Disparate Dynamics of Gene Body and cis-Regulatory Element Evolution Illustrated for the Senescence-Associated Cysteine Protease Gene SAG12 of Plants
Abstract
:1. Introduction
2. Results
2.1. The Arabidopsis thaliana SAG12 Promoter
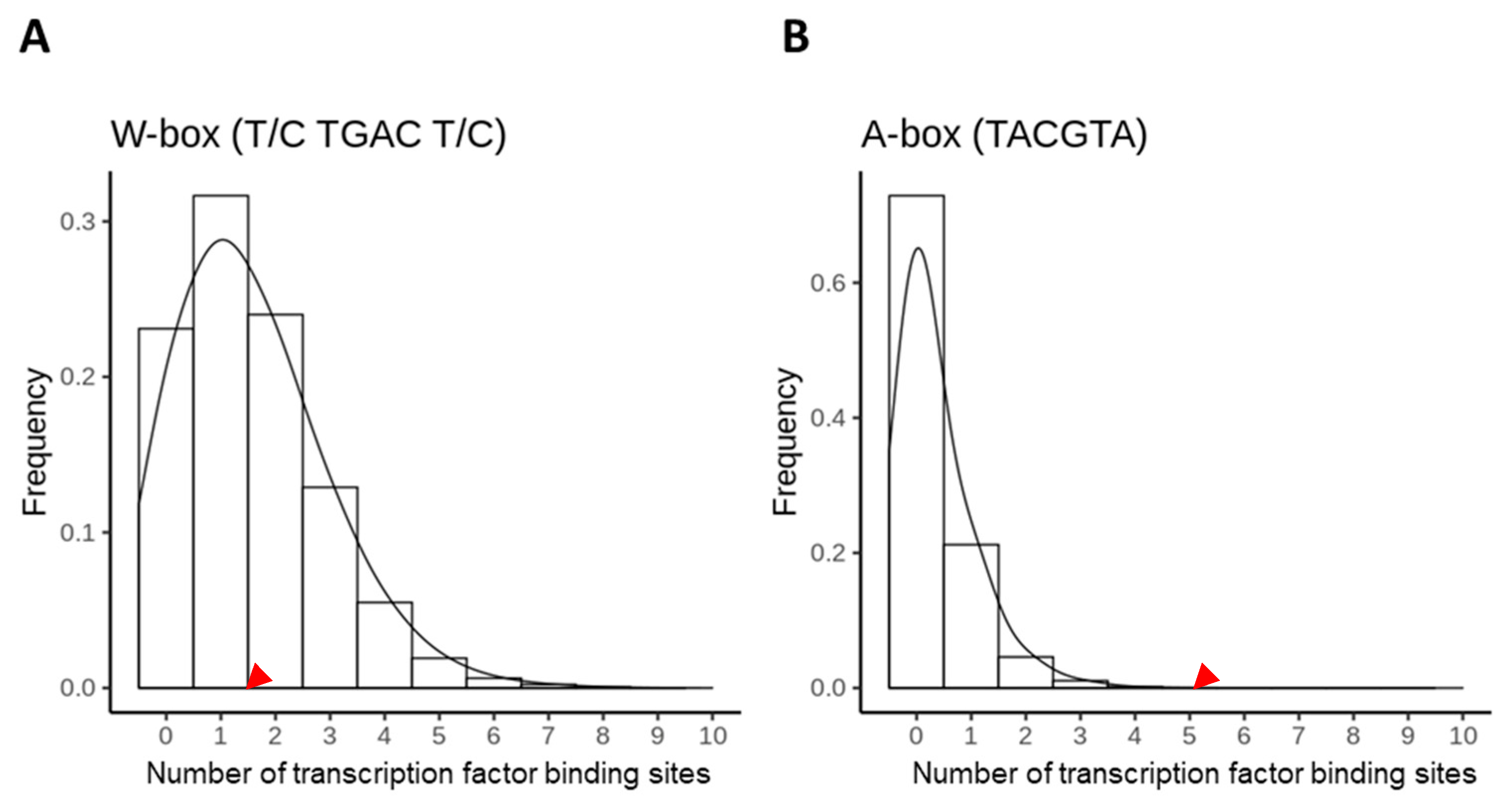
2.2. Random and Non-Random Distribution of A- and W-Boxes in the AtSAG12 Promoter Region
2.3. Evolution of the SAG12 Promoter across the Plant Kingdom
2.4. Intron–Exon Structure of the AtSAG12 Orthologs
3. Discussion
4. Materials and Methods
4.1. Sequence Analyses
4.2. Phylogentic Trees
4.3. cis-Element Analyses
4.4. Recourses Used for Gene Expression Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sade, N.; Rubio-Wilhelmi, M.D.M.; Umnajkitikorn, K.; Blumwald, E. Stress-induced senescence and plant tolerance to abiotic stress. J. Exp. Bot. 2018, 69, 845–853. [Google Scholar] [CrossRef]
- Zentgraf, U.; Jobst, J.; Kolb, D.; Rentsch, D. Senescence-related gene expression profiles of rosette leaves of Arabidopsis thaliana: Leaf age versus plant age. Plant Biol. 2004, 6, 178–183. [Google Scholar] [CrossRef]
- Breeze, E.; Harrison, E.; McHattie, S.; Hughes, L.; Hickman, R.; Hill, C.; Kiddle, S.; Kim, Y.; Penfold, C.A.; Jenkins, D.; et al. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 2011, 23, 873–894. [Google Scholar] [CrossRef] [Green Version]
- Jones, D.M.; Vandepoele, K. Identification and evolution of gene regulatory networks: Insights from comparative studies in plants. Curr. Opin. Plant Biol. 2020, 54, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Signor, S.A.; Nuzhdin, S.V. The evolution of gene expression in cis and trans. Trends Genet. 2018, 34, 532–544. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ngu, D.W.; Carvalho, D.; Liang, Z.; Qiu, Y.; Roston, R.L.; Schnable, J.C. Differentially regulated orthologs in sorghum and the subgenomes of maize. Plant Cell 2017, 29, 1938–1951. [Google Scholar] [CrossRef] [Green Version]
- Zhou, P.; Li, Z.; Magnusson, E.; Gomez Cano, F.; Crisp, P.A.; Noshay, J.M.; Grotewold, E.; Hirsch, C.N.; Briggs, S.P.; Springer, N.M. Meta Gene Regulatory Networks in Maize Highlight Functionally Relevant Regulatory Interactions. Plant Cell 2020, 32, 1377–1396. [Google Scholar] [CrossRef] [Green Version]
- Lohman, K.N.; Gan, S.; John, M.C.; Amasino, R.M. Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiol. Plant 1994, 92, 322–328. [Google Scholar] [CrossRef]
- Weaver, L.M.; Gan, S.; Quirino, B.; Amasino, R.M. A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol. Biol. 1998, 37, 455–469. [Google Scholar] [CrossRef] [PubMed]
- James, M.; Poret, M.; Masclaux-Daubresse, C.; Marmagne, A.; Coquet, L.; Jouenne, T.; Chan, P.; Trouverie, J.; Etienne, P. SAG12, a major cysteine protease involved in nitrogen allocation during senescence for seed production in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 2052–2063. [Google Scholar] [CrossRef] [Green Version]
- Otegui, M.S.; Noh, Y.S.; Martínez, D.E.; Vila Petroff, M.G.; Andrew Staehelin, L.; Amasino, R.M.; Guiamet, J.J. Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J. 2005, 41, 831–844. [Google Scholar] [CrossRef]
- Carrión, C.A.; Costa, M.L.; Martínez, D.E.; Mohr, C.; Humbeck, K.; Guiamet, J.J. In vivo inhibition of cysteine proteases provides evidence for the involvement of ‘senescence-associated vacuoles’ in chloroplast protein degradation during dark-induced senescence of tobacco leaves. J. Exp. Bot. 2013, 64, 4967–4980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gan, S.; Amasino, R.M. Inhibition of leaf senescence by autoregulated production of cytokinin. Science 1995, 270, 1986–1988. [Google Scholar] [CrossRef] [PubMed]
- Noh, Y.S.; Amasino, R.M. Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Mol. Biol. 1999, 41, 181–194. [Google Scholar] [CrossRef]
- Miao, Y.; Laun, T.; Zimmermann, P.; Zentgraf, U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 2004, 55, 853–867. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xiang, S.; Chen, Y.; Li, D.; Yu, D. Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol. Plant 2017, 10, 1174–1189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Cai, Z.; Gan, S. Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 2004, 27, 521–549. [Google Scholar] [CrossRef]
- Kim, H.J.; Nam, H.G.; Lim, P.O. Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 2016, 33, 48–56. [Google Scholar] [CrossRef]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Ferrier, T.; Matus, J.T.; Jin, J.; Riechmann, J.L. Arabidopsis paves the way: Genomic and network analyses in crops. Curr. Opin. Biotechnol. 2011, 22, 260–270. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Hu, M.; Wang, Q.; Cheng, L.; Zhang, Z. Role of Papain-Like Cysteine Proteases in Plant Development. Front. Plant Sci. 2018, 9, 1717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, Y.Y.; Ichida, H.; Matsui, M.; Obokata, J.; Sakurai, T.; Satou, M.; Seki, M.; Shinozaki, K.; Abe, T. Identification of plant promoter constituents by analysis of local distribution of short sequences. BMC Genom. 2007, 8, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rushton, P.J.; Torres, J.T.; Parniske, M.; Wernert, P.; Hahlbrock, K.; Somssich, I.E. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 1996, 15, 5690–5700. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. DNA-binding specificity and molecular functions of NAC transcription factors. Plant Sci. 2005, 169, 785–797. [Google Scholar] [CrossRef]
- Dröge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family-an update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef]
- Solovyev, V.V.; Shahmuradov, I.A.; Salamov, A.A. Identification of promoter regions and regulatory sites. Methods Mol. Biol. 2010, 674, 57–83. [Google Scholar]
- Shahmuradov, I.; Solovyev, V. Nsite, NsiteH and NsiteM computer tools for studying transcription regulatory elements. Bioinformatics 2015, 31, 3544–3545. [Google Scholar] [CrossRef] [Green Version]
- Haudry, A.; Platts, A.E.; Vello, E.; Hoen, D.R.; Leclercq, M.; Williamson, R.J.; Forczek, E.; Joly-Lopez, Z.; Steffen, J.G.; Hazzouri, K.M.; et al. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat. Genet. 2013, 45, 891–898. [Google Scholar] [CrossRef]
- Liu, J.; Jung, C.; Xu, J.; Wang, H.; Deng, S.; Bernad, L.; Arenas-Huertero, C.; Chua, N.H. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 2012, 24, 4333–4345. [Google Scholar] [CrossRef] [Green Version]
- James, M.; Masclaux-Daubresse, C.; Marmagne, A.; Azzopardi, M.; Laîné, P.; Goux, D.; Etienne, P.; Trouverie, J. A new role for SAG12 cysteine protease in roots of Arabidopsis thaliana. Front. Plant Sci. 2019, 9, 1998. [Google Scholar] [CrossRef]
- Lu, K.; Wei, L.; Li, X.; Wang, Y.; Wu, J.; Liu, M.; Zhang, C.; Chen, Z.; Xiao, Z.; Jian, H.; et al. Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, H.; Cui, Y.; Ding, Y.; Mei, J.; Dong, H.; Zhang, W.; Wu, S.; Liang, Y.; Zhang, C.; Li, J.; et al. Time-series analyses of transcriptomes and proteomes reveal molecular networks underlying oil accumulation in canola. Front. Plant Sci. 2017, 7, 2007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, R.; Wang, J.P.; Lin, Y.C.; Li, Q.; Sun, Y.H.; Chen, H.; Sederoff, R.R.; Chiang, V.L. Tissue and cell-type co-expression networks of transcription factors and wood component genes in Populus trichocarpa. Planta 2017, 245, 927–938. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, M.; Takahashi, H.; Iwakawa, H.; Nakagawa, A.; Ishikawa, T.; Tanaka, H.; Matsumura, Y.; Pekker, I.; Eshed, Y.; Vial-Pradel, S.; et al. Dual regulation of ETTIN (ARF3) gene expression by AS1-AS2, which maintains the DNA methylation level, is involved in stabilization of leaf adaxial-abaxial partitioning in Arabidopsis. Development 2013, 140, 1958–1969. [Google Scholar] [CrossRef] [Green Version]
- Safi, A.; Medici, A.; Szponarski, W.; Ruffel, S.; Lacombe, B.; Krouk, G. The world according to GARP transcription factors. Curr. Opin. Plant Biol. 2017, 39, 159–167. [Google Scholar] [CrossRef]
- de Boer, C.G.; Vaishnav, E.D.; Sadeh, R.; Abeyta, E.L.; Friedman, N.; Regev, A. Deciphering eukaryotic gene-regulatory logic with 100 million random promoters. Nat. Biotechnol. 2020, 38, 56–65. [Google Scholar] [CrossRef]
- Honjo, M.N.; Kudoh, H. Arabidopsis halleri: A perennial model system for studying population differentiation and local adaptation. AoB Plants 2019, 11, plz076. [Google Scholar] [CrossRef] [Green Version]
- Thibaud-Nissen, F.; Wu, H.; Richmond, T.; Redman, J.C.; Johnson, C.; Green, R.; Arias, J.; Town, C.D. Development of Arabidopsis whole-genome microarrays and their application to the discovery of binding sites for the TGA2 transcription factor in salicylic acid-treated plants. Plant J. 2006, 47, 152–162. [Google Scholar] [CrossRef]
- Lee, J.; He, K.; Stolc, V.; Lee, H.; Figueroa, P.; Gao, Y.; Tongprasit, W.; Zhao, H.; Lee, I.; Deng, X.W. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 2007, 19, 731–749. [Google Scholar] [CrossRef] [Green Version]
- Noh, Y.S.; Amasino, R.M. Regulation of developmental senescence is conserved between Arabidopsis and Brassica napus. Plant Mol. Biol. 1999, 41, 195–206. [Google Scholar] [CrossRef]
- Hörtensteiner, S.; Chinner, J.; Matile, P.; Thomas, H.; Donnison, I.S. Chlorophyll breakdown in Chlorella protothecoides: Characterization of degreening and cloning of degreening-related genes. Plant Mol. Biol. 2000, 42, 439–450. [Google Scholar] [CrossRef]
- Thomas, H.; Huang, L.; Young, M.; Ougham, H. Evolution of plant senescence. BMC Evol. Biol. 2009, 9, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohno, S. Evolution by Gene Duplication; Springer Science & Business Media: New York, NY, USA, 2013. [Google Scholar]
- Kellis, M.; Birren, B.W.; Lander, E.S. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 2004, 428, 617–624. [Google Scholar] [CrossRef]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 2019, 20, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Wong, G.K.S.; Soltis, D.E.; Leebens-Mack, J.; Wickett, N.J.; Barker, M.S.; de Peer, Y.V.; Graham, S.W.; Melkonian, M. Sequencing and Analyzing the Transcriptomes of a Thousand Species Across the Tree of Life for Green Plants. Annu. Rev. Plant Biol. 2019, 71, 741–765. [Google Scholar] [CrossRef] [PubMed]
- Rogozin, I.B.; Carmel, L.; Csuros, M.; Koonin, E.V. Origin and evolution of spliceosomal introns. Biol. Direct 2012, 7, 1–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knowles, D.G.; McLysaght, A. High rate of recent intron gain and loss in simultaneously duplicated Arabidopsis genes. Mol. Biol. Evol. 2006, 23, 1548–1557. [Google Scholar] [CrossRef]
- Basu, M.K.; Rogozin, I.B.; Deusch, O.; Dagan, T.; Martin, W.; Koonin, E.V. Evolutionary dynamics of introns in plastid-derived genes in plants: Saturation nearly reached but slow intron gain continues. Mol. Biol. Evol. 2008, 25, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; White, M.J.; MacRae, T.H. Transcription factors and their genes in higher plants: Functional domains, evolution and regulation. Eur. J. Biochem. 1999, 262, 247–257. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, L. The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 2005, 5, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Corrêa, L.G.G.; Riaño-Pachón, D.M.; Schrago, C.G.; dos Santos, R.V.; Mueller-Roeber, B.; Vincentz, M. The role of bZIP transcription factors in green plant evolution: Adaptive features emerging from four founder genes. PLoS ONE 2008, 3, e2944. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Rinerson, C.I.; Rabara, R.C.; Tripathi, P.; Shen, Q.J.; Rushton, P.J. The evolution of WRKY transcription factors. BMC Plant Biol. 2015, 15, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Li, C.; Wang, H.; Guo, Z. WRKY transcription factors: Evolution, binding, and action. Phytopathol. Res. 2019, 1, 13. [Google Scholar] [CrossRef]
- Moses, A.M.; Chiang, D.Y.; Kellis, M.; Lander, E.S.; Eisen, M.B. Position specific variation in the rate of evolution in transcription factor binding sites. BMC Evol. Biol. 2003, 3, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shultzaberger, R.K.; Maerkl, S.J.; Kirsch, J.F.; Eisen, M.B. Probing the Informational and regulatory plasticity of a transcription factor DNA–binding domain. PLoS Genet. 2012, 8, e1002614. [Google Scholar] [CrossRef]
- Chiang, D.Y.; Moses, A.M.; Kellis, M.; Lander, E.S.; Eisen, M.B. Phylogenetically and spatially conserved word pairs associated with gene-expression changes in yeasts. Genome Biol. 2003, 4, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shultzaberger, R.K.; Malashock, D.S.; Kirsch, J.F.; Eisen, M.B. The fitness landscapes of cis-acting binding sites in different promoter and environmental contexts. PLoS Genet. 2010, 6, e1001042. [Google Scholar] [CrossRef] [PubMed]
- Lusk, R.W.; Eisen, M.B. Evolutionary mirages: Selection on binding site composition creates the illusion of conserved grammars in Drosophila enhancers. PLoS Genet. 2010, 6, e1000829. [Google Scholar] [CrossRef] [Green Version]
- Bradley, R.K.; Li, X.Y.; Trapnell, C.; Davidson, S.; Pachter, L.; Chu, H.C.; Tonkin, L.A.; Biggin, M.D.; Eisen, M.B. Binding site turnover produces pervasive quantitative changes in transcription factor binding between closely related Drosophila species. PLoS Biol. 2010, 8, e1000343. [Google Scholar] [CrossRef] [Green Version]
- Fay, J.C.; McCullough, H.L.; Sniegowski, P.D.; Eisen, M.B. Population genetic variation in gene expression is associated with phenotypic variation in Saccharomyces cerevisiae. Genome Biol. 2004, 5, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, F.; Wang, M.; Zhang, S. Sedoheptulose-1, 7-bisphosphatase is involved in methyl jasmonate-and dark-induced leaf senescence in tomato plants. Int. J. Mol. Sci. 2018, 19, 3673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forlani, S.; Cozzi, C.; Rosa, S.; Tadini, L.; Masiero, S.; Mizzotti, C. HEBE, a novel positive regulator of senescence in Solanum lycopersicum. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vatov, E.; Ludewig, U.; Zentgraf, U. Disparate Dynamics of Gene Body and cis-Regulatory Element Evolution Illustrated for the Senescence-Associated Cysteine Protease Gene SAG12 of Plants. Plants 2021, 10, 1380. https://doi.org/10.3390/plants10071380
Vatov E, Ludewig U, Zentgraf U. Disparate Dynamics of Gene Body and cis-Regulatory Element Evolution Illustrated for the Senescence-Associated Cysteine Protease Gene SAG12 of Plants. Plants. 2021; 10(7):1380. https://doi.org/10.3390/plants10071380
Chicago/Turabian StyleVatov, Emil, Uwe Ludewig, and Ulrike Zentgraf. 2021. "Disparate Dynamics of Gene Body and cis-Regulatory Element Evolution Illustrated for the Senescence-Associated Cysteine Protease Gene SAG12 of Plants" Plants 10, no. 7: 1380. https://doi.org/10.3390/plants10071380
APA StyleVatov, E., Ludewig, U., & Zentgraf, U. (2021). Disparate Dynamics of Gene Body and cis-Regulatory Element Evolution Illustrated for the Senescence-Associated Cysteine Protease Gene SAG12 of Plants. Plants, 10(7), 1380. https://doi.org/10.3390/plants10071380





