SpCas9-NG | S. pyogenes | 1372 | 2/II | NG | Yes | Blunt/ds or ss | R1335V/L1111R/D1135V/G1218R/ E1219F/A1322R/T1337R | Arabidopsis and rice | [] |
| SpCas9-HF1 | S. pyogenes | 1368 | 2-II | NGG | Enhanced specificity | Blunt/ds or ss | N497A/R661A/Q695A/Q926A | Arabidopsis and rice | [] |
| eHF1-Cas9 | S. pyogenes | 1368 | 2-II | NGG | Enhanced specificity | Blunt/ds or ss | N497A/R661A/Q695A/K848A/ Q926A/K1003A/R1060A | Rice | [] |
| HiFi Cas9 | S. pyogenes | 1368 | 2-II | NGG | Enhanced specificity | Blunt/ds or ss | R691A | Rice | [] |
| XCas9 | S. pyogenes | 1368 | 2-II | NG, GAA & GAT | Enhanced specificity | Blunt/ds or ss | A262T/R324L/S409I/E480K/ E543D/M694I/E1219V | Rice | [] |
| dCas9 | S. pyogenes | 1368 | 2-II | NGG | No | Blunt/ds or ss | D10A/H840A | Arabidopsis and rice | [,] |
| nCas9 | S. pyogenes | 1368 | 2-II | NG, GAA & GAT | Enhanced specificity | Blunt/ds or ss | D10A | Rice, tobacco | [,] |
| SaCas9 | Staphylococcus aureus | 1053 | 2-II | NNGRRT | - | Blunt/ds or ss | - | Tobacco, rice, Arabidopsis and citrus | [] |
| SaCas9-KKH | S. aureus | 1053 | 2-II | NNNRRT | Enhanced specificity | Blunt/ds or ss | E782K/N968K/R1015H | Tobacco, rice, Arabidopsis and citrus | [] |
| BlatCas9 | Brevibacillus laterosporus | 1092 | 2-II | NNNNCND | Staggered/ds | - | Maize | [] | |
| FnCas9 | Francisella novicida | 1629 | 2B-II | NGG | - | Staggered/ds | - | Arabidopsis | [] |
| Cpf1 (Cas12a) | Prevotella & Franscisella | 1300 | 2-V | TTTN | - | Staggered/ds | - | Many plant species | [,] |
| AsCas12a RR | Acidaminococcus | 1307 | 2-V | TYCV & CCCC | Yes | Staggered/ds | S542R/K607R | - | [,] |
| AsCas12a RVR | Acidaminococcus | 1307 | 2-V | TATV | Yes | Staggered/ds | S542R/K548V/N552R | - | [] |
| LbCas12a | Lachnospiraceae bacterium | 1228 | 2-V | TTTV | - | Staggered/ds | - | Many plant species | [] |
| LbCas12a RR | Lachnospiraceae bacterium | 1228 | 2-V | TYCV & CCCC | Yes | Staggered/ds | G532R/K595R | Rice | [] |
| LbCas12a RVR | Lachnospiraceae bacterium | 1228 | 2-V | TATV | Yes | Staggered/ds | G532R/K538V/Y542R | Rice | [] |
| FnCas12a | F. novicida | 1300 | 2-V | TTV, TTTV & KYTV | - | Staggered/ds | - | Rice | [] |
| FnCas12a RR | F. novicida | 1300 | 2-V | TYCV & TCTV | Yes | Staggered/ds | N607R/K671R | Rice | [] |
| FnCas12a RVR | F. novicida | 1300 | 2-V | TWTV | Yes | Staggered/ds | N607R/K613V/N617R | Rice | [] |
| Cas12b | Alicyclobacillus, Acidoterrestris, Bacillus Thermoamylovorans, A. acidiphilus | 1100–1300 | 2-VB | TTTN | - | Staggered/ds | - | Many plant species | [,] |
| Cas12X | Deltaproteobacteria | <1000 | 2-V | TTCN | - | Staggered/ds | - | - | [,] |
The presence of the integrated endogenic banana streak virus (eBSV) in the B plantain genome (AAB) is a limiting factor as it becomes activated under water stress conditions, rendering the plants unsuitable for breeding and dissemination. The eBSV was targeted via CRISPR/Cas9 at a 27% editing efficiency in regenerated plantlets, which remained asymptomatic after exposure to water stress, confirming eBSV obstruction at either transcriptional and/or translational levels []. To yield transgene-free GE bananas, PEG-mediated delivery of two different CRISPR-Cas9 systems—a CRISPR-Cas12a plasmid and a CRISPR-Cas9-RNP system—was evaluated in banana protoplast to target PDS genes. The results of deep amplicon sequencing revealed that the CRISPR/Cas9 system has higher editing efficiency compared to the other two systems used []. The MaACO1 gene regulates ethylene synthesis and fruit ripening in bananas. The disruption of MaACO1 via CRISPR-Cas9 in Musa acuminata (AAA group, cv. Brazilian) banana delayed fruit ripening both in the field and post-harvest storage conditions up to 60 days [].
The β-carotene-enriched banana cv. Cavendish was developed using a CRISPR/Cas9 after targeting the fifth exon of the lycopene epsilon-cyclase (LCYε) gene. Sequence analysis of the edited plants revealed multiple indels in the LCYε gene, up to sixfold enhanced accumulation of β-carotene, a severe reduction in α-carotene and lutein levels, without any substantial perturbed agro-morphological traits []. To examine the functions of five MaGA20ox2 genes, implicated in having a role in gibberellic acid biosynthesis and plant height, in banana cv. Gros Michel were edited via CRISPR-Cas9 using embryonic cell suspension cultures. The resultant 152 independent modified transgenic lines contained indels as a major mutation type, low transcription levels of these five genes and the modified banana plants exhibited a significant reduction in height compared to wild-types [].
5.5. CRISPR-Mediated Genome Editing in Citrus
Citrus is one of the top three fruit crops across the globe and a source of many nutrients, principally vitamin C. However, it is susceptible to a plethora of stresses, both biotic and abiotic. Due to its long juvenility, polyploidy, long crossing life cycle and high heterozygosity, conventional breeding techniques have often proved time-consuming and tedious.
The first reported execution of the CRISPR-Cas9 system to target the PDS gene in the leaves of sweet orange yielded a mutation efficient of 3.2 to 3.9% with no quantifiable off-targets [,]. Subsequently, the same researchers successfully employed CRISPR-LbCas12a to edit the PDS in citrus with improved editing efficiency []. Modification of the CsLOB1 canker susceptibility gene in Duncan grapefruit via CRISPR-Cas9 was achieved at a mutation frequency of 23.80 to 89.36% with no off-targets. The regenerated six edited plant lines were inoculated with the pathogen Xanthomonas citri ssp. citri, then two of the four edited lines exhibited comparable canker symptoms to wild grapefruit, while the remaining four remained asymptomatic at the beginning, but showed very mild symptoms at the later stage [].
5.6. CRISPR-Mediated Genome Editing in Papaya
Papaya is a tropical fruit of commercial importance due to its high nutritional and medicinal value. In 2019, papaya was grown on 462,552 ha, with a current total world production of 13,735,086 tons [].
The papaya produces a unique cysteine protease (papain), via regulating PpalEPIC8 gene to counter the invading oomycete, Phytophthora palmivora. Homozygous PpalEPIC8 mutants were produced via the CRISPR/Cas9, which suggested that PpalEPIC8 does indeed play a role in P. palmivora virulence by inhibiting papain []. Another similar study on P. palmivora characterized a glycoprotein, Ppal15kDa, of P. palmivora that is highly induced during infection in papaya plants. Six Ppal15kDa mutants produced through CRISPR/Cas9 having homozygous mutation completely lost the pathogenicity, while the heterozygous mutants exhibited varying levels of infection, suggesting that Ppal15kDa plays an important role in the normal development of P. palmivora infection. Overall, a unique component with a role in the pathogenicity and development of P. palmivora infection or possibly other Phytophthora spp. was demonstrated in this study [].
5.7. CRISPR-Mediated Genome Editing in Pear
The area under pear cultivation is 1,379,387 ha, with annual production of 13,919,075 tons, which includes both Asian pears (Pyrus sp.) and European pears (P. communis L.) [].
Charrier et al. (2019) knocked out TFL1 genes in pear, which led to early flowering in 9% of the transgenic lines. Sequencing of the target region of transgenic lines revealed that mutations were induced at varying frequencies and insertion mutation was dominant over deletions and substitutions. Nonetheless, the most frequently demonstrated edition pattern of TFL1 genes was biallelic chimeric. The high frequency of chimerism is a problem, and this was solved by including an adventitious regeneration step from leaves of T0 transgenic pears. In addition, CRISPR-Cas9 BE was executed to induce C-to-T base substitution in pears by co-editing ALS and PDS genes, which yielded albino and chlorsulfuron lines in pear [].
In the dwarf pear (Pyrus bretschneideri), the tree achieved enhanced yield; the loss-of-function mutant of the PbPAT14 gene was generated by the CRISPR/Cas9 system. Sequence analysis revealed that out of 22 dwarf yellow lines, six were homozygous mutants and had an elevated level of endogenous abscisic acid (ABA) [].
5.8. Mitigation of Off-Target Mutations in GE Trees
The mitigation of off-target mutations is critical in tree species breeding to bring genetic improvement without disrupting the genetic background of the parent tree. A systematic approach to the design, execution and delivery of the best results is required for successful implementation of the CRISPR-Cas system, without overloading off-targets. In the subsequent sections, key points are addressed.
5.8.1. GC Content of sgRNA
The gRNA structure and its GC contents play a key role in determining the specificity of the CRISPR-Cas system. Ideal GC contents of 40 to 60% in gRNA sequence form a stable DNA:RNA duplex, destabilize off-target binding and ultimately enhance on-target activity []. In addition, purine residues at the end of four nucleotides in gRNAs—in particular, guanine at the 20th position and cytosine at the 16th position—improve editing efficiency [,]. A positive correlation has been identified between PAM-proximate GC % and gRNA secondary structure.
5.8.2. gRNA Length and Mismatches
The length of the gRNA determines its functionality and the level of off-target activity. Different lengths, 16 to 20 nucleotides long, of gRNAs were evaluated for GE efficiency and off-targets, of which 17-nucleotides-long gRNAs yielded higher GE efficiency compared to 18- to 20-bp-long gRNAs, but 20-bp-long gRNAs did not exhibit any unwanted mutations [,]. Dead RNA off-target suppression (dOTS), the new strategy employing dead truncated gRNA, has resulted in reduced off-target activity and increased on-target activity by 40 times []. General guidelines for mitigating off-targets have been formulated as: (a) more than three mismatches within the first 7 to 10 bp of the PAM; and (b) gRNA bulges within the first 12 bp of the PAM, which should be avoided []. Lee et al. [] looked at off-target mutations in rice using four sgRNAs and found that the highest off-target mutation rate (67.5%) occurred in the presence of two mismatched bases between the target site and sgRNA; it became severely compromised (2.5%), with six mismatches. Using more than one mismatched RNA base pair to target a sequence can prevent unintended changes in rice plants.
5.8.3. Chemical Modification of gRNA
Chemical modification of gRNA has the potential to improve GE efficiency. A 40- to 120-fold reduction in off-target activity was observed after the incorporation of 20-O-methyl-30-phosphonoacetate into the gRNA ribose-phosphate backbone []. The modification of hairpin structure at 50 bp upstream of gRNA enhances the specificity of Cas proteins by reducing off-target effects by up to 55-fold [].
5.8.4. Concentration of Cas Protein/gRNA
Controlled and low expressions of Cas protein/gRNA could effectively reduce off-target levels. Compared to the constitutive (CaMV35S) promoter, Cas9 expression under an inducible (egg-cell) promoter yielded a high on-target efficiency in Arabidopsis plants []. Similarly, Cas9 expression under embryo-specific promoters (YAO) yielded improved GE efficiency in Citrus sinesis at the reproductive stage []. The expression of the Cas9 protein in monocots under the control of plant endogenous promoters resulted in higher on-target mutations than the constitutive CaMV35S promoter [,,,]. Likewise, comparative studies were conducted to assess the Cas9 expression under endogenous and constitutive promoters; the results showed that the endogenous promoter yielded improved heritability and on-target efficacy [,,,]. Soya bean promoter (U6-10) and Arabidopsis Ubi (AtUbi) promoter-mediated expression of Cas9 protein was investigated in Glycine max; a two- to fourfold improvement in on-target efficacy was achieved by the U6-10 promoter compared to the AtUbi promoter [].
5.8.5. Cas Protein Variants
Aside from the known Cas protein variants, several new versions have been developed through protein engineering to improve the on-target efficiency. Two of the most used Cas proteins—Cas9 and Cas12a—have been shown to be highly efficient, with Cas9 efficiency exceeding 90% and Cas12a efficiency at about 60% []. Two naturally occurring Cas9 variants—SaCas9 (S. aureus) and StCas9 (Streptococcus thermophilus)—recognize longer PAM sequences, including NNGRRT and NNAGAAW, respectively, that can ultimately enhance their on-target efficiency. SpCas9, however, demonstrated higher on-target specificity and expression levels compared to SaCas9 in A. thaliana []. Two engineered versions of the Cas9—SpCas9-VQR and SpCas9-EQR (Table 3)—were tested in plants for their on-target efficiency and found to be more efficient than the conventional Cas9 [].
Another simple yet robust approach to mitigate the off-targets is to use a Cas9 mutant paired with a nickase (HNH or RuvC-like). The ability of Cas9-paired nickase to reduce unwanted mutations is its main advantage over Cas9. Other variants of Cas proteins referred to as ‘deactivated/dead (d) Cas’ have been developed by mutating the nuclease domain, and these ‘dead’ variants have been widely used in GE []. The dCas protein variants bind to the target sequence to block transcription elongation []. The recently developed base editors (ABE and CBE) can convert G to A and C to T in the target genome, while the CRISPR system equipped with deaminases can regulate gene expression [].

Table 3.
Execution of CRISPR-Cas systems in different fruit trees (correct to alpha order of tree species).
Table 3.
Execution of CRISPR-Cas systems in different fruit trees (correct to alpha order of tree species).
| Tree Species | Gene Target | Trait Modified | CRISPR-Cas System | Reference |
|---|---|---|---|---|
| Apple | PDS | Albino phenotype | CRISPR-Cas9 | [,,] |
| DIPM-1 DIPM-2 DIPM-4 | Fire blight | CRISPR-Cas9 | [,] | |
| TFL1 | Early flowering | CRISPR-Cas9 | [] | |
| ALS | CRISPR-Cas9 | [] | ||
| CNGC2 | B. dothidea resistance | CRISPR-Cas9 | [] | |
| Detection of viruses and viroids | CRISPR-Cas12 | [] | ||
| Banana | PDS | Albino phenotype | CRISPR-Cas9 | [,] |
| PDS | eBSV resistance | CRISPR-Cas9 | [] | |
| PDS | Albino phenotype | CRISPR-Cas9 | [] | |
| PDS | Albino phenotype | CRISPR-Cas12a | [] | |
| ACO1 | Fruit ripening | CRISPR-Cas9 | [] | |
| LCYε | β-carotene | CRISPR-Cas9 | [] | |
| GA20ox2 | Gibberlic acid biosynthesis | CRISPR-Cas9 | [] | |
| Cacao | TcNPR3 | Resistance to Phytophthora tropicalis | CRISPR-Cas9 | [] |
| Citrus | PDS | Albino phenotype | CRISPR-Cas9 | [,] |
| LOB1 | Canker resistance | CRISPR-Cas9 | [] | |
| PDS | Albino phenotype | CRISPR-Cas12 | [] | |
| Papaya | Papain (PpalEPIC8) | Cysteine protease, P. palmivora resistance | CRISPR-Cas9 | [] |
| Ppal15kDa | P. palmivora resistance | CRISPR-Cas9 | [] | |
| Pear | TFL1 | Early flowering | CRISPR-Cas9 | [] |
| PDS and ALS | Albino and chlorsulfuron | CRISPR-Cas9 C-to-T BE | [] | |
| PbPAT14 | Dwarf and yellowing | CRISPR-Cas9 | [] |
5.9. CRISPR Delivery Techniques and Vectors
Delivery of the CRISPR-Cas system is one of the crucial elements for its successful execution. Several transformation techniques to deliver the CRISPR system into plants are practiced: Agrobacterium-mediated transformation, biolistic transformation, RNP-complex, polyethylene glycol (PEG)-mediated transformation, lipid and polymer transformation, and viral vectors.
PEG-mediated transformation of the CRISPR-Cas system was initially achieved in maize []. Since then, several plant species have been successfully transformed []. The main challenges of using PEG-mediated delivery systems are the isolation of suspension cells and protoplasts.
Agrobacterium-mediated transformation is the most common method of delivering the CRISPR-Cas system into plants. This technique has offered improved transformation efficiency rate (40% to 100%) compared to particle bombardment in the plant species [,,]. Agrobacterium-mediated transformation via the floral dip method has also been executed in A. thaliana [], Brassica rapa, flax, tomato, radish, Setaria viridis and wheat [,,,].
The second most common method of CRISPR-Cas transformation into plants is by biolistic means, which has been executed in a variety of plant species such as brassica [], maize [], potato [], soybean [] and wheat []. However, the regeneration of transformed tissues, optimization of selection pressure, time-consumption, less cost-effective and low transformation efficiency are the challenges associated with this technique. For example, in maize, merely 2.4 to 9.7% GE efficiency could be achieved via biolistic inoculation [].
The recently used RNP, a technique for achieving GE plants, includes apple [], banana [], brassica [], capsicum [], rice [], potato [], and lettuce []. In RNP-mediated delivery, Cas9 swiftly becomes degraded after cleaving the target site, thus reducing the off-targets vulnerability and GMO/ethical concerns. Six polyphenol oxidase genes of mushrooms were edited successfully via RNP-mediated CRISPR delivery, and transgene-free GE mushroom had a 30% reduction in the enzyme activity responsible for browning and also escapes US regulations []. This technique is useful for vegetatively propagated trees, where removing transgenes from GE plants via backcrosses is almost impossible.
Viral vectors, both DNA and RNA, have successfully conveyed the CRISPR/Cas9 system to plants. The DNA viruses employed include bean yellow dwarf virus (BeYDV, [,], wheat dwarf virus (WDV, [] and cabbage leaf curl virus (CabLCuV [], but also RNA viruses including tobacco rattle virus (TRV, []. BeYDV vectors yielded a 12-fold improved on-target efficiency in wheat, whereas WDV vectors yielded a 10-fold enhanced on-target efficiency in wheat. The TRV-based vectors lead to 15% fewer off-target mutations.
Several other delivery methods using CRISPR have been reported including cell-penetrating peptides [], DNA nanoclews [], Cas9En-arginine nano-assemblies [], and polyethylene imine (PEI)-based nanocarrier []. Future delivery methods based on lipids and polymers will shape CRISPR-Cas technology in the coming years.
5.10. Genome Editing Tools: Base, Prime and RNA Editors
The base editing tool is a new innovation in the CRISPR-Cas precise engineering toolbox. Such base editor (BEs) systems use facets of DNA modifying enzymes (such as deaminases) to substitute a nucleotide base. Various versions of CRISPR-Cas systems are available for all four transition mutations, such as C to T, G to A, A to G, and T to C. BEs are categorized mainly as CBEs, which can convert C to T; ABEs, which convert A to G; and RBEs, which convert A to I, or C to U. BEs are comprised of four main components—gRNA, nCas or dCas, a deaminase, and a uracil DNA glycosylase inhibitor (UGI). The function of the gRNA is to guide a CRISPR-Cas9 to bind to the target sequence, after which the Cas9 catalyzes the base conversion [], while UGI is a bacteriophage-derived 83-residue protein that blocks the uracil DNA glycosylase activity. During substituting a base via the CRISPR-Cas system, DSB is not induced, so the cell’s DNA repair mechanisms are not activated, resulting in considerably fewer off- and on-target indels [].
5.10.1. Base Editors
Adenine base editors (ABEs) yield much cleaner DNA products than cytosine-BE and they are less prone to insertions and have virtually no inversions. A·T to G·C conversions have been successfully mediated in various plant species [,,,]. In high-quality sequencing of ABE-edited wheat and rice target DNA, the ABE-P1 system led to no undesired off- and on-target base editing [,]. Similarly, four cotton genes for GhCLA and GhPEBP were targeted using a unique base editor Gossypium hirsutum (Gh) BE3, resulting in cleavage efficiencies ranging from 27 to 58% with only 0.1% off-target activity [].
An adenine base editor (Adenine base Editor14) based on nCas9 (D10A) that is guided by TadA:TadA7.10 heterodimer was engineered to achieve A·T to G·C conversion in OsMPK6, OsSERK2 and OsWRKY45 in rice with editing efficiencies of 16.7, 32.1, and 62.3%, respectively []. A new plant-based ABE (based on an evolved tRNA adenosine deaminase fused to the nCas9) enabled A·T to G·C editing in protoplasts and regenerated rice and wheat at frequencies up to 7.5 and 59.1%, respectively. A rice mutation ACC-T1 with C2186R mutation conferred herbicide tolerance []. Four chimeric ecTadAs were made after fusing E. coli TadAs, which had different modification, with D10A. The plant ABE-P1 (plant version) produced by fusing recombinant ecTadA*7.10 protein to the N terminus of nCas9 (D10A) yielded 26 and 12.5% editing efficiency in the OsSPL14 and OsSLR1 gene, respectively. In addition, this system has enabled multiplex base editing in Arabidopsis and B. napus with respective efficiencies of up to 4.1 and 8.8% []. New ABEs were developed by engineering the SpCas9 variants to expand the target sites. This led to an increase in OsSPL14 and OsSPL17 editing efficiency of 25 and 45%, respectively, of the rice genome [].
The high frequency of SNPs in tree genomes makes them good candidates to execute BE systems. A majority of plant resistance genes are allelic and may differ by a single or a few nts. Resistance to several pathogens can be engineered by substituting these nts via BEs. Likewise, BEs can be employed to engineer the susceptibility (S) genes to generate resistant alleles. However, such modifications may lead to pleiotrophic effects, such as reductions in yield, growth, or other stresses. To circumvent this issue, nts in the promoter region can be engineered though BEs to enhance resistance without compromising its pleiotropic effects.
5.10.2. Prime Editing
Prime editing (PE) is the latest GE technology that has enabled almost all types of edits, including transitions (C→T, G→A, A→G, T→C) and transversion mutations (C→A, C→G, G→C, G→T, A→C, A→T, T→A, T→G), and small indels, without the requirement for inducing DSBs [,]. The prime editing system has two main components: a prime editing guide (peg) RNA and a prime editor. A short 8- to 16-nucleotide primer binding site (PBS) sequence, a corresponding reverse transcriptase (RT) template, and a desired editing sequence serve as the foundation for the construction of a pegRNA. As prime editing offers a great deal of flexibility for achieving a variety of genome edits, it offers tremendous potential for the advancement of superior crops for a wide range of purposes, such as increasing yield, resisting various abiotic and biotic stresses, and improving the quality of plant product.
5.10.3. RNA Editing
RNA editing (RE) is another modified version of BE to regulate RNA splicing pathways. The majority of the eukaryotic mRNA is spliced according to the GU/AG rules, with 5′GU serving as the splice donor site and 3′AG as splice acceptor site. Any mutation at these sites can result in mis-splicing or loss of a certain splice form. By adopting the same strategy, G was substituted for A in the splice donor site via the RE to hamper the excision of an intron to gain hypersensitivity to abscisic acid []. In a similar study, single null mutants of Arabidopsis MTA genes and double null mutants of rice genes OsGL1 and OsNAL1 were generated by mis-splicing []. Therefore, CRISPR-Cas BEs could have a substantial role in tree GE.
PE was used to confer herbicide resistance by targeting three loci of the Acetolactate Synthase gene (OsALS) in rice. The regenerated rice shoots carrying either ALS-PE2 or ALS-PE3 were herbicide resistant, and Sanger sequencing revealed the successful editing []. Mutations induced by PE can be variable and one type of mutation can occur at a higher rate than others. Reportedly the frequency of deletions (6 bp) ranges up to 21.8% [] and insertions (3 bp) range up to 19.8% [], while the mutation frequency ranges from 0.03 to 18.75% in rice []. Mutations in wheat were less common than in rice, particularly at the codon level, which is about 1.4% in comparison with 9.38% in rice. For all cases with 12 base-to-base substitutions, the frequency of edits was between 0.2 and 8% []. In plants, the frequency of indels increases as the length of targeted sequences increases.
The BE system could also be used to understand the role of conserved amino acids in protein structure and function. Using the CBE, the role of four Arabidopsis genes was revalidated as either constitutive splicing or impeding it.
6. Techniques to Estimate and Quantify the Mutation Rate
Despite remarkable progress in GE, current methods to detect mutations induced by CRISPR-Cas systems are still challenging in plants. Detection techniques are extremely important when inbred lines are desired, or when screening a large population. Nevertheless, several methods have been applied to detect both on- and off-target mutations. The methods share certain pros and cons; some of the most commonly used methods are briefly discussed here.
6.1. T7E1 Mismatch Cleavage Assay
The T7E1 mismatch cleavage assays are popular due to their speed, cost, simplicity and effectiveness on single clones and pooled samples. This technique is used to identify clones and clone segments prior to more detailed analysis. This assay relies on the hybridization of modified and wild-type DNA strands to detect mutation. Mismatches are detected and cleaved by the nuclease enzyme, and the resulting DNA fragments are resolved by gel electrophoresis. However, the assay shares certain limitations, including the lack of sequence information, missing SNPs and small indels [], possibly requiring optimization, and needing a PCR step to detect homozygous mutations. This technique has been successfully executed in different plant species to detect CRISPR-mediated mutagenesis [,,,].
6.2. High-Resolution Melting Assay
High-resolution melting (HRM) analysis is based on the post-real time PCR (fluorescent dye-based) method and involves the analysis of melting curves. Fluorescent intensity is plotted against the melting temperature to provide raw melt curve data, and each type of genome change generates a unique melting curve. The resulting melting curve helps to distinguish between different mutants such as heterozygous, bi-allelic, or homozygous mutations. HRM is sensitive enough to detect even a single-base indel pair with high precision []. HRM analysis requires a simple set-up and allows rapid, high-throughput mutation screening, but requires a dedicated software. This particular technology has been found to work across different plant species [,,,,].
6.3. Sanger Sequencing
This method of identifying induced mutations at the target locus involves amplifying the target region by PCR, followed by a Sanger amplicon sequence. This method is simple, robust, cost-effective and provides information about the type and frequency of mutations. However, in order to identify mutations in all copies of the genome, many colonies must be sequenced. Furthermore, Sanger sequencing can be difficult, laborious, and time-consuming. The Sanger sequencing platform is the most widely opted for mechanism of mutation detection after executing the CRISPR-Cas and has been successfully executed in a variety of plants [,,,,,].
6.4. Next-Generation Sequencing
Next-generation sequencing (NGS) is a highly robust technique that allows indel detection and simultaneous screening of off-target mutations in both mixed populations and clonal cell lines. These abilities have made it a popular choice among researchers. NGS can detect mutations with a high sensitivity of as low as 0.01% [] and can detect the locations of indels and whether a cell population is truly monoclonal. The main limitations of NGS are cost, the need for bioinformatics tools, and the production of short readings that can be missed by larger indels. NGS has been successfully utilized in plant species to achieve a comprehensive understanding of all the on- and off-targets present in the edited genome [,,,,,].
6.5. FLA-PCR (Fragment Analysis)
Fragment analysis is a capillary electrophoresis (CE) method based on the detection of AFLP, multiplex ligation-dependent probe amplification (MLPA) and SNPs. CE is a proven sensitive, high throughput and high-resolution nucleic acid analysis system. Several fragment analysis methods such as IDAA, fluorescent PCR, and CRISPR-STAT have recently been developed in CRISPR/Cas9 genome editing studies. It has been reported that their sensitivity and resolution are comparable to NGS with an indel detection sensitivity of approximately 0.1% []. The bottleneck for fragment analysis methods lies in secondary data analysis, requiring sophisticated software for targeting efficiency calculations for genome editing studies. FLA-PCR has been successfully carried out in some plant species to accurately identify on- and off-targets [,].
7. Critical Assessment of CRISPR-Cas Based GE in Fruit Trees
Despite the broad applications, and unparalleled popularity and acclaim over other GE techniques of CRISPR-Cas, it still poses some limitations in woody plants. These may include a low mutation efficiency/rate, unwanted (off-target) mutations, inefficient gene delivery techniques, in vitro regeneration dependency, persistent activity in subsequent generations, the spread of transgene to other plants and reversion of mutation via cross-pollination, difficulties of execution in woody plants and long-life cycle, genotypic chimerism and strict GMO regulations. Most of these issues can be tackled through a meticulous approach by avoiding erroneous gRNA design, choosing the best Cas protein variant, designing a better expression cassette (including a promoter), employing a highly efficient DNA delivery approach and targeting the right tissue types [,,].
Tree genomes are highly complex due to their high gene copy numbers and ploidy levels, so knocking out all copies of a gene or gene with high homology is a daunting task. To address such issues using conventional genetic manipulation techniques, a series of allelic mutations is performed first, followed by selection in the segregating population []. On the other hand, CRISPR-Cas-mediated mutagenesis has simplified the process of modifying and introducing multiple traits into polyploid plants without introducing any linkage drag. CRISPR/Cas-based knockout mutants have created genome-wide deletion mutants that can be used to study gene function in species that are long lived, cannot be easily self-pollinated, and have low transformation efficiency. The elimination of a binding site for bacterial-coded pathogenesis protein in the citrus genome [], the prevention of floral development [,], and the reduction of lignin biosynthesis [] have opened up new avenues for the development of genes involved in wood structure and chemistry.
To successfully execute CRISPR-Cas in trees, prior knowledge of chromosomal rearrangements, copy number and genetic variations, indels, SNPs, and transposon occurrence are prerequisites. Creating knock-in mutants of a desired gene by deleting a repressor-binding site in the promoter or by mutating a motif involved in rapid degradation is plausible. Such mutants could be generated to increase resistance to herbicides, insects, or pathogens, and thus provide some of the most promising opportunities to generate value traits for forestry. Furthermore, a single insertion into the target loci (hemizygosity) via DSBs may be advantageous in generating the same gene in the corresponding locus of unrelated genotypes. This approach would allow the production of homozygous offspring in just one generation as two hemizygous insertions at the same locus would be crossed. This would streamline inheritance, mitigate linkage drag, and reduce the inbreeding depression that could otherwise occur from repeated use of the original resistance event.
For most tree species, transformation and regeneration remain major bottlenecks after GE. Lowe et al. [] demonstrated effective transformation in extremely difficult plant genotypes by overexpressing the morphogenic genes Baby boom (Bbm) and Wuschel2 (Wus2) from maize. It is anticipated that the same technology could be applied to forest trees after achieving the essential customization. The regeneration bottleneck could be alleviated substantially using transgenes that can boost the regenerability of transgenic cells. The effectiveness of morphogenic genes to induce regeneration in angiosperms was first documented in dicots, where somatic embryos were induced on various explants [,], and these genes exhibited improved regeneration [,].
Although CRISPR is a precise and powerful GE technique, it creates chimeric plants. Chimerism often occurs in regenerated plants during genetic transformation by organogenesis and it is not eliminated by subsequent selfing in most tree plants due to self-incompatibility, which is controlled by a single S-locus []. This phenomenon is further exacerbated by the lengthy and difficult regeneration process during the Cas9 use. One of the important ways to prevent chimerism is to exclude the sexual propagation of transgenic plants []. Unfortunately, this step does not apply to a wide range of tree species. Alternative strategies to combat chimerism include adventitious shoot regeneration [], which has been effectively applied to apples and pears [,].
Another major bottleneck to GE plants is troublesome GMO regulations, people, and market trends. Market limitations pertaining to forest industries are now mostly covered by forest certification research. At present, there is limited commercial utility of GE in trees due to marketing and commercialization obstacles [].
8. Biological and Regulatory Constraints to GE in Trees
Applications of GE in fruit tree breeding have to overcome certain biological and regulatory constraints. The identification of the genetic basis of desirable traits is still a laborious task and involves many forward and reverse genetics tools, along with the execution of whole genome sequence using the NGS approach. After successfully identifying the desired genes, the next challenge is to choose a suitable delivery method of the CRISPR-Cas system and regeneration of GE mutants. The most commonly opted for methods are Agrobacterium or viral-mediated systems. Tissue culturing is the most common method for the transformation and regeneration of GE plants; however, many trees lack an established system for efficient transformation and tissue culture. For example, date palm is a major fruit tree in oasis agriculture and due to its large and complex genomic architecture, the application of GE can be a challenging task. Recently, a generalized GE strategy in date palm, its potential applications and limitations have been discussed in detail []. Another challenging task is eliminating the footprints of foreign DNA fragments (such as T-DNA insertion from the plasmids) from the GE plants in heterozygous or vegetatively propagated plants []. Possibly, the transgene-free mutants can be obtained by high-throughput screening of a large population of the transformants [] (Figure 5). The transgene-free GE can also be executed by in vitro expression of ribonucleoprotein complexes [] and transcripts []. An alternative to in vitro agro-transformation is in planta transformation of plants, which involves targeting in vivo explants (apical meristem, inflorescence, pollen, stigmatic tissue). This method can be further optimized for recalcitrant tree species in classical ways of genetic transformation. In addition, several other key factors can also affect the execution of a GE event in a tree species. These may include the size of sgRNAs and their GC contents, co-expression of sgRNAs and Cas9 protein and the formation of secondary structures during pairing of sgRNAs with the target region []. From a scientific perspective, the use of non-inherited and transiently expressed RNP complex to generate DNA-free GE plants is equivalent to using gamma rays or EMS to induce mutations. It is rather advantageous to use GE in contrast to gamma rays or EMS, which are hazardous to human health.
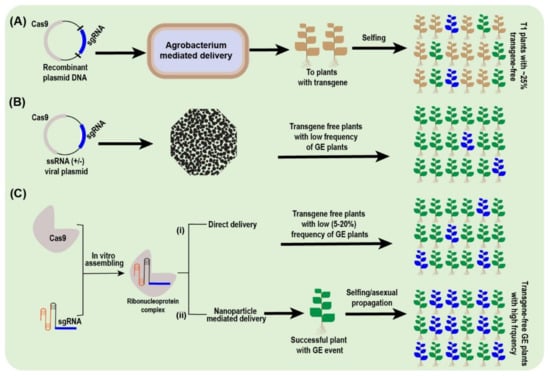
Figure 5.
Potential transgene-free approaches to modify tree plants through CRISPR-Cas system. (A) Agrobacterium-mediated CRISPR-Cas system keep majority of the transgene cargo in T0 generation. Nonetheless, after selfing, a quarter of the T1 progeny will be transgene free if these plants received a single copy of the transgene. As more than one transgene is inserted by this approach, less than 25% of plants will be transgene free in the T1 generation. (B) Viral vector-mediated CRISPR-Cas delivery can lead to yielding transgene-free GE plants. However, their cargo and target specificity limit their usage. (C) Ribonucleoprotein (RNP) complex utilizes in vitro hybridization of expressed Cas9 and sgRNA followed by delivery either by biolistic means (i), or via nanoparticles (ii). The resultant cells (both transformed and untransformed) will form transgene-free seedlings, which can be separated out after laborious screening. The wild-type plants are shown as dark green, GE plants harboring transgene as light brown, and GE and transgene-free plants in dark blue color.
Advancements in GE technologies have enabled researchers to replicate classical breeding outcomes by precisely mutating the genome of many fruit trees (without any off-target loads) and avoiding prolonged cycles of backcrossing and screening. This juridical capability of GE provides an opportunity for GE plants to circumvent the current GMO regulatory framework []. In CRISPR-Cas9-based GE in plants, the predominant repair pathway employs NHEJ, which helps to create transgene-free plants because no foreign elements are involved []. Moreover, it seems absurd to consider interspecific hybridization more natural than precisely editing the genome for a specific trait using wild plant sources, which leaves no genetic footprints from a foreign source. The regulatory framework model is either product-based (Canada) or process-based (EU); thus, there are different perspectives about the regulatory framework of GE plants (Table 4). For example, Canada imposes a pre-market assessment for any GE plant, feed, or food product, which differs from an already available source []. Recently, Canada has announced that it will reconsider its risk assessment policy to exempt GE crops without any foreign DNA footprints from biosafety regulations. Meanwhile, Japan has approved the world’s first GE tomato containing a high level of gamma aminobutyric acid (GABA) (https://www.isaaa.org/kc/cropbiotechupdate/article/; accessed on 7 March 2021). This clearly shows Japan’s policy towards GE crops, which will not be considered as genetically modified. Additionally, Australia permits the use of transgene-free GE crops []. In contrast, the European Union Court of Justice has imposed strict GMO regulations on all GE crops [,]. Thus, now is an appropriate time to review and revise the global scientific consensus and regulatory framework that are limiting the development of GE cultivars.

Table 4.
Genome editing related regulations in the selected countries.
9. Conclusions and Future Perspectives
Mutation breeding has significantly contributed to crop improvement across the globe and led to the commercialization of hundreds of mutated crops with higher yield potential, improved nutritional quality and tolerance to biotic and abiotic stresses. Mutations have generated impressive genetic resources for all major crops worldwide. Stable gene-specific mutations are now very efficient with the discovery of TILLING as a high-throughput mutant screening technique. Mutations can be more precisely detected at specific loci or genes with TILLING screening based upon CEL I-, HRM- and NGS-based approaches. Under such circumstances, the conventional mutation breeding can be comparable to the NBT based upon a CRISPR-Cas approach. Apart from this convergence, GE may surpass TILLING-based spontaneous and induced mutagenesis approaches due to precision and off-target mediation. Nonetheless, breeding of trees poses a crucial bottleneck such as long-life cycle, the ploidy level, occurrence of sequence polymorphisms, nature of parthenocarpic fruit development, and linkage drag. The development of the NBTs with a high-degree of precision, robust selection, and speed breeding, such as CRISPR-Cas and its contemporary versions (BE and PE), could substantially meet the growing food security challenges. Apart from these, several techniques have been devised to mitigate the off-targets and other limitations of NBTs in trees, such as chimeras. CRISPR-mediated tree breeding has the potential to substantially sustain yield with less effort and cost. Although GE has been accomplished for many field crops, its application in tree breeding necessarily requires the identification of major breeding traits, after communicating with all stakeholders. Moreover, the selection of suitable GE reagents and protocols to regenerate plant mutant requires more suitable methods. Given that, we may speculate that such challenges will be properly addressed in the coming period of time.
Author Contributions
Conceptualization, S.M.J., J.M.A.-K., M.N.S. and Z.I.; writing—original draft preparation, M.N.S. and Z.I.; writing—review and editing, J.M.A.-K. and S.M.J.; supervision, J.M.A.-K. and S.M.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors are grateful to Dennis V. Johnson, Cincinnati, OH, USA for linguistic improvement of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Grattapaglia, D.; Silva-Junior, O.B.; Resende, R.T.; Cappa, E.P.; Muller, B.S.F.; Tan, B.Y.; Isik, F.; Ratcliffe, B.; El-Kassaby, Y.A. Quantitative genetics and genomics converge to accelerate forest tree breeding. Front. Plant Sci. 2018, 9, 1693. [Google Scholar] [CrossRef]
- Burdon, R.D.; Klapste, J. Alternative selection methods and explicit or implied economic-worth functions for different traits in tree breeding. Tree Genet. Genom. 2019, 15, 79. [Google Scholar] [CrossRef]
- Hough, L.F.; Shay, J.R.; Dayton, D.F. Apple Scab Resistance from Malus-Floribunda Sieb. Proc. Am. Soc. Hort. Sci. 1953, 62, 341–347. [Google Scholar]
- Schouten, H.J.; Krens, F.A.; Jacobsen, E. Cisgenic plants are similar to traditionally bred plants—International regulations for genetically modified organisms should be altered to exempt cisgenesis. EMBO Rep. 2006, 7, 750–753. [Google Scholar] [CrossRef] [Green Version]
- Semagn, K.; Bjornstad, A.; Ndjiondjop, M.N. Progress and prospects of marker assisted backcrossing as a tool in crop breeding programs. Afr. J. Biotechnol. 2006, 5, 2588–2603. [Google Scholar]
- Moose, S.P.; Mumm, R.H. Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiol. 2008, 147, 969–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omura, M.; Shimada, T. Citrus breeding, genetics and genomics in Japan. Breed. Sci. 2016, 66, 3–17. [Google Scholar] [CrossRef] [Green Version]
- Feng, S.J.; Zhang, J.P.; Mu, Z.H.; Wang, Y.J.; Wen, C.L.; Wu, T.; Yu, C.; Li, Z.; Wang, H.S. Recent progress on the molecular breeding of Cucumis sativus L. in China. Theor. Appl. Genet. 2020, 133, 1777–1790. [Google Scholar] [CrossRef] [PubMed]
- Endo, T.; Fujii, H.; Omura, M.; Shimada, T. Fast-track breeding system to introduce CTV resistance of trifoliate orange into citrus germplasm, by integrating early flowering transgenic plants with marker-assisted selection. BMC Plant Biol. 2020, 20, 224. [Google Scholar] [CrossRef]
- Elo, A.; Lemmetyinen, J.; Novak, A.; Keinonen, K.; Porali, I.; Hassinen, M.; Sopanen, T. BpMADS4 has a central role in inflorescence initiation in silver birch (Betula pendula). Physiol. Plant. 2007, 131, 149–158. [Google Scholar] [CrossRef]
- Schlathölter, I.; Jänsch, M.; Flachowsky, H.; Broggini, G.A.L.; Hanke, M.-V.; Patocchi, A. Generation of advanced fire blight-resistant apple (Malus × domestica) selections of the fifth generation within 7 years of applying the early flowering approach. Planta 2018, 247, 1475–1488. [Google Scholar] [CrossRef] [Green Version]
- Stadler, L.J. Genetic effects of x rays in maize. Proc. Natl. Acad. Sci. USA 1928, 14, 69–75. [Google Scholar] [CrossRef] [Green Version]
- Acquaah, G. Principles of Plant Genetics and Breeding; John Wiley & Sons: Hoboken, NJ, USA, 2009. [Google Scholar]
- Sink, K.; Jain, R.; Chowdhury, J. Somatic cell hybridization. In Distant Hybridization of Crop Plants; Springer: Berlin/Heidelberg, Germany, 1992; pp. 168–198. [Google Scholar]
- Kohli, A.; Christou, P. Stable transgenes bear fruit. Nat. Biotechnol. 2008, 26, 653–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ammann, K. Genomic misconception: A fresh look at the biosafety of transgenic and conventional crops. A plea for a process agnostic regulation. New Biotechnol. 2014, 31, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Twyman, R.M.; Kohli, A.; Stoger, E.; Christou, P. Foreign DNA: Integration and expression in transgenic plants. In Genetic Engineering: Principles and Methods; Setlow, J.K., Ed.; Springer: Boston, MA, USA, 2002; pp. 107–136. [Google Scholar]
- Ghogare, R.; Williamson-Benavides, B.; Ramirez-Torres, F.; Dhingra, A. CRISPR-associated nucleases: The Dawn of a new age of efficient crop improvement. Transgenic Res. 2020, 29, 1–35. [Google Scholar] [CrossRef]
- Zhu, C.; Bortesi, L.; Baysal, C.; Twyman, R.M.; Fischer, R.; Capell, T.; Schillberg, S.; Christou, P. Characteristics of genome editing mutations in cereal crops. Trends Plant Sci. 2017, 22, 38–52. [Google Scholar] [CrossRef]
- Monsur, M.B.; Shao, G.; Lv, Y.; Ahmad, S.; Wei, X.; Hu, P.; Tang, S. Base editing: The ever expanding clustered regularly interspaced short palindromic repeats (CRISPR) tool kit for precise genome editing in plants. Genes 2020, 11, 466. [Google Scholar] [CrossRef]
- Quispe-Huamanquispe, D.G.; Gheysen, G.; Kreuze, J.F. Horizontal gene transfer contributes to plant evolution: The case of Agrobacterium T-DNAs. Front. Plant Sci. 2017, 8, 2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isabel, N.; Holliday, J.A.; Aitken, S.N. Forest genomics: Advancing climate adaptation, forest health, productivity, and conservation. Evol. Appl. 2020, 13, 3–10. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, M.J.P.; Lewis, S.L.; Affum-Baffoe, K.; Castilho, C.; Costa, F.; Sanchez, A.C.; Ewango, C.E.N.; Hubau, W.; Marimon, B.; Monteagudo-Mendoza, A.; et al. Long-term thermal sensitivity of Earth’s tropical forests. Science 2020, 368, 869–874. [Google Scholar] [CrossRef] [PubMed]
- Cortes, A.J.; Restrepo-Montoya, M.; Bedoya-Canas, L.E. Modern strategies to assess and breed forest tree adaptation to changing climate. Front. Plant Sci. 2020, 11, 583323. [Google Scholar] [CrossRef] [PubMed]
- Neale, D.B.; Kremer, A. Forest tree genomics: Growing resources and applications. Nat. Rev. Genet. 2011, 12, 111–122. [Google Scholar] [CrossRef] [PubMed]
- White, T.L.; Adams, W.T.; Neale, D.B. Forest Genetics; Cabi: Wallingford, UK, 2007. [Google Scholar]
- Seehausen, O. Hybridization and adaptive radiation. Trends Ecol. Evol. 2004, 19, 198–207. [Google Scholar] [CrossRef]
- Burkhart, H.E.; Brunner, A.M.; Stanton, B.J.; Shuren, R.A.; Amateis, R.L.; Creighton, J.L. An assessment of potential of hybrid poplar for planting in the Virginia Piedmont. New For. 2017, 48, 479–490. [Google Scholar] [CrossRef]
- Cipollini, M.; Dingley, N.R.; Felch, P.; Maddox, C. Evaluation of phenotypic traits and blight-resistance in an American chestnut backcross orchard in Georgia. Glob. Ecol. Conserv. 2017, 10, 1–8. [Google Scholar] [CrossRef]
- Badenes, M.L.; Fernández, I.; Martí, A.; Ríos, G.; Rubio-Cabetas, M.J. Application of genomic technologies to the breeding of trees. Front. Genet. 2016, 7, 198. [Google Scholar] [CrossRef] [Green Version]
- Lebedev, V.G.; Lebedeva, T.N.; Chernodubov, A.I.; Shestibratov, K.A. Genomic selection for forest tree improvement: Methods, achievements and perspectives. Forests 2020, 11, 1190. [Google Scholar] [CrossRef]
- Thistlethwaite, F.R.; Ratcliffe, B.; Klápště, J.; Porth, I.; Chen, C.; Stoehr, M.U.; El-Kassaby, Y.A. Genomic selection of juvenile height across a single-generational gap in Douglas-fir. Heredity 2019, 122, 848–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isik, F.; Bartholomé, J.; Farjat, A.; Chancerel, E.; Raffin, A.; Sanchez, L.; Plomion, C.; Bouffier, L. Genomic selection in maritime pine. Plant Sci. 2016, 242, 108–119. [Google Scholar] [CrossRef]
- Kaiser, N.; Douches, D.; Dhingra, A.; Glenn, K.C.; Herzig, P.R.; Stowe, E.C.; Swarup, S. The role of conventional plant breeding in ensuring safe levels of naturally occurring toxins in food crops. Trends Food Sci. Technol. 2020, 100, 51–66. [Google Scholar] [CrossRef]
- Iwata, H.; Minamikawa, M.F.; Kajiya-Kanegae, H.; Ishimori, M.; Hayashi, T. Genomics-assisted breeding in fruit trees. Breed. Sci. 2016, 66, 100–115. [Google Scholar] [CrossRef] [Green Version]
- Te Beest, M.; Le Roux, J.J.; Richardson, D.M.; Brysting, A.K.; Suda, J.; Kubesova, M.; Pysek, P. The more the better? The role of polyploidy in facilitating plant invasions. Ann. Bot. 2012, 109, 19–45. [Google Scholar] [CrossRef] [Green Version]
- Lamo, K.; Bhat, D.J.; Kour, K.; Solanki, S.P.S. Mutation studies in fruit crops: A review. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 3620–3633. [Google Scholar] [CrossRef]
- Zonneveld, B.J.M.; Pollock, W.I. Flow cytometric analysis of somaclonal variation in lineages of Hosta sports detects polyploidy and aneuploidy chimeras. Plant Biol. 2012, 14, 972–979. [Google Scholar] [CrossRef]
- Malabarba, J.; Chevreau, E.; Dousset, N.; Veillet, F.; Moizan, J.; Vergne, E. New strategies to overcome present CRISPR/Cas9 limitations in apple and pear: Efficient dechimerization and base editing. Int. J. Mol. Sci. 2020, 22, 319. [Google Scholar] [CrossRef]
- Gambino, G.; Moine, A.; Boccacci, P.; Perrone, I.; Pagliarani, C. Somatic embryogenesis is an effective strategy for dissecting chimerism phenomena in Vitis vinifera cv Nebbiolo. Plant Cell Rep. 2021, 40, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Kumawat, S.; Rana, N.; Bansal, R.; Vishwakarma, G.; Mehetre, S.T.; Das, B.K.; Kumar, M.; Yadav, S.K.; Sonah, H.; Sharma, T.R.; et al. Expanding avenue of fast neutron mediated mutagenesis for crop improvement. Plants 2019, 8, 164. [Google Scholar] [CrossRef] [Green Version]
- Predieri, S. Mutation induction and tissue culture in improving fruits. Plant Cell Tissue Organ Cult. 2001, 64, 185–210. [Google Scholar] [CrossRef]
- Lokko, Y.; Amoatey, H. Improvement of Pineapple Using in Vitro and Mutation Breeding Techniques; Biotechnology and Nuclear Agriculture Research Institute: Legon-Accra, Ghana, 2001; pp. 25–29. [Google Scholar]
- International Atomic Energy Agency. In Vitro Techniques for Selection of Radiation Induced Mutations Adapted to Adverse Environmental Conditions. In Proceedings of a Final Research Coordination Meeting; Joint FAO/IAEA Division of Nuclear Techniques in Food and Agriculture: Vienna, Austria, 2001; pp. 1–96. [Google Scholar]
- Mba, C.; Afza, R.; Bado, S.; Mohan Jain, S. Induced mutagenesis in plants using physical and chemical agents. Plant Cell Cult. Essent. Methods 2010, 20, 111–130. [Google Scholar]
- Elhiti, M.; Wang, H.Y.; Austin, R.S.; Chen, B.; Brown, D.; Wang, A.M. Generation of chemically induced mutations using in vitro propagated shoot tip tissues for genetic improvement of fruit trees. Plant Cell Tissue Organ Cult. 2016, 124, 447–452. [Google Scholar] [CrossRef]
- Alvarez, D.; Cerda-Bennasser, P.; Stowe, E.; Ramirez-Torres, F.; Capell, T.; Dhingra, A.; Christou, P. Fruit crops in the era of genome editing: Closing the regulatory gap. Plant Cell Rep. 2021, 40, 915–930. [Google Scholar] [CrossRef] [PubMed]
- Sikora, P.; Chawade, A.; Larsson, M.; Olsson, J.; Olsson, O. Mutagenesis as a tool in plant genetics, functional genomics, and breeding. Int. J. Plant Genom. 2011, 2011, 314829. [Google Scholar] [CrossRef] [Green Version]
- Kumawat, S.; Rana, N.; Bansal, R.; Vishwakarma, G.; Mehetre, S.; Das, B.K.; Kumar, M.; Yadav, S.; Sonah, H.; Sharma, T.R.; et al. Fast neutron mutagenesis in plants: Advances, applicability and challenges. Plant Sci. 2019. [Google Scholar] [CrossRef]
- Spengler, R.N. Origins of the apple: The role of megafaunal mutualism in the domestication of Malus and rosaceous trees. Front. Plant Sci. 2019, 10, 617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.A.; Prochnik, S.; Jenkins, J.; Salse, J.; Hellsten, U.; Murat, F.; Perrier, X.; Ruiz, M.; Scalabrin, S.; Terol, T.; et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 2014, 32, 656–662. [Google Scholar] [CrossRef]
- Crosby, J.A.; Janick, J.; Pecknold, P.C.; Korban, S.S.; Oconnor, P.A.; Ries, S.M.; Goffreda, J.; Voordeckers, A. Breeding apples for scab resistance: 1945–1990. Fruit Var. J. 1992, 46, 145–166. [Google Scholar] [CrossRef]
- Mikulic-Petkovsek, M.; Ivancic, A.; Schmitzer, V.; Veberic, R.; Stampar, F. Comparison of major taste compounds and antioxidative properties of fruits and flowers of different Sambucus species and interspecific hybrids. Food Chem. 2016, 200, 134–140. [Google Scholar] [CrossRef]
- Rajan, R.P.; Singh, G. A review on application of somaclonal variation in important horticulture crops. Plant Cell Biotechnol. Mol. Biol. 2021, 22, 161–175. [Google Scholar]
- Ranghoo-Sanmukhiya, V.M. Somaclonal variation and methods used for its detection. In Propagation and Genetic Manipulation of Plants; Siddique, I., Ed.; Springer: Singapore, 2021; pp. 1–18. [Google Scholar]
- Orbović, V.; Ćalović, M.; Viloria, Z.; Nielsen, B.; Gmitter, F.G.; Castle, W.S.; Grosser, J.W. Analysis of genetic variability in various tissue culture-derived lemon plant populations using RAPD and flow cytometry. Euphytica 2008, 161, 329–335. [Google Scholar] [CrossRef]
- Cevallos-Cevallos, J.M.; Jines, C.; Maridueña-Zavala, M.G.; Molina-Miranda, M.J.; Ochoa, D.E.; Flores-Cedeno, J.A. GC-MS metabolite profiling for specific detection of dwarf somaclonal variation in banana plants. Appl. Plant Sci. 2018, 6, e01194. [Google Scholar] [CrossRef] [Green Version]
- Henao-Ramírez, A.M.; Salazar Duque, H.J.; Calle Tobón, A.F.; Urrea Trujillo, A.I. Determination of genetic stability in cacao plants (Theobroma Cacao L.) derived from somatic embryogenesis using microsatellite molecular markers (SSR). Int. J. Fruit Sci. 2021, 21, 284–298. [Google Scholar] [CrossRef]
- Mba, C. Induced mutations unleash the potentials of plant genetic resources for food and agriculture. Agronomy 2013, 3, 200–231. [Google Scholar] [CrossRef] [Green Version]
- McCallum, C.M.; Comai, L.; Greene, E.A.; Henikoff, S. Targeted screening for induced mutations. Nat. Biotechnol. 2000, 18, 455–457. [Google Scholar] [CrossRef] [PubMed]
- Jankowicz-Cieslak, J.; Mba, C.; Till, B.J. Mutagenesis for crop breeding and functional genomics. In Biotechnologies for Plant Mutation Breeding; Springer: Cham, Switzerland, 2017; pp. 3–18. [Google Scholar]
- Sato, Y.; Sato, K.; Nishio, T. Interspecific pairs of class II S haplotypes having different recognition specificities between Brassica oleracea and Brassica rapa. Plant Cell Physiol. 2006, 47, 340–345. [Google Scholar] [CrossRef] [Green Version]
- Acevedo-Garcia, J.; Spencer, D.; Thieron, H.; Reinstädler, A.; Hammond-Kosack, K.; Phillips, A.L.; Panstruga, R. mlo-based powdery mildew resistance in hexaploid bread wheat generated by a non-transgenic TILLING approach. Plant Biotechnol. J. 2017, 15, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.; Howell, T.; Nitcher, R.; Missirian, V.; Watson, B.; Ngo, K.J.; Lieberman, M.; Fass, J.; Uauy, C.; Tran, R.K.; et al. Discovery of rare mutations in populations: TILLING by sequencing. Plant Physiol. 2011, 156, 1257–1268. [Google Scholar] [CrossRef] [Green Version]
- Taheri, S.; Abdullah, T.L.; Jain, S.M.; Sahebi, M.; Azizi, P. TILLING, high-resolution melting (HRM), and next-generation sequencing (NGS) techniques in plant mutation breeding. Mol. Breed. 2017, 37, 40. [Google Scholar] [CrossRef]
- Gilchrist, E.J.; Sidebottom, C.H.D.; Koh, C.S.; MacInnes, T.; Sharpe, A.G.; Haughn, G.W. A mutant Brassica napus (Canola) population for the identification of new genetic diversity via TILLING and next generation sequencing. PLoS ONE 2013, 8, e84303. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Hao, L.; Parry, M.A.J.; Phillips, A.L.; Hu, Y.-G. Progress in TILLING as a tool for functional genomics and improvement of crops. J. Integrat. Plant Biol. 2014, 56, 425–443. [Google Scholar] [CrossRef] [Green Version]
- Freeman, J.S.; Potts, B.M.; Downes, G.M.; Pilbeam, D.; Thavamanikumar, S.; Vaillancourt, R.E. Stability of quantitative trait loci for growth and wood properties across multiple pedigrees and environments in Eucalyptus globulus. New Phytol. 2013, 198, 1121–1134. [Google Scholar] [CrossRef] [Green Version]
- Osakabe, Y.; Sugano, S.S.; Osakabe, K. Genome engineering of woody plants: Past, present and future. J. Wood Sci. 2016, 62, 217–225. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Li, S.; Zhang, D.; Han, M.; Jin, X.; Zhao, C.; Wang, S.; Xing, L.; Ma, J.; Ji, J.; et al. Sequencing of a wild apple (Malus baccata) genome unravels the differences between cultivated and wild apple species regarding disease resistance and cold tolerance. G3-Genes Genom. Genet. 2019, 9, 2051–2060. [Google Scholar] [CrossRef] [Green Version]
- Torales, S.L.; El Mujtar, V.; Marcucci-Poltri, S.; Pomponio, F.; Soliani, C.; Villalba, P.; Estravis-Barcala, M.; Klein, L.; García, M.; Pentreath, V.; et al. Application of high-throughput sequencing technologies in native forest tree species in Argentina: Implications for breeding. In Low Intensity Breeding of Native Forest Trees in Argentina: Genetic Basis for their Domestication and Conservation; Pastorino, M.J., Marchelli, P., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 455–482. [Google Scholar]
- Ricroch, A.E.; Henard-Damave, M.C. Next biotech plants: New traits, crops, developers and technologies for addressing global challenges. Crit. Rev. Biotechnol. 2016, 36, 675–690. [Google Scholar] [CrossRef] [PubMed]
- Yabor, L.; Perez, L.; Gomez, D.; Villalobos-Olivera, A.; Mendoza, J.R.; Martinez, J.; Escalante, D.; Garro, G.; Hajari, E.; Lorenzo, J.C. Histological evaluation of pineapple transgenic plants following 8 years of field growth. Euphytica 2020, 216, 23. [Google Scholar] [CrossRef]
- Ault, K.; Viswanath, V.; Jayawickrama, J.; Ma, C.; Eaton, J.; Meilan, R.; Beauchamp, G.; Hohenschuh, W.; Murthy, G.; Strauss, S.H. Improved growth and weed control of glyphosate-tolerant poplars. New For. 2016, 47, 653–667. [Google Scholar] [CrossRef] [Green Version]
- Gonsalves, D.; Tripathi, S.; Carr, J.B.; Suzuki, J.Y. Papaya ringspot virus. Plant Health Inst. 2010, 10, 1094. [Google Scholar] [CrossRef]
- Carter, N. Petition for Determination of Nonregulated Status: Arctic™ Apple (Malus × domestica) Events GD743 and GS784; United States Department of Agriculture—Animal and Plant Health Inspection Service: Summerland, BC, Canada, 2012.
- Stowe, E.; Dhingra, A. Development of the Arctic® Apple. Plant Breed. Rev. 2021, 44, 273–296. [Google Scholar]
- Scorza, R.; Callahan, A.; Ravelonandro, M.; Braverman, M. Development and regulation of the Plum pox virus resistant transgenic plum “HoneySweet”. In Regulation of Agricultural Biotechnology: The United States and Canada; Springer: Berlin/Heidelberg, Germany, 2012; pp. 269–280. [Google Scholar]
- Xu, K. An overview of Arctic apples: Basic facts and characteristics. NY Fruit Q. 2013, 21, 8–10. [Google Scholar]
- Food and Drug Administration (FDA). Biotechnology Consultations on Food from GE Plant Varieties; Food and Drug Administration (FDA): Silver Spring, MD, USA, 2017.
- Hu, L.; Lu, H.; Liu, Q.; Chen, X.; Jiang, X. Overexpression of mtlD gene in transgenic Populus tomentosa improves salt tolerance through accumulation of mannitol. Tree Physiol. 2005, 25, 1273–1281. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.L.; Su, X.H.; Zhang, B.Y.; Huang, Q.J.; Zhang, X.H.; Huang, R.F. Expression of jasmonic ethylene responsive factor gene in transgenic poplar tree leads to increased salt tolerance. Tree Physiol. 2009, 29, 273–279. [Google Scholar] [CrossRef] [Green Version]
- Randle, M.; Tennant, P. Transgenic Papaya. In Genetically Modified Crops: Current Status, Prospects and Challenges; Kavi Kishor, P.B., Rajam, M.V., Pullaiah, T., Eds.; Springer: Singapore, 2021; Volume 2, pp. 129–160. [Google Scholar]
- Vadlamudi, T.; Patil, B.L.; Kaldis, A.; Sai Gopal, D.V.R.; Mishra, R.; Berbati, M.; Voloudakis, A. DsRNA-mediated protection against two isolates of Papaya ringspot virus through topical application of dsRNA in papaya. J. Virol. Meth. 2020, 275, 113750. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, E.; Nanto, K.; Oishi, M.; Ebinuma, H.; Morishita, Y.; Sakurai, N.; Suzuki, H.; Shibata, D.; Shimada, T. Agrobacterium-mediated transformation of Eucalyptus globulus using explants with shoot apex with introduction of bacterial choline oxidase gene to enhance salt tolerance. Plant Cell Rep. 2012, 31, 225–235. [Google Scholar] [CrossRef]
- Kweon, J.; Kim, D.-E.; Jang, A.-H.; Kim, Y. CRISPR/Cas-based customization of pooled CRISPR libraries. PLoS ONE 2018, 13, e0199473. [Google Scholar] [CrossRef]
- Veillet, F.; Durand, M.; Kroj, T.; Cesari, S.; Gallois, J.-L. Precision Breeding made real with CRISPR: Illustration through genetic resistance to pathogens. Plant Commun. 2020, 1, 100102. [Google Scholar] [CrossRef]
- Horvath, P.; Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef] [Green Version]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR–Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Koonin, E.V. Annotation and classification of CRISPR-Cas systems. In Methods in Molecular Biology; Lundgren, M., Emmanuelle, C., Peter, F., Eds.; Springer Science+Business Media: New York, NY, USA, 2015; Volume 1311, pp. 47–75. [Google Scholar]
- Hille, F.; Richter, H.; Wong, S.P.; Bratovič, M.; Ressel, S.; Charpentier, E. The biology of CRISPR-Cas: Backward and forward. Cell 2018, 172, 1239–1259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Deveau, H.; Barrangou, R.; Garneau, J.E.; Labonté, J.; Fremaux, C.; Boyaval, P.; Romero, D.A.; Horvath, P.; Moineau, S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008, 190, 1390–1400. [Google Scholar] [CrossRef] [Green Version]
- Karvelis, T.; Gasiunas, G.; Young, J.; Bigelyte, G.; Silanskas, A.; Cigan, M.; Siksnys, V. Rapid characterization of CRISPR-Cas9 protospacer adjacent motif sequence elements. Genome Biol. 2015, 16, 253. [Google Scholar] [CrossRef] [Green Version]
- Symington, L.S.; Gautier, J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011, 45, 247–271. [Google Scholar] [CrossRef] [PubMed]
- Budman, J.; Chu, G. Processing of DNA for nonhomologous end-joining by cell-free extract. EMBO J. 2005, 24, 849–860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, C.; Bongiorno, P.; Martins, A.; Stephanou, N.C.; Zhu, H.; Shuman, S.; Glickman, M.S. Mechanism of nonhomologous end-joining in mycobacteria: A low-fidelity repair system driven by Ku, ligase D and ligase C. Nat. Struct. Mol. Biol. 2005, 12, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Burstein, D.; Harrington, L.B.; Strutt, S.C.; Probst, A.J.; Anantharaman, K.; Thomas, B.C.; Doudna, J.A.; Banfield, J.F. New CRISPR-Cas systems from uncultivated microbes. Nature 2017, 542, 237–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shmakov, S.; Smargon, A.; Scott, D.; Cox, D.; Pyzocha, N.; Yan, W.; Abudayyeh, O.O.; Gootenberg, J.S.; Makarova, K.S.; Wolf, Y.I.; et al. Diversity and evolution of class 2 CRISPR–Cas systems. Nat. Rev. Microbiol. 2017, 15, 169–182. [Google Scholar] [CrossRef] [Green Version]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef]
- FAOSTAT. Food and Agriculture Organization of the United Nations Statistics Division. 2021. Available online: http://faostat3.fao.org/home/E (accessed on 28 January 2021).
- Nishitani, C.; Hirai, N.; Komori, S.; Wada, M.; Okada, K.; Osakabe, K.; Yamamoto, T.; Osakabe, Y. Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 2016, 6, 1–8. [Google Scholar]
- Malnoy, M.; Viola, R.; Jung, M.H.; Koo, O.J.; Kim, S.; Kim, J.S.; Velasco, R.; Nagamangala Kanchiswamy, C. DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 2016, 7, 1904. [Google Scholar] [CrossRef]
- Osakabe, Y.; Liang, Z.; Ren, C.; Nishitani, C.; Osakabe, K.; Wada, M.; Komori, S.; Malnoy, M.; Velasco, R.; Poli, M.; et al. CRISPR–Cas9-mediated genome editing in apple and grapevine. Nat. Protoc. 2018, 13, 2844–2863. [Google Scholar] [CrossRef] [PubMed]
- Chevreau, E.; Dousset, N.; Joffrion, C.; Richer, A.; Charrier, A.; Vergne, E. Agroinfiltration is a key factor to improve the efficiency of apple and pear transformation. Sci. Hortic. 2019, 251, 150–154. [Google Scholar] [CrossRef]
- Charrier, A.; Vergne, E.; Dousset, N.; Richer, A.; Petiteau, A.; Chevreau, E. Efficient targeted mutagenesis in apple and first time edition of pear using the CRISPR-Cas9 system. Front. Plant Sci. 2019, 10, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pompili, V.; Dalla Costa, L.; Piazza, S.; Pindo, M.; Malnoy, M. Reduced fire blight susceptibility in apple cultivars using a high-efficiency CRISPR/Cas9-FLP/FRT-based gene editing system. Plant Biotechnol. J. 2020, 18, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Bai, S.; Wang, N.; Sun, X.; Zhang, Y.; Zhu, J.; Dong, C. CRISPR/Cas9-mediated mutagenesis of MdCNGC2 in apple callus and VIGS-mediated silencing of MdCNGC2 in fruits improve resistance to Botryosphaeria dothidea. Front. Plant Sci. 2020, 11, 575477. [Google Scholar] [CrossRef]
- Jiao, J.; Kong, K.; Han, J.; Song, S.; Bai, T.; Song, C.; Wang, M.; Yan, Z.; Zhang, H.; Zhang, R.; et al. Field detection of multiple RNA viruses/viroids in apple using a CRISPR/Cas12a-based visual assay. Plant Biotechnol. J. 2021, 19, 394–405. [Google Scholar] [CrossRef] [PubMed]
- Picq, C. Bananas; International Network for the Improvement of Banana and Plantain (INIBAP): Montpellier, France, 2000; Volume 14, p. 14. [Google Scholar]
- Perrier, X.; De Langhe, E.; Donohue, M.; Lentfer, C.; Vrydaghs, L.; Bakry, F.; Carreel, F.; Hippolyte, I.; Horry, J.-P.; Jenny, C.; et al. Multidisciplinary perspectives on banana (Musa spp.) domestication. Proc. Natl. Acad. Sci. USA 2011, 108, 11311–11318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dash, P.K.; Rai, R. Translating the “banana genome” to delineate stress resistance, dwarfing, parthenocarpy and mechanisms of fruit ripening. Front. Plant Sci. 2016, 7, 1543. [Google Scholar] [CrossRef] [Green Version]
- Kaur, N.; Alok, A.; Shivani, M.; Pandey, P.; Awasthi, P.; Tiwari, S. CRISPR/Cas9-mediated efficient editing in phytoene desaturase (PDS) demonstrates precise manipulation in banana cv. Rasthali genome. Funct. Integr. Genom. 2018, 18, 89–99. [Google Scholar] [CrossRef]
- Naim, F.; Dugdale, B.; Kleidon, J.; Brinin, A.; Shand, K.; Waterhouse, P.; Dale, J. Gene editing the phytoene desaturase alleles of Cavendish banana using CRISPR/Cas9. Transgenic Res. 2018, 27, 451–460. [Google Scholar] [CrossRef] [Green Version]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef] [Green Version]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1713. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [Green Version]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Topkar, V.V.; Nguyen, N.T.; Zheng, Z.; Gonzales, A.P.W.; Li, Z.; Peterson, R.T.; Yeh, J.-R.J.; et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015, 523, 481–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, C.; Bargsten, K.; Jinek, M. Structural Plasticity of PAM Recognition by Engineered Variants of the RNA-Guided Endonuclease Cas9. Mol. Cell 2016, 61, 895–902. [Google Scholar] [CrossRef] [Green Version]
- Nishimasu, H.; Shi, X.; Ishiguro, S.; Gao, L.; Hirano, S.; Okazaki, S.; Noda, T.; Abudayyeh, O.O.; Gootenberg, J.S.; Mori, H.; et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 2018, 361, 1259–1262. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef] [Green Version]
- Liang, Z.; Chen, K.; Yan, Y.; Zhang, Y.; Gao, C. Genotyping genome-edited mutations in plants using CRISPR ribonucleoprotein complexes. Plant Biotechnol. J. 2018, 16, 2053–2062. [Google Scholar] [CrossRef] [PubMed]
- Vakulskas, C.A.; Dever, D.P.; Rettig, G.R.; Turk, R.; Jacobi, A.M.; Collingwood, M.A.; Bode, N.M.; McNeill, M.S.; Yan, S.; Camarena, J.; et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med. 2018, 24, 1216–1224. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.H.; Miller, S.M.; Geurts, M.H.; Tang, W.; Chen, L.; Sun, N.; Zeina, C.M.; Gao, X.; Rees, H.A.; Lin, Z.; et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 2018, 556, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Piatek, A.; Ali, Z.; Baazim, H.; Li, L.; Abulfaraj, A.; Al-Shareef, S.; Aouida, M.; Mahfouz, M.M. RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol. J. 2015, 13, 578–589. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, D.; Xiong, X.; Yan, B.; Xie, W.; Sheen, J.; Li, J.-F. A potent Cas9-derived gene activator for plant and mammalian cells. Nat. Plants 2017, 3, 930–936. [Google Scholar] [CrossRef] [Green Version]
- Mikami, M.; Toki, S.; Endo, M. Precision targeted mutagenesis via Cas9 paired nickases in rice. Plant Cell Physiol. 2016, 57, 1058–1068. [Google Scholar] [CrossRef] [Green Version]
- Hsu, C.-T.; Cheng, Y.-J.; Yuan, Y.-H.; Hung, W.-F.; Cheng, Q.-W.; Wu, F.-H.; Lee, L.-Y.; Gelvin, S.B.; Lin, C.-S. Application of Cas12a and nCas9-activation-induced cytidine deaminase for genome editing and as a non-sexual strategy to generate homozygous/multiplex edited plants in the allotetraploid genome of tobacco. Plant Mol. Biol. 2019, 101, 355–371. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Mikami, M.; Endo, A.; Kaya, H.; Itoh, T.; Nishimasu, H.; Nureki, O.; Toki, S. Genome editing in plants by engineered CRISPR–Cas9 recognizing NG PAM. Nat. Plants 2019, 5, 14–17. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Topkar, V.V.; Zheng, Z.; Joung, J.K. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 2015, 33, 1293–1298. [Google Scholar] [CrossRef] [PubMed]
- Hirano, H.; Gootenberg, J.S.; Horii, T.; Abudayyeh, O.O.; Kimura, M.; Hsu, P.D.; Nakane, T.; Ishitani, R.; Hatada, I.; Zhang, F.; et al. Structure and engineering of Francisella novicida Cas9. Cell 2016, 164, 950–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endo, A.; Toki, S. FnCpf1-Mediated targeted mutagenesis in plants. Meth. Mol. Biol. 2018, 1795, 223–239. [Google Scholar]
- Begemann, M.B.; Gray, B.N.; January, E.; Gordon, G.C.; He, Y.; Liu, H.; Wu, X.; Brutnell, T.P.; Mockler, T.C.; Oufattole, M. Precise insertion and guided editing of higher plant genomes using Cpf1 CRISPR nucleases. Sci. Rep. 2017, 7, 11606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zetsche, B.; Gootenberg Jonathan, S.; Abudayyeh Omar, O.; Slaymaker Ian, M.; Makarova Kira, S.; Essletzbichler, P.; Volz Sara, E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Cox, D.B.T.; Yan, W.X.; Manteiga, J.C.; Schneider, M.W.; Yamano, T.; Nishimasu, H.; Nureki, O.; Crosetto, N.; Zhang, F. Engineered Cpf1 variants with altered PAM specificities. Nat. Biotechnol. 2017, 35, 789–792. [Google Scholar] [CrossRef] [Green Version]
- Tóth, E.; Czene, B.C.; Kulcsár, P.I.; Krausz, S.L.; Tálas, A.; Nyeste, A.; Varga, É.; Huszár, K.; Weinhardt, N.; Ligeti, Z.; et al. Mb- and FnCpf1 nucleases are active in mammalian cells: Activities and PAM preferences of four wild-type Cpf1 nucleases and of their altered PAM specificity variants. Nucleic Acids Res. 2018, 46, 10272–10285. [Google Scholar] [CrossRef]
- Shmakov, S.; Abudayyeh, O.O.; Makarova, K.S.; Wolf, Y.I.; Gootenberg, J.S.; Semenova, E.; Minakhin, L.; Joung, J.; Konermann, S.; Severinov, K.; et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell 2015, 60, 385–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Gao, P.; Rajashankar, K.R.; Patel, D.J. PAM-Dependent Target DNA Recognition and cleavage by C2c1 CRISPR-Cas endonuclease. Cell 2016, 167, 1814–1828.e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Patel, D.J. CasX: A new and small CRISPR gene-editing protein. Cell Res. 2019, 29, 345–346. [Google Scholar] [CrossRef]
- Liu, J.-J.; Orlova, N.; Oakes, B.L.; Ma, E.; Spinner, H.B.; Baney, K.L.M.; Chuck, J.; Tan, D.; Knott, G.J.; Harrington, L.B.; et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature 2019, 566, 218–223. [Google Scholar] [CrossRef]
- Tripathi, J.N.; Ntui, V.O.; Ron, M.; Muiruri, S.K.; Britt, A.; Tripathi, L. CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Commun. Biol. 2019, 2, 46. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Zhu, H.; Liu, J.; Yang, Q.; Shao, X.; Bi, F.; Hu, C.; Huo, H.; Chen, K.; Yi, G. Establishment of a PEG-mediated protoplast transformation system based on DNA and CRISPR/Cas9 ribonucleoprotein complexes for banana. BMC Plant Biol. 2020, 20, 425. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Sheng, O.; Deng, G.; He, W.; Dong, T.; Yang, Q.; Dou, T.; Li, C.; Gao, H.; Liu, S.; et al. CRISPR/Cas9-mediated genome editing of MaACO1 (aminocyclopropane-1-carboxylate oxidase 1) promotes the shelf life of banana fruit. Plant Biotechnol. J. 2020. [Google Scholar] [CrossRef]
- Kaur, N.; Alok, A.; Shivani, M.; Kumar, P.; Awasthi, P.; Chaturvedi, S.; Pandey, P.; Pandey, A.; Pandey, A.K.; Tiwari, S. CRISPR/Cas9 directed editing of lycopene epsilon-cyclase modulates metabolic flux for β-carotene biosynthesis in banana fruit. Metab. Eng. 2020, 59, 76–86. [Google Scholar] [CrossRef]
- Shao, X.; Wu, S.; Dou, T.; Zhu, H.; Hu, C.; Huo, H.; He, W.; Deng, G.; Sheng, O.; Bi, F.; et al. Using CRISPR/Cas9 genome editing system to create MaGA20ox2 gene-modified semi-dwarf banana. Plant Biotechnol. J. 2020, 18, 17–19. [Google Scholar] [CrossRef] [Green Version]
- Jia, H.; Wang, N. Xcc-facilitated agroinfiltration of citrus leaves: A tool for rapid functional analysis of transgenes in citrus leaves. Plant Cell Rep. 2014, 33, 1993–2001. [Google Scholar] [CrossRef]
- Jia, H.; Xu, J.; Orbović, V.; Zhang, Y.; Wang, N. Editing citrus genome via SaCas9/sgRNA system. Front. Plant Sci. 2017, 8, 2135. [Google Scholar] [CrossRef]
- Jia, H.; Orbović, V.; Wang, N. CRISPR-LbCas12a-mediated modification of citrus. Plant Biotechnol. J. 2019, 17, 1928–1937. [Google Scholar] [CrossRef] [Green Version]
- Jia, H.; Zhang, Y.; Orbović, V.; Xu, J.; White, F.F.; Jones, J.B.; Wang, N. Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 2017, 15, 817–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gumtow, R.; Wu, D.; Uchida, J.; Tian, M. A Phytophthora palmivora extracellular cystatin-like protease inhibitor targets papain to contribute to virulence on papaya. Mol. Plant Microbe Interact. 2018, 31, 363–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettongkhao, S.; Navet, N.; Schornack, S.; Tian, M.; Churngchow, N. A secreted protein of 15 kDa plays an important role in Phytophthora palmivora development and pathogenicity. Sci. Rep. 2020, 10, 2319. [Google Scholar] [CrossRef] [PubMed]
- Pang, H.; Yan, Q.; Zhao, S.; He, F.; Xu, J.; Qi, B.; Zhang, Y. Knockout of the S-acyltransferase gene, PbPAT14, confers the dwarf yellowing phenotype in first generation pear by ABA accumulation. Int. J. Mol. Sci. 2019, 20, 6347. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014, 32, 947–951. [Google Scholar] [CrossRef]
- Doench, J.G.; Hartenian, E.; Graham, D.B.; Tothova, Z.; Hegde, M.; Smith, I.; Sullender, M.; Ebert, B.L.; Xavier, R.J.; Root, D.E. Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat. Biotechnol. 2014, 32, 1262–1267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, N.; Liu, W.; Wang, X. WU-CRISPR: Characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 2015, 16, 218. [Google Scholar] [CrossRef] [Green Version]
- Sugano, S.S.; Nishihama, R.; Shirakawa, M.; Takagi, J.; Matsuda, Y.; Ishida, S.; Shimada, T.; Hara-Nishimura, I.; Osakabe, K.; Kohchi, T. Efficient CRISPR/Cas9-based genome editing and its application to conditional genetic analysis in Marchantia polymorpha. PLoS ONE 2018, 13, e0205117. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Sander, J.D.; Reyon, D.; Cascio, V.M.; Joung, J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014, 32, 279–284. [Google Scholar] [CrossRef] [Green Version]
- Rose, J.C.; Popp, N.A.; Richardson, C.D.; Stephany, J.J.; Mathieu, J.; Wei, C.T.; Corn, J.E.; Maly, D.J.; Fowler, D.M. Suppression of unwanted CRISPR-Cas9 editing by co-administration of catalytically inactivating truncated guide RNAs. Nat. Commun. 2020, 11, 2697. [Google Scholar] [CrossRef]
- Zhang, F.; LeBlanc, C.; Irish, V.F.; Jacob, Y. Rapid and efficient CRISPR/Cas9 gene editing in Citrus using the YAO promoter. Plant Cell Rep. 2017, 36, 1883–1887. [Google Scholar] [CrossRef]
- Lee, C.M.; Cradick, T.J.; Fine, E.J.; Bao, G. Nuclease target site selection for maximizing on-target activity and minimizing off-target effects in genome editing. Mol. Ther. 2016, 24, 475–487. [Google Scholar] [CrossRef] [Green Version]
- Ryan, D.E.; Taussig, D.; Steinfeld, I.; Phadnis, S.M.; Lunstad, B.D.; Singh, M.; Vuong, X.; Okochi, K.D.; McCaffrey, R.; Olesiak, M.; et al. Improving CRISPR–Cas specificity with chemical modifications in single-guide RNAs. Nucleic Acids Res. 2017, 46, 792–803. [Google Scholar] [CrossRef] [PubMed]
- Kocak, D.D.; Josephs, E.A.; Bhandarkar, V.; Adkar, S.S.; Kwon, J.B.; Gersbach, C.A. Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat. Biotechnol. 2019, 37, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef]
- Xu, R.; Li, H.; Qin, R.; Wang, L.; Li, L.; Wei, P.; Yang, J. Gene targeting using the Agrobacterium tumefaciens-mediated CRISPR-Cas system in rice. Rice 2014, 7, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, J.; Guo, D.; Zhang, J.; Huang, Q.; Qin, G.; Zhang, X.; Wan, J.; Gu, H.; Qu, L.J. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013, 23, 1233–1236. [Google Scholar] [CrossRef] [Green Version]
- Liang, Z.; Zhang, K.; Chen, K.; Gao, C. Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Gen. 2014, 41, 63–68. [Google Scholar] [CrossRef]
- Hyun, Y.; Kim, J.; Cho, S.W.; Choi, Y.; Kim, J.-S.; Coupland, G. Site-directed mutagenesis in Arabidopsis thaliana using dividing tissue-targeted RGEN of the CRISPR/Cas system to generate heritable null alleles. Planta 2015, 241, 271–284. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.-P.; Xing, H.-L.; Dong, L.; Zhang, H.-Y.; Han, C.-Y.; Wang, X.-C.; Chen, Q.-J. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015, 16, 144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.-F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and homologous recombination–mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef]
- Sun, X.; Hu, Z.; Chen, R.; Jiang, Q.; Song, G.; Zhang, H.; Xi, Y. Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci. Rep. 2015, 5, 10342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.; Zhang, Y.; Kleinstiver, B.P.; Guo, J.A.; Aryee, M.J.; Miller, J.; Malzahn, A.; Zarecor, S.; Lawrence-Dill, C.J.; Joung, J.K.; et al. Activities and specificities of CRISPR/Cas9 and Cas12a nucleases for targeted mutagenesis in maize. Plant Biotechnol. J. 2019, 17, 362–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolter, F.; Klemm, J.; Puchta, H. Efficient in planta gene targeting in Arabidopsis using egg cell-specific expression of the Cas9 nuclease of Staphylococcus aureus. Plant J. 2018, 94, 735–746. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, A.; Ishida, T.; Yoshimura, M.; Kimura, Y.; Sawa, S. Developing heritable mutations in Arabidopsis thaliana using a modified CRISPR/Cas9 toolkit comprising PAM-altered Cas9 variants and gRNAs. Plant Cell Physiol. 2019, 60, 2255–2262. [Google Scholar] [CrossRef] [PubMed]
- Brocken, D.J.; Tark-Dame, M.; Dame, R.T. dCas9: A versatile tool for epigenome editing. Curr. Issues Mol. Biol. 2017, 26, 15–32. [Google Scholar]
- Bikard, D.; Jiang, W.; Samai, P.; Hochschild, A.; Zhang, F.; Marraffini, L.A. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013, 41, 7429–7437. [Google Scholar] [CrossRef] [Green Version]
- Lowder, L.G.; Zhang, D.; Baltes, N.J.; Paul, J.W.; Tang, X.; Zheng, X.; Voytas, D.F.; Hsieh, T.-F.; Zhang, Y.; Qi, Y. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 2015, 169, 971–985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fister, A.S.; Landherr, L.; Maximova, S.N.; Guiltinan, M.J. Transient expression of CRISPR/Cas9 machinery targeting TcNPR3 enhances defense response in Theobroma cacao. Front. Plant Sci. 2018, 9, 268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandhya, D.; Jogam, P.; Allini, V.R.; Abbagani, S.; Alok, A. The present and potential future methods for delivering CRISPR/Cas9 components in plants. J. Genet. Eng. Biotechnol. 2020, 18, 25. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.-l.; Liang, H.-z.; Wang, S.-f.; Lian, Y.; Wei, Y.-l.; Wang, T.-f. Research progress and commercialization on transgenic soybean in china. Soybean Sci. 2010, 29, 143–150. [Google Scholar]
- Li, S.; Cong, Y.; Liu, Y.; Wang, T.; Shuai, Q.; Chen, N.; Gai, J.; Li, Y. Optimization of Agrobacterium-mediated transformation in soybean. Front. Plant Sci. 2017, 8, 246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castel, B.; Tomlinson, L.; Locci, F.; Yang, Y.; Jones, J.D.G. Optimization of T-DNA architecture for Cas9-mediated mutagenesis in Arabidopsis. PLoS ONE 2019, 14, e0204778. [Google Scholar] [CrossRef] [Green Version]
- Zale, J.M.; Agarwal, S.; Loar, S.; Steber, C.M. Evidence for stable transformation of wheat by floral dip in Agrobacterium tumefaciens. Plant Cell Rep. 2009, 28, 903–913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bastaki, N.K.; Cullis, C.A. Floral-dip transformation of flax (Linum usitatissimum) to generate transgenic progenies with a high transformation rate. J. Visual. Exp. 2014, 19, 52189. [Google Scholar] [CrossRef] [Green Version]
- Curtis, I.S.; Nam, H.G. Transgenic radish (Raphanus sativus L. longipinnatus Bailey) by floral-dip method-plant development and surfactant are important in optimizing transformation efficiency. Transgenic Res. 2001, 10, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Sharada, M.S.; Kumari, A.; Pandey, A.K.; Sharma, S.; Sharma, P.; Sreelakshmi, Y.; Sharma, R. Generation of genetically stable transformants by Agrobacterium using tomato floral buds. Plant Cell Tissue Org. Cult. 2017, 129, 299–312. [Google Scholar] [CrossRef]
- Murovec, J.; Guček, K.; Bohanec, B.; Avbelj, M.; Jerala, R. DNA-free genome editing of Brassica oleracea and B. rapa protoplasts using CRISPR-Cas9 ribonucleoprotein complexes. Front. Plant Sci. 2018, 9, 1594. [Google Scholar] [CrossRef]
- Svitashev, S.; Young, J.K.; Schwartz, C.; Gao, H.; Falco, S.C.; Cigan, A.M. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 2015, 169, 931–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, M.; Turesson, H.; Olsson, N.; Fält, A.-S.; Ohlsson, P.; Gonzalez, M.N.; Samuelsson, M.; Hofvander, P. Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol. Plant. 2018, 164, 378–384. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Liu, Z.-B.; Xing, A.; Moon, B.P.; Koellhoffer, J.P.; Huang, L.; Ward, R.T.; Clifton, E.; Falco, S.C.; Cigan, A. Cas9-guide RNA directed genome editing in soybean. Plant Physiol. 2015, 169, 960–970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; Choi, J.; Won, K.-H. A stable DNA-free screening system for CRISPR/RNPs-mediated gene editing in hot and sweet cultivars of Capsicum annuum. BMC Plant Biol. 2020, 20, 449. [Google Scholar] [CrossRef]
- Foster, A.J.; Martin-Urdiroz, M.; Yan, X.; Wright, H.S.; Soanes, D.M.; Talbot, N.J. CRISPR-Cas9 ribonucleoprotein-mediated co-editing and counterselection in the rice blast fungus. Sci. Rep. 2018, 8, 14355. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Choi, S.; Park, S.; Yoon, J.; Park, A.Y.; Choe, S. DNA-free genome editing via ribonucleoprotein (RNP) delivery of CRISPR/Cas in lettuce. In Plant Genome Editing with CRISPR Systems: Methods and Protocols; Qi, Y., Ed.; Springer: New York, NY, USA, 2019; pp. 337–354. [Google Scholar]
- Waltz, E. Gene-edited CRISPR mushroom escapes US regulation. Nature 2016, 532, 293. [Google Scholar] [CrossRef] [Green Version]
- Baltes, N.J.; Gil-Humanes, J.; Cermak, T.; Atkins, P.A.; Voytas, D.F. DNA replicons for plant genome engineering. Plant Cell 2014, 26, 151–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cermak, T.; Baltes, N.; Cegan, R.; Zhang, Y.; Voytas, D. High-frequency, precise modification of the tomato genome. Genome Biol. 2015, 16, 232–246. [Google Scholar] [CrossRef] [Green Version]
- Gil-Humanes, J.; Wang, Y.; Liang, Z.; Shan, Q.; Ozuna, C.V.; Sánchez-León, S.; Baltes, N.J.; Starker, C.; Barro, F.; Gao, C.; et al. High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 2017, 89, 1251–1262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, K.; Han, T.; Liu, G.; Chen, T.; Wang, Y.; Yu, A.Y.L.; Liu, Y. A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci. Rep. 2015, 5, 14926. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Abul-faraj, A.; Piatek, M.; Mahfouz, M.M. Activity and specificity of TRV-mediated gene editing in plants. Plant Signal. Behav. 2015, 10, e1044191. [Google Scholar] [CrossRef]
- Ramakrishna, S.; Kwaku Dad, A.-B.; Beloor, J.; Gopalappa, R.; Lee, S.-K.; Kim, H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014, 24, 1020–1027. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Ji, W.; Hall, J.M.; Hu, Q.; Wang, C.; Beisel, C.L.; Gu, Z. Self-assembled DNA nanoclews for the efficient delivery of CRISPR–Cas9 for genome editing. Angew. Chem. Int. Ed. 2015, 54, 12029–12033. [Google Scholar] [CrossRef] [PubMed]
- Mout, R.; Ray, M.; Yesilbag Tonga, G.; Lee, Y.-W.; Tay, T.; Sasaki, K.; Rotello, V.M. Direct cytosolic delivery of CRISPR/Cas9-ribonucleoprotein for efficient gene editing. ACS Nano 2017, 11, 2452–2458. [Google Scholar] [CrossRef] [Green Version]
- Gori, J.L.; Hsu, P.D.; Maeder, M.L.; Shen, S.; Welstead, G.G.; Bumcrot, D. Delivery and specificity of CRISPR/Cas9 genome editing technologies for human gene therapy. Hum. Gene Ther. 2015, 26, 443–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molla, K.A.; Yang, Y. CRISPR/Cas-mediated base editing: Technical considerations and practical applications. Trends Biotechnol. 2019, 37, 1121–1142. [Google Scholar] [CrossRef]
- Rees, H.A.; Komor, A.C.; Yeh, W.-H.; Caetano-Lopes, J.; Warman, M.; Edge, A.S.B.; Liu, D.R. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat. Commun. 2017, 8, 15790. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Zong, Y.; Wang, Y.; Jin, S.; Zhang, D.; Song, Q.; Zhang, R.; Gao, C. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 2018, 19, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, B.-C.; Yun, J.-Y.; Kim, S.-T.; Shin, Y.; Ryu, J.; Choi, M.; Woo, J.W.; Kim, J.-S. Precision genome engineering through adenine base editing in plants. Nat. Plants 2018, 4, 427–431. [Google Scholar] [CrossRef]
- Hua, K.; Tao, X.; Yuan, F.; Wang, D.; Zhu, J.-K. Precise A·T to G·C base editing in the rice genome. Mol. Plant 2018, 11, 627–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hua, K.; Tao, X.; Zhu, J.-K. Expanding the base editing scope in rice by using Cas9 variants. Plant Biotechnol. J. 2019, 17, 499–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, L.; Li, J.; Wang, Q.; Xu, Z.; Sun, L.; Alariqi, M.; Manghwar, H.; Wang, G.; Li, B.; Ding, X.; et al. High-efficient and precise base editing of C•G to T•A in the allotetraploid cotton (Gossypium hirsutum) genome using a modified CRISPR/Cas9 system. Plant Biotechnol. J. 2020, 18, 45–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, F.; Kuang, Y.; Ren, B.; Wang, J.; Zhang, D.; Lin, H.; Yang, B.; Zhou, X.; Zhou, H. Highly efficient A.T to G.C base editing by Cas9n-guided tRNA adenosine deaminase in rice. Mol. Plant 2018, 11, 631–634. [Google Scholar] [CrossRef] [Green Version]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Yan, J.; Cirincione, A.; Adamson, B. Prime editing: Precision genome editing by reverse transcription. Mol. Cell 2020, 77, 210–212. [Google Scholar] [CrossRef]
- Xue, C.; Zhang, H.; Lin, Q.; Fan, R.; Gao, C. Manipulating mRNA splicing by base editing in plants. Sci. China Life Sci. 2018, 61, 1293–1300. [Google Scholar] [CrossRef]
- Li, Z.; Xiong, X.; Wang, F.; Liang, J.; Li, J.-F. Gene disruption through base editing-induced messenger RNA missplicing in plants. New Phytol. 2019, 222, 1139–1148. [Google Scholar] [CrossRef] [PubMed]
- Butt, H.; Rao, G.S.; Sedeek, K.; Aman, R.; Kamel, R.; Mahfouz, M. Engineering herbicide resistance via prime editing in rice. Plant Biotechnol. J. 2020, 18, 2370–2372. [Google Scholar] [CrossRef]
- Tang, X.; Sretenovic, S.; Ren, Q.; Jia, X.; Li, M.; Fan, T.; Yin, D.; Xiang, S.; Guo, Y.; Liu, L.; et al. Plant prime editors enable precise gene editing in rice cells. Mol. Plant 2020, 13, 667–670. [Google Scholar] [CrossRef]
- Xu, R.; Li, J.; Liu, X.; Shan, T.; Qin, R.; Wei, P. Development of plant prime-editing systems for precise genome editing. Plant Commun. 2020, 1, 100043. [Google Scholar] [CrossRef]
- Li, H.; Li, J.; Chen, J.; Yan, L.; Xia, L. Precise modifications of both exogenous and endogenous genes in rice by prime editing. Mol. Plant 2020, 13, 671–674. [Google Scholar] [CrossRef] [PubMed]
- Vouillot, L.; Thélie, A.; Pollet, N. Comparison of T7E1 and surveyor mismatch cleavage assays to detect mutations triggered by engineered nucleases. G3 Genes Genomes Genet. 2015, 5, 407–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, K.; Yang, Y. RNA-guided genome editing in plants using A CRISPR-Cas system. Mol. Plant 2013, 6, 1975–1983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sentmanat, M.F.; Peters, S.T.; Florian, C.P.; Connelly, J.P.; Pruett-Miller, S.M. A survey of validation strategies for CRISPR-Cas9 editing. Sci. Rep. 2018, 8, 888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richter, J.; Watson, J.M.; Stasnik, P.; Borowska, M.; Neuhold, J.; Berger, M.; Stolt-Bergner, P.; Schoft, V.; Hauser, M.-T. Multiplex mutagenesis of four clustered CrRLK1L with CRISPR/Cas9 exposes their growth regulatory roles in response to metal ions. Sci. Rep. 2018, 8, 12182. [Google Scholar] [CrossRef]
- Pan, C.; Ye, L.; Qin, L.; Liu, X.; He, Y.; Wang, J.; Chen, L.; Lu, G. CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci. Rep. 2016, 6, 24765. [Google Scholar] [CrossRef]
- Denbow, C.J.; Lapins, S.; Dietz, N.; Scherer, R.; Nimchuk, Z.L.; Okumoto, S. Gateway-compatible CRISPR-Cas9 vectors and a rapid detection by high-resolution melting curve analysis. Front. Plant Sci. 2017, 8, 1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samarut, É.; Lissouba, A.; Drapeau, P. A simplified method for identifying early CRISPR-induced indels in zebrafish embryos using High Resolution Melting analysis. BMC Genom. 2016, 17, 547. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Li, K.; Jin, L.; Xu, R.; Miao, K.; Yang, F.; Qi, C.; Zhang, L.; Botella, J.R.; Wang, R.; et al. A simple and cost-effective method for screening of CRISPR/Cas9-induced homozygous/biallelic mutants. Plant Meth. 2018, 14, 40. [Google Scholar] [CrossRef]
- Chen, L.; Li, W.; Katin-Grazzini, L.; Ding, J.; Gu, X.; Li, Y.; Gu, T.; Wang, R.; Lin, X.; Deng, Z.; et al. A method for the production and expedient screening of CRISPR/Cas9-mediated non-transgenic mutant plants. Hort. Res. 2018, 5, 13. [Google Scholar] [CrossRef]
- Li, S.; Liu, S.; Liu, Y.; Lu, H.; Tan, Y.; Huang, J.; Wei, P.; Shu, Q.Y. HRM-facilitated rapid identification and genotyping of mutations induced by CRISPR/Cas9 mutagenesis in rice. Crop Breed. Appl. Biotechnol. 2018, 18, 184–191. [Google Scholar] [CrossRef] [Green Version]
- Rocafort, M.; Arshed, S.; Hudson, D.; Singh, J.; Bowen, J.K.; Plummer, K.M.; Bradshaw, R.E.; Johnson, R.D.; Johnson, L.J.; Mesarich, C.H. CRISPR-Cas9 gene editing and rapid detection of gene-edited mutants using high-resolution melting in the apple scab fungus, Venturia inaequalis. bioRxiv 2021. [Google Scholar] [CrossRef]
- Bewg, W.P.; Ci, D.; Tsai, C.-J. Genome editing in trees: From multiple repair pathways to long-term stability. Front. Plant Sci. 2018, 9, 1732. [Google Scholar] [CrossRef]
- Ren, Q.; Zhong, Z.; Wang, Y.; You, Q.; Li, Q.; Yuan, M.; He, Y.; Qi, C.; Tang, X.; Zheng, X.; et al. Bidirectional promoter-based CRISPR-Cas9 systems for plant genome editing. Front. Plant Sci. 2019, 10, 1173. [Google Scholar] [CrossRef] [PubMed]
- Bruegmann, T.; Deecke, K.; Fladung, M. Evaluating the efficiency of gRNAs in CRISPR/Cas9 mediated genome editing in poplars. Int. J. Mol. Sci. 2019, 20, 3623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalla Costa, L.; Piazza, S.; Pompili, V.; Salvagnin, U.; Cestaro, A.; Moffa, L.; Vittani, L.; Moser, C.; Malnoy, M. Strategies to produce T-DNA free CRISPRed fruit trees via Agrobacterium tumefaciens stable gene transfer. Sci. Rep. 2020, 10, 20155. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Chen, Y.; Ma, Y.; Wang, H.; Wei, J. Effective reduction in chimeric mutants of poplar trees produced by CRISPR/Cas9 through a second round of shoot regeneration. Plant Biotechnol. Rep. 2020, 14, 549–558. [Google Scholar] [CrossRef]
- Hendel, A.; Fine, E.J.; Bao, G.; Porteus, M.H. Quantifying on- and off-target genome editing. Trends Biotechnol. 2015, 33, 132–140. [Google Scholar] [CrossRef] [Green Version]
- Macovei, A.; Sevilla, N.R.; Cantos, C.; Jonson, G.B.; Slamet-Loedin, I.; Čermák, T.; Voytas, D.F.; Choi, I.-R.; Chadha-Mohanty, P. Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol. J. 2018, 16, 1918–1927. [Google Scholar] [CrossRef] [Green Version]
- de Pater, S.; Klemann, B.; Hooykaas, P.J.J. True gene-targeting events by CRISPR/Cas-induced DSB repair of the PPO locus with an ectopically integrated repair template. Sci. Rep. 2018, 8, 3338. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wu, J.-J.; Tang, T.; Liu, K.-D.; Dai, C. CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci. Rep. 2017, 7, 7489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minkenberg, B.; Xie, K.; Yang, Y. Discovery of rice essential genes by characterizing a CRISPR-edited mutation of closely related rice MAP kinase genes. Plant J. 2017, 89, 636–648. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zhang, J.; Wei, P.; Zhang, B.; Gou, F.; Feng, Z.; Mao, Y.; Yang, L.; Zhang, H.; Xu, N.; et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 2014, 12, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Fernandez i Marti, A.; Dodd, R.S. Using CRISPR as a gene editing tool for validating adaptive gene function in tree landscape genomics. Front. Ecol. Evol. 2018, 6. [Google Scholar] [CrossRef] [Green Version]
- Lonowski, L.A.; Narimatsu, Y.; Riaz, A.; Delay, C.E.; Yang, Z.; Niola, F.; Duda, K.; Ober, E.A.; Clausen, H.; Wandall, H.H.; et al. Genome editing using FACS enrichment of nuclease-expressing cells and indel detection by amplicon analysis. Nat. Protoc. 2017, 12, 581–603. [Google Scholar] [CrossRef]
- Tsai, C.J.; Xue, L.J. CRISPRing into the woods. GM Crops Food 2015, 6, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wu, H.; Chen, Y.; Yin, T. Efficient CRISPR/Cas9-mediated gene editing in an interspecific hybrid poplar with a highly heterozygous genome. Front. Plant Sci. 2020, 11, 996. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.S.; Sattar, M.N.; Iqbal, Z.; Raza, A.; Al-Sadi, A.M. Next-generation sequencing and the CRISPR-Cas nexus: A molecular plant virology perspective. Front. Microbiol. 2021, 11, 3456. [Google Scholar] [CrossRef] [PubMed]
- Montecillo, J.A.V.; Chu, L.L.; Bae, H. CRISPR-Cas9 system for plant genome editing: Current approaches and emerging developments. Agronomy 2020, 10, 1033. [Google Scholar] [CrossRef]
- Chang, S.; Mahon, E.L.; MacKay, H.A.; Rottmann, W.H.; Strauss, S.H.; Pijut, P.M.; Powell, W.A.; Coffey, V.; Lu, H.; Mansfield, S.D.; et al. Genetic engineering of trees: Progress and new horizons. In Vitro Cell. Develop. Biol. Plant 2018, 54, 341–376. [Google Scholar] [CrossRef]
- Vats, S.; Kumawat, S.; Kumar, V.; Patil, G.B.; Joshi, T.; Sonah, H.; Sharma, T.R.; Deshmukh, R. Genome editing in plants: Exploration of technological advancements and challenges. Cells 2019, 8, 1386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, Y.; Xin, S.; Dai, X.; Yang, X.; Huang, H.; Hua, Y. Efficient genome editing of rubber tree (Hevea brasiliensis) protoplasts using CRISPR/Cas9 ribonucleoproteins. Ind. Crops Prod. 2020, 146, 112146. [Google Scholar] [CrossRef]
- Tsai, C.J.; Xu, P.; Xue, L.J.; Hu, H.; Nyamdari, B.; Naran, R.; Zhou, X.; Goeminne, G.; Gao, R.; Gjersing, E.; et al. Compensatory guaiacyl lignin biosynthesis at the expense of syringyl lignin in 4CL1-knockout poplar. Plant Physiol. 2020, 183, 123–136. [Google Scholar] [CrossRef] [Green Version]
- Lowe, K.; Wu, E.; Wang, N.; Hoerster, G.; Hastings, C.; Cho, M.-J.; Scelonge, C.; Lenderts, B.; Chamberlin, M.; Cushatt, J.; et al. Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 2016, 28, 1998–2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lotan, T.; Ohto, M.-A.; Yee, K.M.; West, M.A.L.; Lo, R.; Kwong, R.W.; Yamagishi, K.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 1998, 93, 1195–1205. [Google Scholar] [CrossRef] [Green Version]
- Stone, S.L.; Braybrook, S.A.; Paula, S.L.; Kwong, L.W.; Meuser, J.; Pelletier, J.; Hsieh, T.-F.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: Implications for somatic embryogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 3151–3156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srinivasan, C.; Liu, Z.; Heidmann, I.; Supena, E.D.J.; Fukuoka, H.; Joosen, R.; Lambalk, J.; Angenent, G.; Scorza, R.; Custers, J.B.M.; et al. Heterologous expression of the BABY BOOM AP2/ERF transcription factor enhances the regeneration capacity of tobacco (Nicotiana tabacum L.). Planta 2006, 225, 341. [Google Scholar] [CrossRef]
- Deng, W.; Luo, K.; Li, Z.; Yang, Y. A novel method for induction of plant regeneration via somatic embryogenesis. Plant Sci. 2009, 177, 43–48. [Google Scholar] [CrossRef]
- Bisi, R.B.; Pio, R.; da Hora Farias, D.; Locatelli, G.; de Alcântara Barbosa, C.M.; Pereira, W.A. Molecular characterization of the S-alleles and compatibility among hybrid pear tree cultivars for subtropical regions. Hort. Sci. 2019, 54, 2104–2110. [Google Scholar] [CrossRef] [Green Version]
- Peng, C.; Wang, H.; Xu, X.; Wang, X.; Chen, X.; Wei, W.; Lai, Y.; Liu, G.; Godwin, I.D.; Li, J.; et al. High-throughput detection and screening of plants modified by gene editing using quantitative real-time polymerase chain reaction. Plant J. 2018, 95, 557–567. [Google Scholar] [CrossRef]
- Abu-Qaoud, H.; Skirvin, R.M.; Chevreau, E. In vitro separation of chimeral pears into their component genotypes. Euphytica 1990, 48, 189–196. [Google Scholar] [CrossRef]
- Strauss, S.H.; Costanza, A.; Séguin, A. Genetically engineered trees: Paralysis from good intentions. Science 2015, 349, 794–795. [Google Scholar] [CrossRef] [PubMed]
- Sattar, M.N.; Iqbal, Z.; Tahir, M.N.; Shahid, M.S.; Khurshid, M.; Al-Khateeb, A.A.; Al-Khateeb, S.A. CRISPR/Cas9: A practical approach in date palm genome editing. Front. Plant Sci. 2017, 8, 1469. [Google Scholar] [CrossRef] [Green Version]
- Nadakuduti, S.S.; Buell, C.R.; Voytas, D.F.; Starker, C.G.; Douches, D.S. Genome editing for crop improvement—Applications in clonally propagated polyploids with a focus on potato (Solanum tuberosum L.). Front. Plant Sci. 2018, 9, 1607. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liang, Z.; Zong, Y.; Wang, Y.; Liu, J.; Chen, K.; Qiu, J.L.; Gao, C. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016, 7, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Liang, Z.; Chen, K.; Li, T.; Zhang, Y.; Wang, Y.; Zhao, Q.; Liu, J.; Zhang, H.; Liu, C.; Ran, Y.; et al. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 2017, 8, 14261. [Google Scholar] [CrossRef]
- Hu, N.; Xian, Z.; Li, N.; Liu, Y.; Huang, W.; Yan, F.; Su, D.; Chen, J.; Li, Z. Rapid and user-friendly open-source CRISPR/Cas9 system for single- or multi-site editing of tomato genome. Hort. Res. 2019, 6, 7. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Araki, M. A future scenario of the global regulatory landscape regarding genome-edited crops. GM Crops Food 2017, 8, 44–56. [Google Scholar] [CrossRef] [Green Version]
- Ellens, K.W.; Levac, D.; Pearson, C.; Savoie, A.; Strand, N.; Louter, J.; Tibelius, C. Canadian Regulatory Aspects of Gene Editing Technologies. Transgenic Res. 2019, 28, 165–168. [Google Scholar] [CrossRef]
- Mallapaty, S. Australian gene-editing rules adopt“middle ground”. Nature 2019. [Google Scholar] [CrossRef]
- Callaway, E. CRISPR plants now subject to tough GM laws in European Union. Nature 2018, 560, 16–17. [Google Scholar] [CrossRef] [PubMed]
- Wight, A.J. Strict EU ruling on gene-edited crops squeezes science. Nature 2018, 563, 15–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).