Differences in Grain Microstructure and Proteomics of a Broad Bean (Vicia faba L.) Landrace Cixidabaican in China Compared with Lingxiyicun Introduced from Japan
Abstract
:1. Introduction
2. Results and Discussion
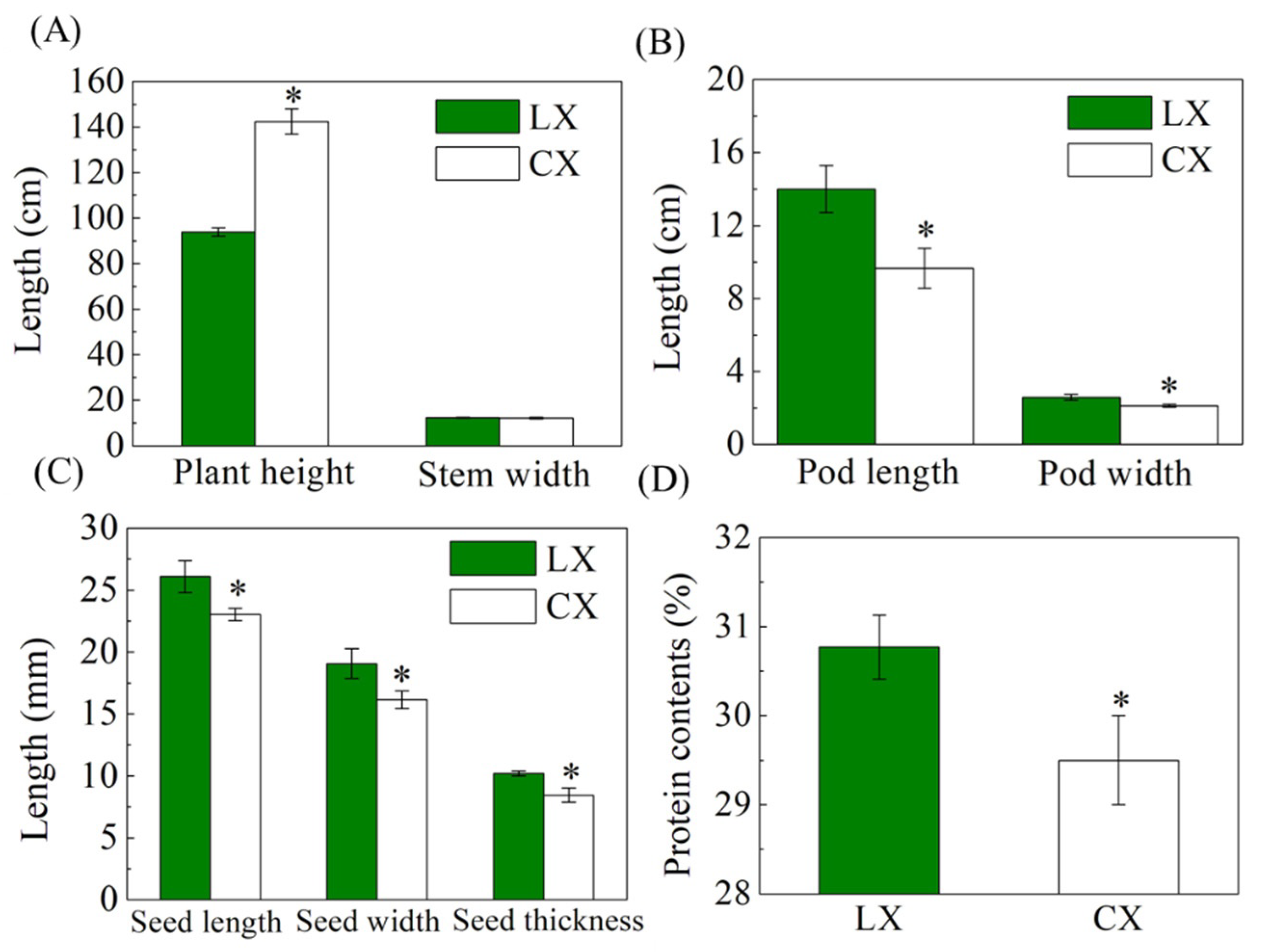
2.1. Differences in Plant Morphology, Flowers, Stem, Roots, Pods, and Seeds between CX and LX
2.2. Pods, Seeds, and Yield per Plant of CX Were Higher, While the Sizes of Pods and Seeds Were Smaller Than LX
2.3. Amino Acid Contents Were High but Showed No Differences between CX and LX
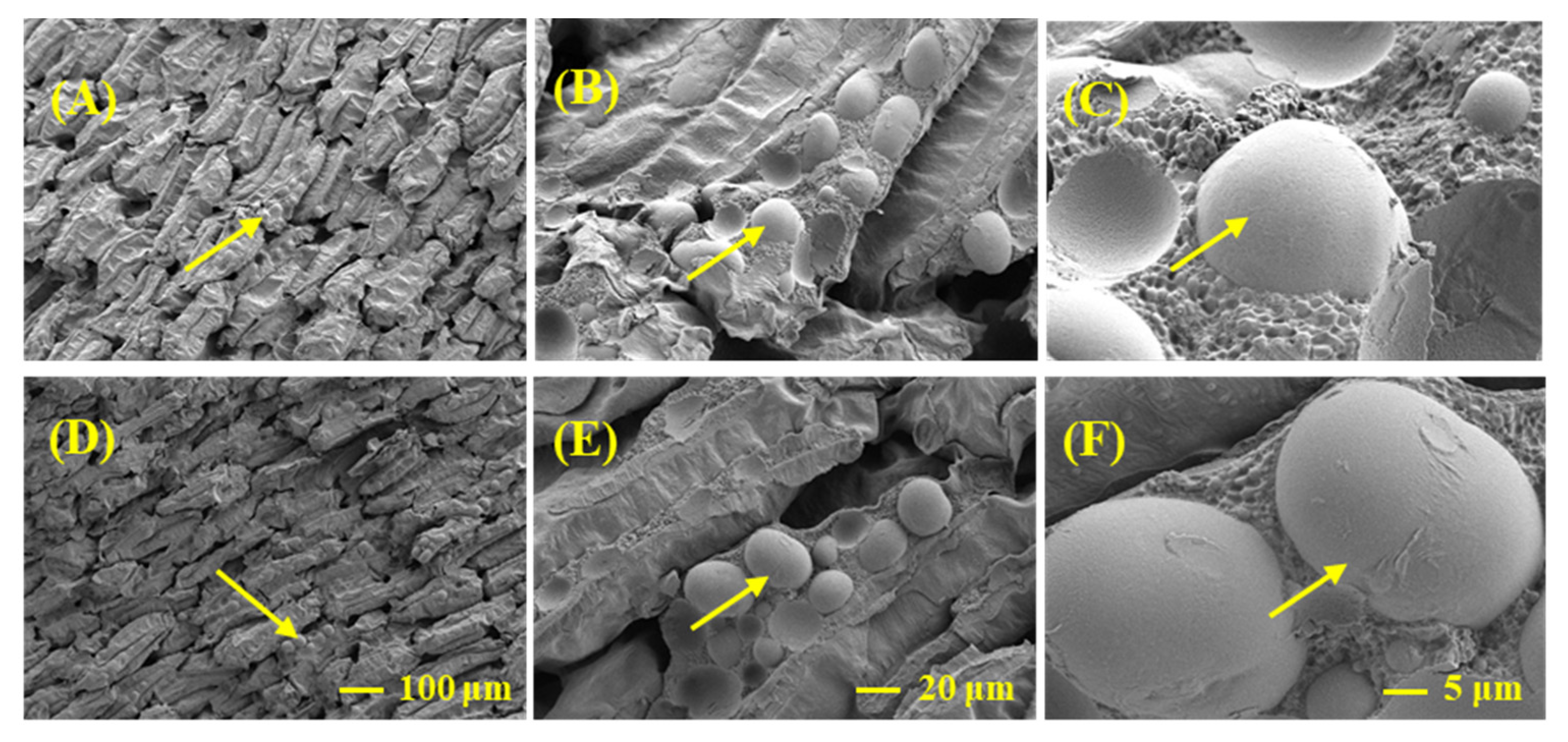
2.4. Seed Crude Protein Contents and Granules Were Lower in CX Than LX
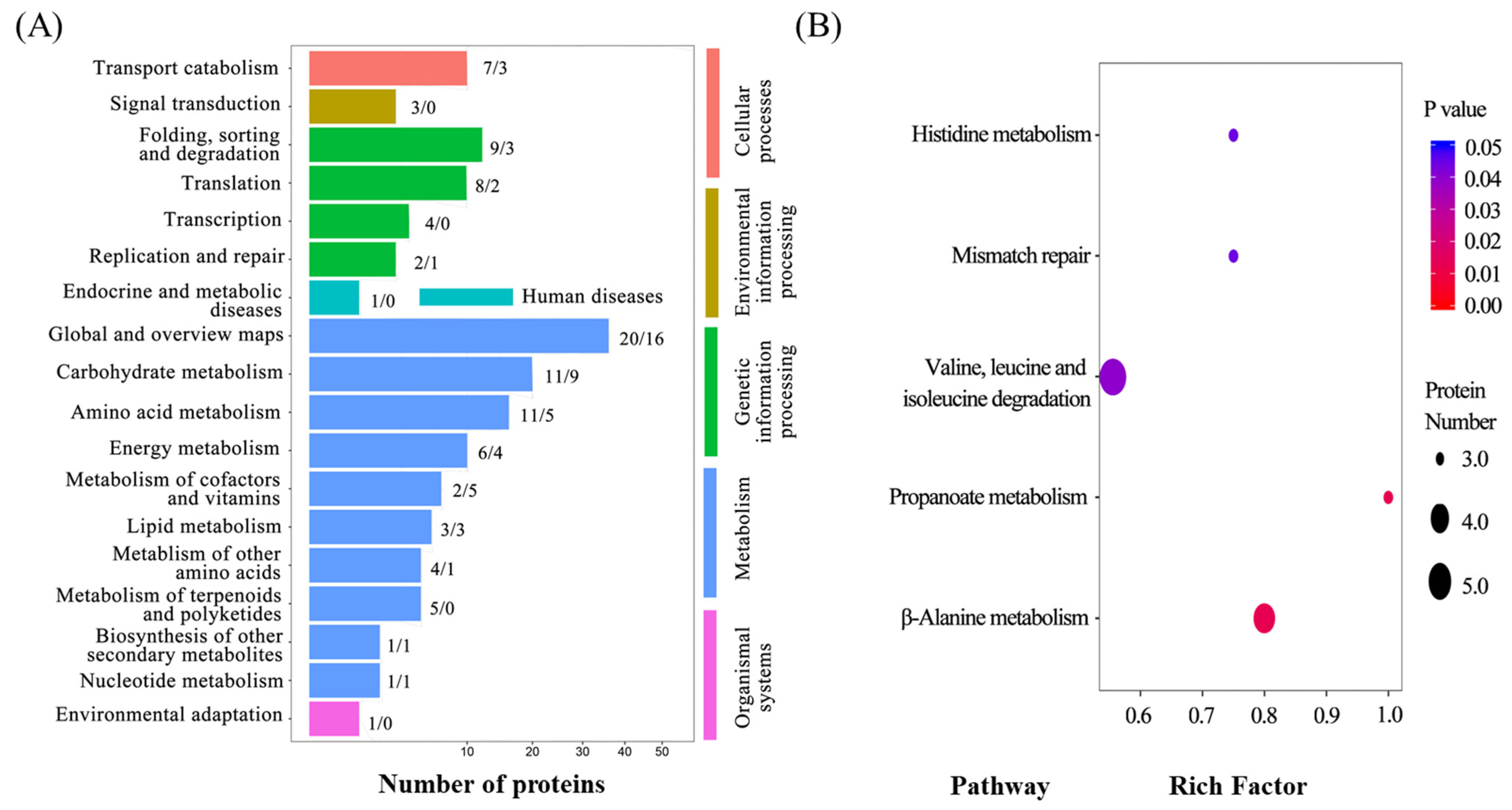
2.5. Identification of Seed Protein Levels and Differentially Abundant Proteins (DAPs) Significantly Enriched in Carbohydrate and Amino Acid Metabolism
3. Materials and Methods
3.1. Experiment Design, Plant Materials, and Harvest
3.2. Determination of Seed Protein and Amino Acid Contents
3.3. Ultrastructure Observation of Broad Bean Cotyledon Cross-Sections
3.4. Proteomics Analysis
3.5. Statistical Analysis
4. Conclusions
- (1)
- CX grew taller than LX, but the seed size of CX was smaller than LX, and seed weight was lower.
- (2)
- Seed yield of CX was higher due to a greater number of pods, but the protein content was lower than LX.
- (3)
- Proteomics analysis showed that, as a result of long-term adaption to the local environment, heat shock proteins, L-ascorbate peroxidase, catalase, EDS1 protein, thioredoxins, and STICHEL protein were upregulated, which could indicate increased environmental adaption ability.
- (4)
- Downregulated LOX activity in CX can also be a useful trait for future work on the alleviation of off-flavors.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Karkanis, A.; Ntatsi, G.; Lepse, L.; Fernández, J.A.; Vågen, I.M.; Rewald, B.; Alsina, I.; Kronberga, A.; Balliu, A.; Olle, M.; et al. Faba bean cultivation-revealing novel managing practices for more sustainable and competitive European cropping systems. Front. Plant Sci. 2018, 9, 1115. [Google Scholar] [CrossRef] [PubMed]
- Caracuta, V.; Barzilai, O.; Khalaily, H.; Milevski, L.; Paz, Y.; Vardi, J.; Regev, L.; Boaretto, E. The onset of faba bean farming in the southern levant. Sci. Rep. 2015, 5, 14370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, E.; Peoples, M.B.; Hauggaard-Nielsen, H. Faba bean in cropping systems. Field Crop. Res. 2010, 115, 203–216. [Google Scholar] [CrossRef] [Green Version]
- O’Donovan, J.T.; Grant, C.A.; Blackshaw, R.E.; Harker, K.N.; Johnson, E.N.; Gan, Y.T. Rotational effects of legumes and non-legumes on hybrid canola and malting. Agron. J. 2014, 106, 1921–1932. [Google Scholar] [CrossRef] [Green Version]
- Lizarazo, C.I.; Lampi, A.M.; Sontag-Strohm, T.; Liu, J.; Piironen, V.; Stoddard, F.L. Nutritive quality and protein production from grain legumes in a boreal climate. J. Sci. Food Agric. 2015, 95, 2053–2064. [Google Scholar] [CrossRef] [Green Version]
- Longobardi, F.; Sacco, D.; Casiello, G.; Ventrella, A.; Sacco, A. Chemical profile of the carpino broad bean by conventional and innovative physicochemical analyses. J. Food Qual. 2015, 38, 273–284. [Google Scholar] [CrossRef] [Green Version]
- Landry, E.J.; Fuchs, S.J.; Hu, J. Carbohydrate composition of mature and immature faba bean seeds. J. Food Compos. Anal. 2016, 50, 55–60. [Google Scholar] [CrossRef] [Green Version]
- Lisiewaka, Z.; Kmiecik, W.; Slupski, J. Content of amino acids in raw and frozen broad beans (Vicia faba var. major) seeds at milk maturity stage, depending on the processing method. Food Chem. 2007, 105, 1468–1473. [Google Scholar] [CrossRef]
- Samaei, S.P.; Ghorbani, M.; Tagliazucchi, D.; Martini, S.; Gotti, R.; Themelis, T.; Tesini, F.; Gianotti, A.; Toschi, T.G.; Babini, E. Functional, nutritional, antioxidant, sensory properties and comparative peptidomic profile of faba bean (Vicia faba L.) seed protein hydrolysates and fortified apple juice. Food Chem. 2020, 330, 127120. [Google Scholar] [CrossRef]
- Cazzato, E.; Tufarelli, V.; Ceci, E.; Stellacci, A.M.; Laudadio, V. Quality, yield and nitrogen fixation of faba bean seeds as affected by sulphur fertilization. Acta Agric. Scand. Sect. B Soil Plant Sci. 2012, 62, 732–738. [Google Scholar] [CrossRef]
- Food and Agriculture Organization. FAOSTAT Database. Food and Agriculture Organization of the United Nations. 2017. Available online: www.fao.org/faostat/ (accessed on 11 June 2017).
- Link, W.; Balko, C.; Stoddard, F.L. Winter hardiness in faba bean: Physiology and breeding. Field Crops Res. 2010, 115, 287–296. [Google Scholar] [CrossRef]
- Harlan, J.R. Our vanishing genetic resources. Science 1975, 188, 618–621. [Google Scholar] [CrossRef]
- Cubero, J.I. On the evolution of Vicia faba L. Theor. Appl. Genet. 1974, 45, 47–51. [Google Scholar] [CrossRef]
- Zong, X.; Liu, X.; Guan, J.; Wang, S.; Liu, Q.; Paull, J.G.; Redden, R. Molecular variation among Chinese and global winter faba bean germplasm. Theor. Appl. Genet. 2009, 118, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Damania, A.B. History, achievements, and current status of genetic resources conservation. Agron. J. 2008, 100, 9–21. [Google Scholar] [CrossRef]
- Basheer-Salimia, R.; Camilli, B.; Scacchi, S.; Noli, E.; Awad, M. Genetic diversity among Palestinian faba bean (Vicia faba L.) ecotypes based on single nucleotide polymorphisms. Euro. J. Hort. Sci. 2014, 79, 300–305. [Google Scholar]
- Duc, G.; Agrama, H.; Bao, S.; Berger, J.; Bourion, V.; De Ron, A.M. 2015a. Breeding annual grain legumes for sustainable agriculture: New methods to approach complex traits and target new cultivar ideotypes. Crit. Rev. Plant Sci. 2015, 34, 381–411. [Google Scholar] [CrossRef] [Green Version]
- Lampi, A.M.; Yang, Z.; Mustonen, O.; Piironen, V. Potential of faba bean lipase and lipoxygenase to promote formation of volatile lipid oxidation products in food models. Food Chem. 2020, 311, 125982. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Du, S.; Wang, H.; Cai, M. In vitro antioxidant activity of extracts from common legumes. Food Chem. 2013, 152, 462–466. [Google Scholar] [CrossRef]
- Cernay, C.; Ben-Ari, T.; Pelzer, E.; Meynard, J.M.; Makowski, D. Estimating variability in grain legume yields across Europe and the Americas. Sci. Rep. 2015, 5, 11171. [Google Scholar] [CrossRef]
- Duc, G.; Aleksi´c, J.M.; Marget, P.; Miki´c, A.; Paull, J.; Redden, R.J. Faba bean. In Grain Legumes; Springer: New York, NY, USA, 2015; pp. 141–178. [Google Scholar]
- Katerji, N.; Mastrorilli, M.; Lahmer, F.Z.; Maalouf, F.; Oweis, T. Faba bean productivity in saline-drought conditions. Eur. J. Agron. 2011, 35, 2–12. [Google Scholar] [CrossRef]
- Zhao, J.; Sykacek, P.; Bodner, G.; Rewald, B. Root traits of European Vicia faba cultivars—Using machine learning to explore adaptations to agroclimatic conditions. Plant Cell Environ. 2018, 41, 1984–1996. [Google Scholar] [CrossRef] [Green Version]
- Sharifi, P. Correlation and path coefficient analysis of yield and yield component in some of broad bean (Vicia faba L.) genotypes. Genet. Belgrad. 2014, 469, 905–914. [Google Scholar] [CrossRef]
- Harker, K.N.; O’Donovan, J.T.; Smith, E.G.; Johnson, E.N.; Peng, G.; Willenborg, C.J.; Grenkow, L.A. Seed size and seeding rate effects on canola emergence, development, yield and seed weight. Can. J. Plant Sci. 2015, 95, 1–8. [Google Scholar] [CrossRef]
- Etemadi, F.; Hashemi, M.; Shureshjani, R.A.; Autio, W.R. Application of data envelopment analysis to assess performance efficiency of eight faba bean varieties. Agron. J. 2017, 109, 1225–1231. [Google Scholar] [CrossRef]
- Vioque, J.; Alaiz, M.; Giron-Call, J. Butritional and functional properties of Vicia faba protein isolates and related fractions. Food Chem. 2012, 132, 67–72. [Google Scholar] [CrossRef]
- Abdalla, A.A.; Naim, A.M.E.; Taha, M.B. Quality and nutritive value of Faba bean (Vicia faba L.) as affected by production environment and genotype. World J. Agric. Res. 2019, 7, 21–24. [Google Scholar] [CrossRef]
- Multari, S.; Stewart, D.; Russell, W.R. Potential of fava bean as future protein supply to partially replace meat intake in the human diet. Compr. Rev. Food Sci. Food Saf. 2015, 14, 511–522. [Google Scholar] [CrossRef]
- Crépon, K.; Marget, P.; Peyronnet, C.; Carrouée, B.; Arese, P.; Duc, G. Nutritional value of faba bean (Vicia faba L.) seeds for feed and food. Field Crop Res. 2010, 115, 329–339. [Google Scholar] [CrossRef]
- Li, P.; Zhang, Y.X.; Wu, X.X.; Liu, Y.J. Drought stress impact on leaf proteome variations of faba bean (Vicia faba L.) in the Qinghai-Tibet Plateau of China. 3 Biotech 2018, 8, 110. [Google Scholar] [CrossRef] [Green Version]
- Hao, P.F.; Qiu, C.W.; Ding, G.H.; Vince, E.; Zhang, G.P.; Zhang, Y.S.; Wu, F.B. Agriculture organic wastes fermentation CO2 enrichment in greenhouse and the fermentation residues can improve growth, yield and fruit quality in tomato. J. Clean. Prod. 2020, 275, 123885. [Google Scholar] [CrossRef]
- Wu, F.B.; Zhang, G.P.; Dominy, P. Four barley genotypes respond differently to cadmium: Lipid peroxidation and activities of antioxidant capacity. Environ. Exp. Bot. 2003, 50, 67–78. [Google Scholar] [CrossRef]
- Clarke, J.D.; Aarts, N.; Feys, B.J.; Dong, X.N.; Parker, J.E. Constitutive disease resistance requires EDS1 in the Arabidopsis mutants cpr1 and cpr6 and is partially EDS1-dependent in cpr5. Plant J. 2001, 26, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.W.; Jiang, G.X.; Yan, H.L.; Xiao, L.; Liang, H.Z.; Zhang, D.D.; Jiang, Y.M.; Duan, X.W. Redox regulation of glutathione peroxidase by thioredoxin in longan fruit in relation to senescence and quality deterioration. Food Chem. 2021, 345, 128664. [Google Scholar] [CrossRef] [PubMed]
- Ilgenfritz, H.; Bouyer, D.; Schnittger, A.; Mathur, J.; Kirik, V.; Schwab, B.; Chua, N.; Jürgens, G.; Hülskamp, M. The Arabidopsis STICHEL gene is a regulator of trichome branch number and encodes a novel protein. Plant Physiol. 2003, 131, 643–655. [Google Scholar] [CrossRef] [Green Version]
- Day, L. Proteins form land plants—Potential resources for human nutrition and food security. Trends Food Sci. Technol. 2013, 32, 25–42. [Google Scholar] [CrossRef]
- Roland, W.S.U.; Pouvreau, L.; Curran, J.; van de Valde, F.; de Kok, P.M.T. Flavor aspects of pulse ingredients. Cereal Chem. 2017, 94, 58–65. [Google Scholar] [CrossRef] [Green Version]
- Zhu, D.; Damodaran, S. Removal of off-flavour-causing precursors in soy protein by concurrent treatment with phospholipase A2 and cyclodextrins. Food Chem. 2018, 264, 319–325. [Google Scholar] [CrossRef]
- Chen, Y.S.; Shan, S.R.; Cao, D.M.; Tang, D. Steam flash explosion pretreatment enhances soybean seed coat phenolic profiles and antioxidant activity. Food Chem. 2020, 319, 126552. [Google Scholar] [CrossRef]
- Zeng, W.Y.; Sun, Z.D.; Cai, Z.Y.; Chen, H.Z.; Lai, Z.G.; Yang, S.Z.; Tang, X.M. Proteomic analysis by iTRAQ-MRM of soybean resistance to Lamprosema Indicate. BMC Genom. 2017, 18, 444. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Chen, T.; Wu, S.; Yang, C.; Bai, M.; Shu, K.; Li, K.; Zhang, G.; Jin, Z.; He, F.; et al. iProX: An integrated proteome resource. Nucleic Acids Res. 2019, 47, D1211–D1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, L.; Zhou, W.H.; Dai, H.X.; Cao, F.B.; Zhang, G.P.; Wu, F.B. Selenium reduces cadmium uptake and mitigates cadmium toxicity in rice. J. Hazard. Mater. 2012, 235, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Karaköy, T.; Baloch, F.S.; Toklu, F.; Özkan, H. Variation for selected morphological and quality-related traits among 178 faba bean landraces collected from Turkey. Plant Genet. Res. 2014, 12, 5–13. [Google Scholar] [CrossRef]


| Protein_ID | Fold Change | Description | MW (Da) | AASC (%) | Score |
|---|---|---|---|---|---|
| XP_006581893.1 | 3.46 | Protein STICHEL-like 2 | 109,238.8 | 1.1 | 652 |
| XP_025981111.1 | 2.55 | Probable ATP synthase 24 kDa subunit, mitochondrial | 20,383.14 | 6.2 | n |
| XP_003523436.1 | 2.46 | Aldehyde dehydrogenase family 3 member H1 | 53,299.19 | 3.3 | 695 |
| NP_001236192.2 | 2.38 | Profilin-2 | 14,167 | 9.9 | 223 |
| XP_003537765.3 | 2 | 50S ribosomal protein L12, chloroplastic | 19,744.65 | 4.3 | 195 |
| XP_006577714.1 | 1.88 | Peroxiredoxin-2E-1, chloroplastic-like | 22,543.65 | 5.3 | 204 |
| XP_025984179.1 | 1.81 | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 | 158,613.8 | 0.6 | 2165 |
| XP_003517606.1 | 1.81 | Pre-sequence protease 2, chloroplastic/mitochondrial | 122,069.5 | 1.1 | 1672 |
| XP_003525659.1 | 1.75 | Nucleoside diphosphate kinase 2, chloroplastic | 25,211.09 | 4.0 | 296 |
| YP_538747.1 | 1.73 | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 53,014.61 | 4.6 | n |
| XP_003529252.1 | 1.69 | Probable N-acetyl-gamma-glutamyl-phosphate reductase | 43,088.54 | 2.8 | 513 |
| XP_003527042.1 | 1.68 | Protein EDS1L | 70,738.43 | 1.0 | n |
| NP_001341836.1 | 1.62 | 2-cys peroxiredoxin | 28,437.57 | 2.3 | 398 |
| XP_006575386.1 | 1.59 | 60S ribosomal protein L10 | 25,486.29 | 2.7 | 409 |
| XP_014629812.1 | 1.56 | Eukaryotic translation initiation factor 5A3 isoform X1 | 14,929.39 | 5.9 | 240 |
| NP_001336714.1 | 1.54 | 40S ribosomal protein S20-2 | 13,738.35 | 9.8 | 224 |
| NP_001235587.1 | 1.53 | L-ascorbate peroxidase 2 | 27,147.82 | 3.6 | n |
| XP_014619949.1 | 1.52 | Putative bZIP domain class transcription factor isoform X1 | 53,525.58 | 2.0 | n |
| NP_001235130.2 | 1.52 | Heat shock 22 kDa protein, mitochondrial isoform 2 | 23,722.07 | 5.2 | 230 |
| XP_003539349.1 | 1.49 | CBS domain-containing protein CBSX1, chloroplastic | 25,106.35 | 4.4 | 50.1 |
| XP_014634736.1 | 1.45 | Probable aldo-keto reductase 2 | 37,788.32 | 4.1 | 564 |
| NP_001343409.1 | 1.45 | Putative 18.5 kDa class I heat shock protein | 17,364.88 | 21.1 | 201 |
| XP_003537679.1 | 1.43 | Phosphoserine aminotransferase 1, chloroplastic | 45,349.77 | 2.7 | 664 |
| XP_003525794.1 | 1.42 | Hsp70-Hsp90 organizing protein 1-like | 63,722.89 | 3.4 | 841 |
| NP_001237987.2 | 1.37 | Aspartate aminotransferase isoform X1 | 50,703.81 | 2.8 | 778 |
| NP_001341770.1 | 1.36 | Putative triosephosphate isomerase | 27,391.21 | 5.1 | 429 |
| XP_006599411.1 | 1.35 | V-type proton ATPase subunit C | 42,660.65 | 2.4 | 642 |
| XP_003548332.1 | 1.35 | Thioredoxin H9 isoform X2 | 15,563.91 | 8.0 | 199 |
| NP_001341767.1 | 1.35 | Phosphoglycerate kinase | 42,347.63 | 14.5 | 723 |
| NP_001240021.1 | 1.34 | Catalase | 57,024.44 | 13.4 | 926 |
| XP_003550821.1 | 0.81 | Leghemoglobin reductase-like | 53,178.63 | 2.6 | 860 |
| XP_003525164.1 | 0.8 | Ribonuclease TUDOR 1-like | 108,891.7 | 1.0 | 1453 |
| NP_001237229.1 | 0.79 | 40S ribosomal protein S13 | 17,168.51 | 11.3 | 280 |
| XP_003540396.1 | 0.79 | Ketol-acid reductoisomerase, chloroplastic | 63,668.43 | 4.3 | n |
| NP_001235189.1 | 0.79 | Lipoxygenase | 96,336.78 | 3.0 | n |
| XP_003528976.1 | 0.79 | Dihydropyrimidine dehydrogenase (NADP (+)) | 46,554.31 | 10.8 | 706 |
| XP_003517743.1 | 0.79 | Polygalacturonase 1 beta-like protein 3 | 68,652.63 | 2.2 | n |
| NP_001235936.2 | 0.78 | Superoxide dismutase | 15,322.56 | 19.7 | 254 |
| XP_003531110.3 | 0.78 | 1-Cys peroxiredoxin | 24,523.66 | 4.6 | 325 |
| NP_001241357.1 | 0.78 | Phosphoenolpyruvate carboxylase isoform X1 | 111,129.3 | 1.6 | n |
| XP_003520940.1 | 0.78 | Frataxin, mitochondrial | 21,874.16 | 5.8 | 169 |
| XP_003531426.1 | 0.77 | Thioredoxin-like protein Clot | 15,204.9 | 6.1 | 166 |
| XP_003526464.3 | 0.77 | Oleosin 1 | 17,516.01 | 3.6 | n |
| XP_003542149.1 | 0.77 | Mitochondrial import inner membrane translocase subunit Tim13 | 9538.618 | 11.6 | 87 |
| XP_003543938.1 | 0.76 | α-1,4 glucan phosphorylase L isozyme | 110,522.1 | 7.9 | 1528 |
| XP_003543443.1 | 0.76 | Probable histone H2B.3 | 14,656.14 | 8.3 | 216 |
| XP_003526742.1 | 0.75 | Probable 6-phosphogluconolactonase 4, chloroplastic | 27,821.5 | 3.5 | 342 |
| XP_003519681.1 | 0.75 | Mitochondrial import inner membrane translocase subunit TIM10 | 9895.614 | 20.2 | 134 |
| XP_003554323.1 | 0.73 | Hypersensitive-induced response protein 2-like isoform X1 | 31,797.14 | 9.8 | 506 |
| XP_003555839.1 | 0.72 | Probable prefoldin subunit 5 | 16,900.77 | 7.7 | 223 |
| XP_003538225.2 | 0.71 | Subtilisin inhibitor CLSI-I | 13,362.84 | 6.7 | n |
| NP_001237169.1 | 0.71 | Seed maturation protein PM41 | 8211.974 | 20.5 | n |
| XP_003522597.1 | 0.7 | DNA mismatch repair protein MLH1 | 82,357.54 | 4.8 | 1066 |
| XP_006600683.1 | 0.7 | Delta-1-pyrroline-5-carboxylate dehydrogenase 12A1 | 50,380.97 | 4.7 | 762 |
| XP_003529967.1 | 0.68 | Ubiquitin carboxyl-terminal hydrolase 6 isoform X2 | 54,583.49 | 1.7 | 730 |
| XP_003521095.1 | 0.68 | α-L-arabinofuranosidase 1 | 74,340.09 | 1.9 | n |
| NP_001238008.1 | 0.68 | Glycinin G4 precursor | 64,196.62 | 3.0 | n |
| XP_014625942.1 | 0.68 | Histone H4, partial | 11,068.2 | 22 | 163 |
| NP_001347984.1 | 0.65 | Glyceraldehyde-3-phosphate dehydrogenase | 36,914.17 | 16.6 | 618 |
| NP_001340170.1 | 0.6 | Alcohol dehydrogenase family protein | 41,619.82 | 7.1 | 677 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, P.; Zhu, Y.; Feng, Q.; Jin, Z.; Wu, F. Differences in Grain Microstructure and Proteomics of a Broad Bean (Vicia faba L.) Landrace Cixidabaican in China Compared with Lingxiyicun Introduced from Japan. Plants 2021, 10, 1385. https://doi.org/10.3390/plants10071385
Hao P, Zhu Y, Feng Q, Jin Z, Wu F. Differences in Grain Microstructure and Proteomics of a Broad Bean (Vicia faba L.) Landrace Cixidabaican in China Compared with Lingxiyicun Introduced from Japan. Plants. 2021; 10(7):1385. https://doi.org/10.3390/plants10071385
Chicago/Turabian StyleHao, Pengfei, Yaming Zhu, Qidong Feng, Zhuqun Jin, and Feibo Wu. 2021. "Differences in Grain Microstructure and Proteomics of a Broad Bean (Vicia faba L.) Landrace Cixidabaican in China Compared with Lingxiyicun Introduced from Japan" Plants 10, no. 7: 1385. https://doi.org/10.3390/plants10071385
APA StyleHao, P., Zhu, Y., Feng, Q., Jin, Z., & Wu, F. (2021). Differences in Grain Microstructure and Proteomics of a Broad Bean (Vicia faba L.) Landrace Cixidabaican in China Compared with Lingxiyicun Introduced from Japan. Plants, 10(7), 1385. https://doi.org/10.3390/plants10071385







