Genome-Wide Association Studies for Sex Determination and Cross-Compatibility in Water Yam (Dioscorea alata L.)
Abstract
1. Introduction
2. Results
2.1. Sex and Cross-Compatibility Indices of D. alata Clones Used for GWAS Analyses
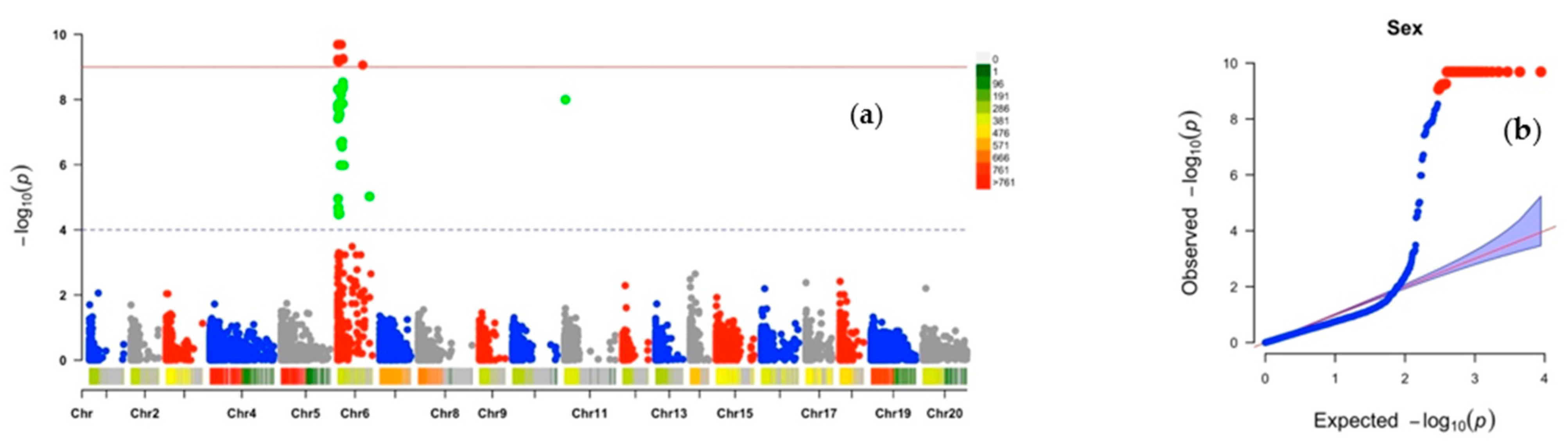
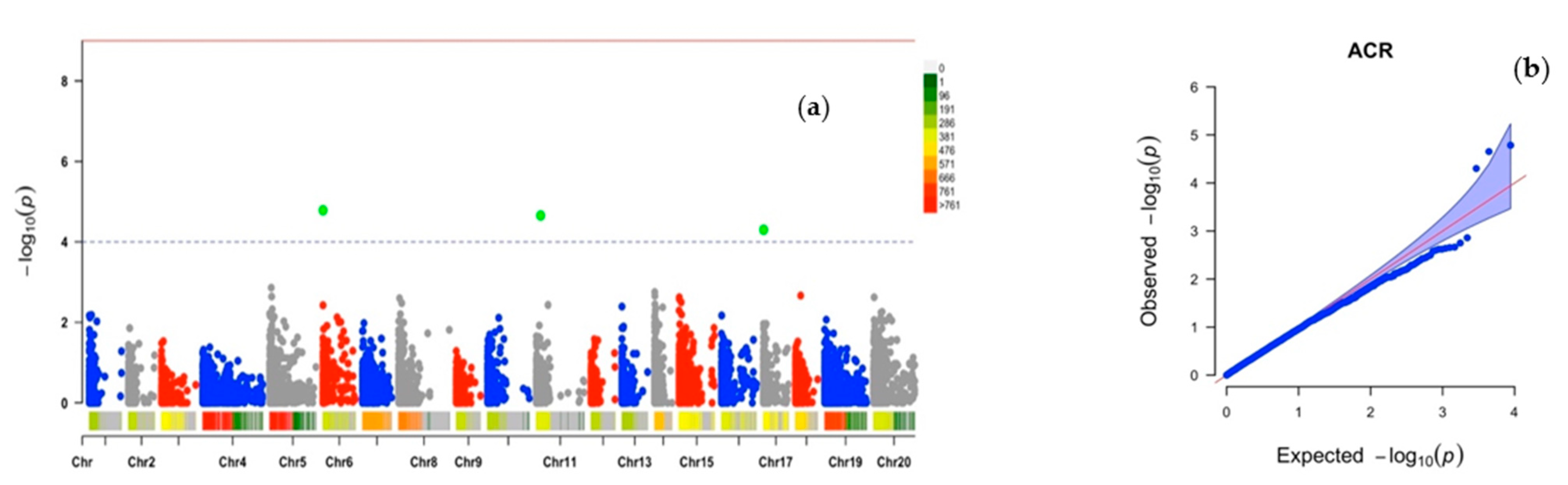
2.2. Chromosomic Regions Linked to D. alata Sex Determination and Cross-Compatibility
2.3. Analysis of the Sex Determination System
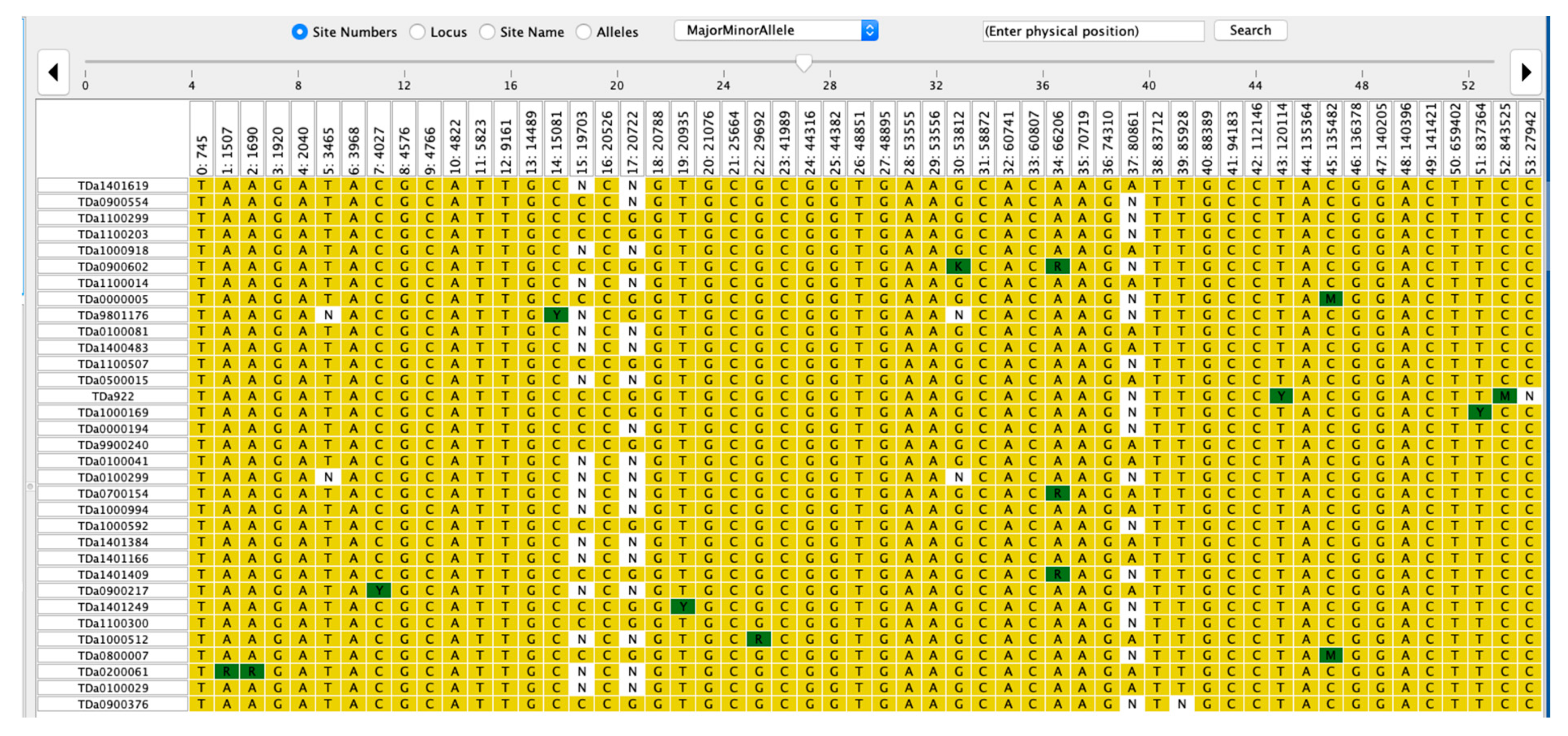
2.4. Haplotype Segregation for ACR and PHC
2.5. Putative Gene Annotation Linked to Flower Sex and Cross-Pollination
3. Discussion
3.1. Genomic Regions Controlling Sex Determination, ACR and PHC Are on the Same Chromosomes
3.2. Dioscorea alata Flower Sex Is Controlled by a Male Heterogametic System
3.3. Gene Annotation Showed the Presence of Gene/Protein Families with Links to Plant Sex and Cross-Pollination
4. Materials and Methods
4.1. Plant Materials and Breeding Sites
4.2. Phenotypic Data Collection
4.2.1. Flower Sex Phenotyping
4.2.2. Genotypes’ ACR and PHC Assessment
4.3. DNA Extraction, Library Construction and SNP Calling
4.3.1. Genotypic Data Analysis
4.3.2. GWAS Analysis and Identification of Putative Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Asiedu, R.; Sartie, A. Crops that feed the World 1. Yams. Food Secur. 2010, 2, 305–315. [Google Scholar] [CrossRef]
- Darkwa, K.; Olasanmi, B.; Asiedu, R.; Asfaw, A. Review of empirical and emerging breeding methods and tools for yam (Dioscorea spp.) improvement: Status and prospects. Plant Breed. 2019, 139, 474–497. [Google Scholar] [CrossRef]
- Cormier, F.; Martin, G.; Vignes, H.; Lachman, L.; Cornet, D.; Faure, Y.; Maledon, E.; Mournet, P.; Arnau, G.; Chaïr, H. Genetic control of flowering in greater yam (Dioscorea alata L.). BMC Plant Biol. 2021, 21, 1–12. [Google Scholar] [CrossRef]
- Sartie, A.; Asiedu, R. Segregation of vegetative and reproductive traits associated with tuber yield and quality in water yam (Dioscorea alata L.). Afr. J. Biotechnol. 2014, 13, 2807–2818. [Google Scholar] [CrossRef]
- Cormier, F.; Lawac, F.; Maledon, E.; Gravillon, M.C.; Nudol, E.; Mournet, P.; Vignes, H.; Arnau, G. A reference high-density genetic map of greater yam (Dioscorea alata L.). Theor. Appl. Genet. 2019, 132, 1733–1744. [Google Scholar] [CrossRef] [PubMed]
- Frossard, E.; Aighewi, B.A.; Aké, S.; Barjolle, D.; Baumann, P.; Bernet, T.; Dao, D.; Diby, L.N.; Floquet, A.; Hgaza, V.K.; et al. The Challenge of Improving Soil Fertility in Yam Cropping Systems of West Africa. Front. Plant Sci. 2017, 8, 1953. [Google Scholar] [CrossRef]
- Matsumoto, R.; Ishikawa, H.; Asfaw, A.; Asiedu, R. Low soil nutrient tolerance and mineral fertilizer response in White Guinea Yam (Dioscorea rotundata) genotypes. Front. Plant Sci. 2021, 12, 223. [Google Scholar] [CrossRef] [PubMed]
- Mondo, J.; Agre, P.; Edemodu, A.; Adebola, P.; Asiedu, R.; Akoroda, M.; Asfaw, A. Floral Biology and Pollination Efficiency in Yam (Dioscorea spp.). Agriculture 2020, 10, 560. [Google Scholar] [CrossRef]
- Tamiru, M.; Natsume, S.; Takagi, H.; White, B.; Yaegashi, H.; Shimizu, M.; Yoshida, K.; Uemura, A.; Oikawa, K.; Abe, A.; et al. Genome sequencing of the staple food crop white Guinea yam enables the development of a molecular marker for sex determination. BMC Biol. 2017, 15, 86. [Google Scholar] [CrossRef]
- Agre, P.; Nwachukwu, C.; Olasanmi, B.; Obidiegwu, J.; Nwachukwu, E.; Adebola, P.; De Koeyer, D.; Asrat, A. Sample Preservation and Plant Sex Prediction in White Guinea yam (Dioscorea rotundata Poir.). J. Appl. Biotechnol. Rep. 2020, 7, 145–151. [Google Scholar] [CrossRef]
- Mondo, J.; Agre, P.; Asiedu, R.; Akoroda, M.; Asfaw, A. Optimized Protocol for In Vitro Pollen Germination in Yam (Dioscorea spp.). Plants 2021, 10, 795. [Google Scholar] [CrossRef] [PubMed]
- Grant, S.; Houben, A.; Vyskot, B.; Siroky, J.; Pan, W.-H.; Macas, J.; Saedler, H. Genetics of sex determination in flowering plants. Dev. Genet. 1994, 15, 214–230. [Google Scholar] [CrossRef]
- Vyskot, B.; Hobza, R. The genomics of plant sex chromosomes. Plant Sci. 2015, 236, 126–135. [Google Scholar] [CrossRef]
- Kumar, S.; Kumari, R.; Sharma, V. Genetics of dioecy and causal sex chromosomes in plants. J. Genet. 2014, 93, 241–277. [Google Scholar] [CrossRef] [PubMed]
- Montalvão, A.P.L.; Kersten, B.; Fladung, M.; Müller, N.A. The Diversity and Dynamics of Sex Determination in Dioecious Plants. Front. Plant Sci. 2021, 11, 2280. [Google Scholar] [CrossRef]
- Martin, F.W. Sex Ratio and Sex Determination in Dioscorea. J. Hered. 1966, 57, 95–99. [Google Scholar] [CrossRef]
- Terauchi, R.; Kahl, G. Mapping of the Dioscorea tokoro genome: AFLP markers linked to sex. Genome 1999, 42, 757–762. [Google Scholar] [CrossRef]
- Bhat, B.K.; Bindroo, B.B. Sex chromosomes in Dioscorea deltoidea wall. Cytologia 1980, 45, 739–742. [Google Scholar] [CrossRef]
- Barrett, S.C.; Hough, J. Sexual dimorphism in flowering plants. J. Exp. Bot. 2013, 64, 67–82. [Google Scholar] [CrossRef]
- Denadi, N.; Gandonou, C.; Missihoun, A.A.; Zoundjihékpon, J.; Quinet, M. Plant Sex Prediction Using Genetic Markers in Cultivated Yams (Dioscorea rotundata Poir.) in Benin. Agronomy 2020, 10, 1521. [Google Scholar] [CrossRef]
- Petit, J.; Salentijn, E.M.J.; Paulo, M.-J.; Denneboom, C.; Trindade, L.M. Genetic Architecture of Flowering Time and Sex Determination in Hemp (Cannabis sativa L.): A Genome-Wide Association Study. Front. Plant Sci. 2020, 11, 1704. [Google Scholar] [CrossRef]
- Mondo, J.M.; Agre, P.A.; Edemodu, A.; Asiedu, R.; Akoroda, M.O.; Asfaw, A. Cross-compatibility Analysis in Intra- and Interspecific Breeding Relationships in Yam (Dioscorea spp.). Front. Plant Sci. 2021. submitted. [Google Scholar]
- Guo, L.; Qiu, F.; Gandhi, H.; Kadaru, S.; De Asis, E.J.; Zhuang, J.-Y.; Xie, F. Genome-wide association study of outcrossing in cytoplasmic male sterile lines of rice. Sci. Rep. 2017, 7, 3223. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Hu, K.; Zhang, Z.; Guan, C.; Chen, S.; Hua, W.; Li, J.; Wen, J.; Yi, B.; Shen, J.; et al. Genome-wide association study reveals the genetic architecture of flowering time in rapeseed (Brassica napus L.). DNA Res. 2015, 23, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Raman, H.; Raman, R.; Qiu, Y.; Yadav, A.S.; Sureshkumar, S.; Borg, L.; Rohan, M.; Wheeler, D.; Owen, O.; Menz, I.; et al. GWAS hints at pleiotropic roles for FLOWERING LOCUS T in flowering time and yield-related traits in canola. BMC Genom. 2019, 20, 636. [Google Scholar] [CrossRef] [PubMed]
- Jaccoud, D.; Peng, K.; Feinstein, D.; Kilian, A. Diversity arrays: A solid state technology for sequence information independent genotyping. Nucleic Acids Res. 2001, 29, 25. [Google Scholar] [CrossRef]
- Gatarira, C.; Agre, P.; Matsumoto, R.; Edemodu, A.; Adetimirin, V.; Bhattacharjee, R.; Asiedu, R.; Asfaw, A. Genome-Wide Association Analysis for Tuber Dry Matter and Oxidative Browning in Water Yam (Dioscorea alata L.). Plants 2020, 9, 969. [Google Scholar] [CrossRef]
- Agre, P.; Asibe, F.; Darkwa, K.; Edemodu, A.; Bauchet, G.; Asiedu, R.; Adebola, P.; Asfaw, A. Phenotypic and molecular assessment of genetic structure and diversity in a panel of winged yam (Dioscorea alata) clones and cultivars. Sci. Rep. 2019, 9, 18221. [Google Scholar] [CrossRef]
- Bredeson, J.V.; Lyons, J.B.; Oniyinde, I.O.; Okereke, N.R.; Kolade, O.; Nnabue, I.; Nwadili, C.O.; Hřibová, E.; Parker, M.; Nwogha, J.; et al. Chromosome evolution and the genetic basis of agronomically important traits in greater yam. BioRxiv 2021. [Google Scholar] [CrossRef]
- Chen, Z.J. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 2007, 58, 377–406. [Google Scholar] [CrossRef]
- Sardos, J.; Rouard, M.; Hueber, Y.; Cenci, A.; Hyma, K.E.; Van Den Houwe, I.; Hribova, E.; Courtois, B.; Roux, N. A Genome-Wide Association Study on the Seedless Phenotype in Banana (Musa spp.) Reveals the Potential of a Selected Panel to Detect Candidate Genes in a Vegetatively Propagated Crop. PLoS ONE 2016, 11, e0154448. [Google Scholar] [CrossRef]
- García, A.; Aguado, E.; Garrido, D.; Martínez, C.; Jamilena, M. Two androecious mutations reveal the crucial role of ethylene receptors in the initiation of female flower development in Cucurbita pepo. Plant J. 2020, 103, 1548–1560. [Google Scholar] [CrossRef]
- García, A.; Aguado, E.; Martínez, C.; Loska, D.; Beltran, S.; Valenzuela, J.L.; Garrido, D.; Jamilena, M. The ethylene receptors CpETR1A and CpETR2B cooperate in the control of sex determination in Cucurbita pepo. J. Exp. Bot. 2020, 71, 154–167. [Google Scholar] [CrossRef]
- Girma, G.; Natsume, S.; Carluccio, A.V.; Takagi, H.; Matsumura, H.; Uemura, A.; Muranaka, S.; Takagi, H.; Stavolone, L.; Gedil, M.; et al. Identification of candidate flowering and sex genes in white Guinea yam (D. rotundata Poir.) by SuperSAGE transcriptome profiling. PLoS ONE 2019, 14, e0216912. [Google Scholar] [CrossRef]
- Munné-Bosch, S. Sex ratios in dioecious plants in the framework of global change. Environ. Exp. Bot. 2015, 109, 99–102. [Google Scholar] [CrossRef]
- Shchennikova, A.V.; Shulga, O.A.; Kochieva, E.; Beletsky, A.V.; Filyushin, M.; Ravin, N.V.; Skryabin, K.G. Homeobox genes encoding WOX transcription factors in the flowering parasitic plant Monotropa hypopitys. Russ. J. Genet. Appl. Res. 2017, 7, 781–788. [Google Scholar] [CrossRef]
- Hamès, C.; Ptchelkine, D.; Grimm, C.; Thevenon, E.; Moyroud, E.; Gérard, F.; Martiel, J.L.; Benlloch, R.; Parcy, F.; Müller, C.W. Structural basis for LEAFY floral switch function and similarity with helix-turn-helix proteins. EMBO J. 2008, 27, 2628–2637. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Lan, S.; Guy, K.M.; Yang, J.; Zhang, M.; Hu, Z. Global Expressions Landscape of NAC Transcription Factor Family and Their Responses to Abiotic Stresses in Citrullus lanatus. Sci. Rep. 2016, 6, 30574. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Peng, J.; Ma, J.; Tang, Y.; Chen, R.; Mysore, K.; Wen, J. NO APICAL MERISTEM (MtNAM) regulates floral organ identity and lateral organ separation in Medicago truncatula. New Phytol. 2012, 195, 71–84. [Google Scholar] [CrossRef]
- Radkova, M.; Revalska, M.; Kertikova, D.; Iantcheva, A. Zinc finger CCHC-type protein related with seed size in model legume species Medicago truncatula. Biotechnol. Biotechnol. Equip. 2019, 33, 278–285. [Google Scholar] [CrossRef]
- Gachomo, E.W.; Jimenez-Lopez, J.C.; Baptiste, L.J.; Kotchoni, O.S. GIGANTUS1 (GTS1), a member of Transducin/WD40 protein superfamily, controls seed germination, growth and biomass accumulation through ribosome-biogenesis protein interactions in Arabidopsis thaliana. BMC Plant Biol. 2014, 14, 37. [Google Scholar] [CrossRef]
- Park, B.S.; Eo, H.J.; Jang, I.-C.; Kang, H.-G.; Song, J.T.; Seo, H.S. Ubiquitination of LHY by SINAT5 regulates flowering time and is inhibited by DET1. Biochem. Biophys. Res. Commun. 2010, 398, 242–246. [Google Scholar] [CrossRef]
- Singh, S.K.; Kumar, V.; Srinivasan, R.; Ahuja, P.S.; Bhat, S.R.; Sreenivasulu, Y. The TRAF Mediated Gametogenesis Progression (TRAMGaP) Gene Is Required for Megaspore Mother Cell Specification and Gametophyte Development. Plant Physiol. 2017, 175, 1220–1237. [Google Scholar] [CrossRef]
- Cucinotta, M.; DI Marzo, M.; Guazzotti, A.; De Folter, S.; Kater, M.M.; Colombo, L. Gynoecium size and ovule number are interconnected traits that impact seed yield. J. Exp. Bot. 2020, 71, 2479–2489. [Google Scholar] [CrossRef]
- Book, A.J.; Smalle, J.; Lee, K.-H.; Yang, P.; Walker, J.M.; Casper, S.; Holmes, J.H.; Russo, L.A.; Buzzinotti, Z.W.; Jenik, P.D.; et al. The RPN5 Subunit of the 26s Proteasome Is Essential for Gametogenesis, Sporophyte Development, and Complex Assembly in Arabidopsis. Plant Cell 2009, 21, 460–478. [Google Scholar] [CrossRef]
- Zarkower, D.; Hodgkin, J. Zinc fingers in sex determination: Only one of the two C. elegans Tra-1 proteins binds DNA in vitro. Nucleic Acids Res. 1993, 21, 3691–3698. [Google Scholar] [CrossRef] [PubMed]
- Massonnet, M.; Cochetel, N.; Minio, A.; VonDras, A.M.; Lin, J.; Muyle, A.; Garcia, J.F.; Zhou, Y.; Delledonne, M.; Riaz, S.; et al. The genetic basis of sex determination in grapes. Nat. Commun. 2020, 11, 2902. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Zhang, D. Roles of jasmonate signalling in plant inflorescence and flower development. Curr. Opin. Plant Biol. 2015, 27, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Pan, B.-Z.; Li, L.; Yang, C.-X.; Xu, Z.-F. Developmental basis for flower sex determination and effects of cytokinin on sex determination in Plukenetia volubilis (Euphorbiaceae). Plant Reprod. 2020, 33, 21–34. [Google Scholar] [CrossRef]
- Li, S.-F.; Zhang, G.-J.; Zhang, X.-J.; Yuan, J.-H.; Deng, C.-L.; Gao, W.-J. Comparative transcriptome analysis reveals differentially expressed genes associated with sex expression in garden asparagus (Asparagus officinalis). BMC Plant Biol. 2017, 17, 143. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, X. Identification of the Sex-Biased Gene Expression and Putative Sex-Associated Genes in Eucommia ulmoides Oliver Using Comparative Transcriptome Analyses. Molecules 2017, 22, 2255. [Google Scholar] [CrossRef]
- Hardenack, S.; Ye, D.; Saedler, H.; Grant, S. Comparison of MADS box gene expression in developing male and female flowers of the dioecious plant white campion. Plant Cell 1994, 6, 1775–1787. [Google Scholar] [CrossRef]
- Li, H.-Y.; E Gray, J. Pollination-enhanced expression of a receptor-like protein kinase related gene in tobacco styles. Plant Mol. Biol. 1997, 33, 653–665. [Google Scholar] [CrossRef]
- Murase, K.; Shigenobu, S.; Fujii, S.; Ueda, K.; Murata, T.; Sakamoto, A.; Wada, Y.; Yamaguchi, K.; Osakabe, Y.; Osakabe, K.; et al. MYB transcription factor gene involved in sex determination in Asparagus officinalis. Genes Cells 2017, 22, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Devani, R.S.; Chirmade, T.; Sinha, S.; Bendahmane, A.; Dholakia, B.B.; Banerjee, A.K.; Banerjee, J. Flower bud proteome reveals modulation of sex-biased proteins potentially associated with sex expression and modification in dioecious Coccinia grandis. BMC Plant Biol. 2019, 19, 330. [Google Scholar] [CrossRef] [PubMed]
- Ventura, J. Characterization of Maize Sex-Determination Gene Orthologs in Rice (Oryza sativa L. japonica cv. Nipponbare). Master’s Thesis, University of Rhode Island, Kingston, RI, USA, 2012; p. 94. Available online: https://digitalcommons.uri.edu/theses/718 (accessed on 1 June 2021).
- Wang, L.; Yin, H.; Qian, Q.; Yang, J.; Huang, C.-F.; Hu, X.; Luo, D. NECK LEAF 1, a GATA type transcription factor, modulates organogenesis by regulating the expression of multiple regulatory genes during reproductive development in rice. Cell Res. 2009, 19, 598–611. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Ding, L.; Jiang, J.; Shentu, Y.; Zhao, W.; Zhao, K.; Zhang, X.; Song, A.; Chen, S.; Chen, F. Overexpression of the CmJAZ1-like gene delays flowering in Chrysanthemum morifolium. Hortic. Res. 2021, 8, 87. [Google Scholar] [CrossRef]
- Pawełkowicz, M.; Pryszcz, L.; Skarzyńska, A.; Wóycicki, R.; Posyniak, K.; Rymuszka, J.; Przybecki, Z.; Plader, W. Comparative transcriptome analysis reveals new molecular pathways for cucumber genes related to sex determination. Plant Reprod. 2019, 32, 193–216. [Google Scholar] [CrossRef]
- Footitt, S.; Dietrich, D.; Fait, A.; Fernie, A.R.; Holdsworth, M.; Baker, A.; Theodoulou, F.L. The COMATOSE ATP-Binding Cassette Transporter Is Required for Full Fertility in Arabidopsis. Plant Physiol. 2007, 144, 1467–1480. [Google Scholar] [CrossRef]
- Arnaud, N.; Pautot, V. Ring the BELL and tie the KNOX: Roles for TALEs in gynoecium development. Front. Plant Sci. 2014, 5, 93. [Google Scholar] [CrossRef]
- Yang, H.W.; Akagi, T.; Kawakatsu, T.; Tao, R. Gene networks orchestrated by MeGI: A single-factor mechanism underlying sex determination in persimmon. Plant J. 2019, 98, 97–111. [Google Scholar] [CrossRef]
- Carmichael, S.N.; Bekaert, M.; Taggart, J.; Christie, H.R.L.; Bassett, D.I.; Bron, J.; Skuce, P.J.; Gharbi, K.; Skern-Mauritzen, R.; Sturm, A. Identification of a Sex-Linked SNP Marker in the Salmon Louse (Lepeophtheirus salmonis) Using RAD Sequencing. PLoS ONE 2013, 8, e77832. [Google Scholar] [CrossRef] [PubMed]
- Graham, P.L.; Yanowitz, J.; Penn, J.K.M.; Deshpande, G.; Schedl, P. The Translation Initiation Factor eIF4E Regulates the Sex-Specific Expression of the Master Switch Gene Sxl in Drosophila melanogaster. PLoS Genet. 2011, 7, e1002185. [Google Scholar] [CrossRef] [PubMed]
- Aso, K.; Kato, M.; Banks, J.A.; Hasebe, M.; Aso, K.; Kato, M.; Banks, J.A.; Hasebe, M. Characterization of homeodomain-leucine zipper genes in the fern Ceratopteris richardii and the evolution of the homeodomain-leucine zipper gene family in vascular plants. Mol. Biol. Evol. 1999, 16, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Asfaw, A. Standard Operating Protocol for Yam Variety Performance Evaluation Trial; IITA: Ibadan, Nigeria, 2016; 27p. [Google Scholar]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Bi, I.V.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2005, 38, 203–208. [Google Scholar] [CrossRef]
- Turner, S.D. Qqman: An R package for visualizing GWAS results using QQ and manhattan plots. Biorxiv 2014, 005165. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]



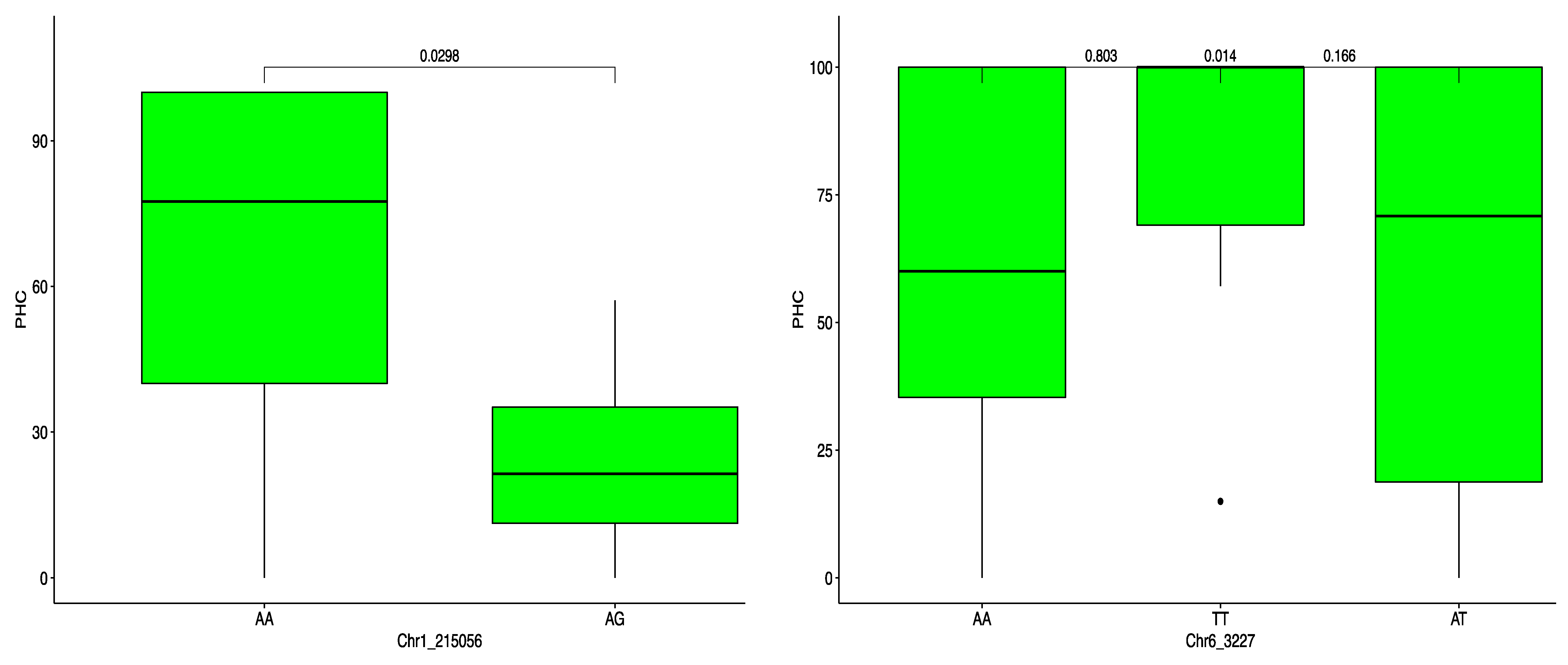

| Clone Name | Type | Sex | ACR (%) | PHC (%) | Cross-Combinations |
|---|---|---|---|---|---|
| TDa0000005 | Breeding line | Female | 28.20 | 38.7 | 38 |
| TDa0000194 | Breeding line | Female | 16.64 | 15.0 | 20 |
| TDa0100004 | Breeding line | Male | 25.05 | 25.0 | 11 |
| TDa0100029 | Breeding line | Female | 7.74 | 0.0 | 10 |
| TDa0100039 | Breeding line | Male | 37.67 | 63.2 | 19 |
| TDa0100041 | Breeding line | Female | 31.19 | 35.3 | 17 |
| TDa0100081 | Breeding line | Female | 41.72 | 57.1 | 21 |
| TDa0100299 | Breeding line | Female | 18.41 | 21.4 | 14 |
| TDa0200012 | Breeding line | Male | 27.74 | 35.3 | 40 |
| TDa0200061 | Breeding line | Female | 46.80 | 60.0 | 5 |
| TDa0500015 | Breeding line | Female | 42.00 | 66.7 | 21 |
| TDa0500056 | Breeding line | Male | 69.12 | 100.0 | 3 |
| TDa0700015 | Breeding line | Male | 73.71 | 100.0 | 3 |
| TDa0700154 | Breeding line | Female | 25.98 | 40.0 | 5 |
| TDa0800007 | Breeding line | Female | 36.30 | 42.9 | 7 |
| TDa0900026 | Breeding line | Male | 28.51 | 35.7 | 14 |
| TDa0900128 | Breeding line | Male | 87.63 | 100.0 | 3 |
| TDa0900146 | Breeding line | Male | 73.21 | 100.0 | 3 |
| TDa0900217 | Breeding line | Female | 32.17 | 47.8 | 23 |
| TDa0900376 | Breeding line | Female | 36.76 | 56.5 | 23 |
| TDa0900554 | Breeding line | Female | 42.54 | 80.0 | 5 |
| TDa0900602 | Breeding line | Female | 57.14 | 100.0 | 3 |
| TDa1000169 | Breeding line | Female | 69.65 | 100.0 | 3 |
| TDa1000365 | Breeding line | Male | 69.29 | 100.0 | 4 |
| TDa1000512 | Breeding line | Female | 66.29 | 71.4 | 7 |
| TDa1000592 | Breeding line | Female | 39.75 | 60.0 | 5 |
| TDa1000918 | Breeding line | Female | 49.54 | 57.1 | 7 |
| TDa1000994 | Breeding line | Female | 56.21 | 87.5 | 8 |
| TDa1100010 | Breeding line | Male | 23.27 | 27.8 | 18 |
| TDa1100014 | Breeding line | Female | 10.56 | 0.0 | 3 |
| TDa1100175 | Breeding line | Male | 84.31 | 100.0 | 3 |
| TDa1100201 | Breeding line | Male | 59.82 | 71.4 | 7 |
| TDa1100202 | Breeding line | Male | 76.32 | 100.0 | 3 |
| TDa1100203 | Breeding line | Female | 51.71 | 50.0 | 3 |
| TDa1100242 | Breeding line | Male | 69.25 | 100.0 | 3 |
| TDa1100295 | Breeding line | Male | 14.77 | 11.1 | 9 |
| TDa1100299 | Breeding line | Female | 63.48 | 80.0 | 5 |
| TDa1100300 | Breeding line | Female | 40.04 | 57.1 | 7 |
| TDa1100302 | Breeding line | Male | 77.10 | 100.0 | 3 |
| TDa1100316 | Breeding line | Male | 45.25 | 66.7 | 9 |
| TDa1100432 | Breeding line | Male | 59.73 | 100.0 | 6 |
| TDa1100507 | Breeding line | Female | 46.12 | 66.7 | 3 |
| TDa1400051 | Breeding line | Male | 64.60 | 100.0 | 3 |
| TDa1400062 | Breeding line | Male | 55.51 | 66.7 | 3 |
| TDa1400064 | Breeding line | Male | 59.50 | 85.7 | 7 |
| TDa1400367 | Breeding line | Male | 73.04 | 100.0 | 3 |
| TDa1400380 | Breeding line | Male | 30.05 | 0.0 | 3 |
| TDa1400432 | Breeding line | Male | 57.37 | 100.0 | 5 |
| TDa1400483 | Breeding line | Female | 82.99 | 100.0 | 4 |
| TDa1400651 | Breeding line | Male | 82.91 | 100.0 | 3 |
| TDa1400911 | Breeding line | Male | 58.11 | 100.0 | 3 |
| TDa1401065 | Breeding line | Male | 65.52 | 75.0 | 4 |
| TDa1401132 | Breeding line | Male | 70.92 | 100.0 | 7 |
| TDa1401162 | Breeding line | Male | 69.12 | 100.0 | 7 |
| TDa1401166 | Breeding line | Female | 68.00 | 100.0 | 3 |
| TDa1401249 | Breeding line | Female | 55.24 | 50.0 | 3 |
| TDa1401253 | Breeding line | Male | 91.04 | 100.0 | 3 |
| TDa1401270 | Breeding line | Male | 65.00 | 100.0 | 4 |
| TDa1401359 | Breeding line | Male | 90.27 | 100.0 | 3 |
| TDa1401384 | Breeding line | Female | 57.50 | 100.0 | 3 |
| TDa1401400 | Breeding line | Male | 56.89 | 80.0 | 5 |
| TDa1401409 | Breeding line | Female | 71.83 | 100.0 | 3 |
| TDa1401619 | Breeding line | Female | 20.93 | 20.0 | 5 |
| TDa1401684 | Breeding line | Male | 71.74 | 100.0 | 3 |
| TDa1402043 | Breeding line | Male | 75.45 | 100.0 | 3 |
| TDa1403882 | Breeding line | Male | 64.33 | 100.0 | 3 |
| TDa291 | Landrace | Male | 3.60 | 0.0 | 3 |
| TDa8500250 | Breeding line | Male | 17.40 | 13.8 | 29 |
| TDa8701091 | Breeding line | Male | 24.63 | 30.4 | 26 |
| TDa922 | Landrace | Female | 12.59 | 0.0 | 7 |
| TDa9801174 | Breeding line | Male | 28.24 | 33.3 | 30 |
| TDa9801176 | Breeding line | Female | 1.59 | 0.0 | 13 |
| TDa98150 | Landrace | Male | 21.57 | 11.1 | 18 |
| TDa9900240 | Breeding line | Female | 32.43 | 40.0 | 45 |
| Traits | SNP Markers | Chr | Position (bp) | MAF | PVE (%) | Effect | LOD |
|---|---|---|---|---|---|---|---|
| Plant sex | Chr6_1920 | 6 | 1920 | 0.27 | 86 | −1.92 | 9.69 |
| Chr6_20526 | 6 | 20,526 | 0.27 | 86 | −1.92 | 9.69 | |
| Chr6_21076 | 6 | 21,076 | 0.27 | 86 | −1.92 | 9.69 | |
| Chr6_3968 | 6 | 3968 | 0.27 | 86 | −1.92 | 9.69 | |
| Chr6_41989 | 6 | 41,989 | 0.27 | 86 | −1.92 | 9.69 | |
| Chr6_44316 | 6 | 44,316 | 0.27 | 86 | −1.92 | 9.69 | |
| Chr6_44382 | 6 | 44,382 | 0.27 | 86 | −1.92 | 9.69 | |
| Chr6_4576 | 6 | 4576 | 0.27 | 86 | −1.92 | 9.69 | |
| Chr6_4766 | 6 | 4766 | 0.27 | 86 | −1.92 | 9.69 | |
| Chr6_4822 | 6 | 4822 | 0.27 | 86 | −1.92 | 9.69 | |
| Chr6_48851 | 6 | 48,851 | 0.27 | 86 | −1.92 | 9.69 | |
| Chr6_48895 | 6 | 48,895 | 0.27 | 86 | −1.92 | 9.69 | |
| Chr6_5823 | 6 | 5823 | 0.27 | 86 | −1.92 | 9.69 | |
| Chr6_58872 | 6 | 58,872 | 0.27 | 86 | −1.92 | 9.69 | |
| Chr6_60741 | 6 | 60,741 | 0.27 | 86 | −1.92 | 9.69 | |
| Chr6_60807 | 6 | 60,807 | 0.27 | 86 | −1.92 | 9.69 | |
| Chr6_70719 | 6 | 70,719 | 0.27 | 86 | −1.92 | 9.69 | |
| Chr6_74310 | 6 | 74,310 | 0.27 | 86 | −1.92 | 9.69 | |
| Chr6_745 | 6 | 745 | 0.27 | 86 | −1.92 | 9.69 | |
| Chr6_83712 | 6 | 83,712 | 0.27 | 86 | −1.92 | 9.69 | |
| Chr6_88389 | 6 | 88,389 | 0.27 | 86 | −1.92 | 9.69 | |
| Chr6_94183 | 6 | 94,183 | 0.27 | 86 | −1.92 | 9.69 | |
| Chr6_140396 | 6 | 140,396 | 0.28 | 83 | 1.74 | 9.26 | |
| Chr6_141421 | 6 | 141,421 | 0.28 | 82 | 1.77 | 9.23 | |
| Chr6_2040 | 6 | 2040 | 0.28 | 82 | 1.77 | 9.23 | |
| Chr6_15081 | 6 | 15,081 | 0.28 | 82 | −1.82 | 9.23 | |
| Chr6_4027 | 6 | 4027 | 0.28 | 82 | −1.82 | 9.17 | |
| Chr6_29692 | 6 | 29,692 | 0.28 | 82 | −1.80 | 9.16 | |
| Chr6_659402 | 6 | 659,402 | 0.26 | 81 | −1.83 | 9.06 | |
| Chr6_135364 | 6 | 135,364 | 0.26 | 77 | 1.63 | 8.53 | |
| Chr6_140205 | 6 | 140,205 | 0.24 | 76 | −1.75 | 8.40 | |
| Chr6_135482 | 6 | 135,482 | 0.28 | 75 | −1.69 | 8.34 | |
| Chr6_1507 | 6 | 1507 | 0.28 | 75 | −1.56 | 8.31 | |
| Chr6_85928 | 6 | 85,928 | 0.28 | 74 | 1.56 | 8.15 | |
| Chr11_27942 | 11 | 27,942 | 0.26 | 72 | −1.56 | 7.99 | |
| Chr6_66206 | 6 | 66,206 | 0.29 | 72 | −1.59 | 7.89 | |
| Chr6_136378 | 6 | 136,378 | 0.34 | 72 | 1.22 | 7.87 | |
| Chr6_20788 | 6 | 20,788 | 0.34 | 72 | 1.22 | 7.87 | |
| Chr6_3465 | 6 | 3465 | 0.43 | 71 | 1.04 | 7.84 | |
| Chr6_53556 | 6 | 53,556 | 0.35 | 71 | 1.13 | 7.80 | |
| Chr6_53555 | 6 | 53,555 | 0.33 | 71 | 1.24 | 7.79 | |
| Chr6_1690 | 6 | 1690 | 0.27 | 70 | −1.46 | 7.73 | |
| Chr6_14489 | 6 | 14,489 | 0.29 | 70 | 1.35 | 7.73 | |
| Chr6_53812 | 6 | 53,812 | 0.43 | 69 | 1.00 | 7.54 | |
| Chr6_20722 | 6 | 20,722 | 0.39 | 68 | −1.02 | 7.46 | |
| Chr6_19703 | 6 | 19,703 | 0.41 | 68 | −1.03 | 7.43 | |
| Chr6_9161 | 6 | 9161 | 0.24 | 68 | −1.48 | 7.42 | |
| Chr6_120114 | 6 | 120,114 | 0.28 | 63 | 1.10 | 6.71 | |
| Chr6_80861 | 6 | 80,861 | 0.40 | 63 | −1.06 | 6.67 | |
| Chr6_112146 | 6 | 112,146 | 0.26 | 62 | 1.18 | 6.55 | |
| Chr6_837364 | 6 | 837,364 | 0.13 | 52 | −1.71 | 5.02 | |
| Chr6_843525 | 6 | 843,525 | 0.13 | 52 | −1.53 | 5.02 | |
| Chr6_20935 | 6 | 20,935 | 0.15 | 49 | −1.69 | 4.52 | |
| Chr6_25664 | 6 | 25,664 | 0.14 | 49 | −1.55 | 4.47 | |
| ACR | Chr6_3161 | 6 | 3161 | 0.25 | 35 | 14.02 | 4.78 |
| Chr11_124789 | 11 | 124,789 | 0.30 | 33 | −17.46 | 4.65 | |
| Chr17_9492 | 17 | 9492 | 0.29 | 32 | −20.47 | 4.30 | |
| PHC | Chr1_215056 | 1 | 215,056 | 0.03 | 29 | −43.11 | 4.01 |
| Chr6_3227 | 6 | 3227 | 0.26 | 29 | −27.36 | 4.04 |
| Trait | Marker | Haplotype | Sequence | Frequency (%) | p-Value | p-Value Adj. Signif. |
|---|---|---|---|---|---|---|
| ACR | Chr6_3161 | Haplotype 1 | CCCG | 39.58 | 0.623 | ns |
| Haplotype 2 | CCGG | 45.14 | 0.063 | ns | ||
| Haplotype 3 | CGGG | 15.28 | 0.156 | ns | ||
| Chr11_124789 | Haplotype 1 | CCTC | 26.39 | 0.709 | ns | |
| Haplotype 2 | CCTT | 25.69 | 0.847 | ns | ||
| Haplotype 3 | TCTT | 47.92 | 0.366 | ns | ||
| Chr17_9492 | Haplotype 1 | CCTC | 23.61 | 0.02 | * | |
| Haplotype 2 | CCTT | 31.94 | 0.002 | ** | ||
| Haplotype 3 | TCTT | 44.44 | 0.04 | * | ||
| PHC | Chr1_215056 | Haplotype 1 | AAAG | 100.00 | 0.03 | * |
| Chr6_3227 | Haplotype 1 | AAAT | 39.58 | 0.803 | ns | |
| Haplotype 2 | AATT | 44.44 | 0.014 | * | ||
| Haplotype 3 | ATTT | 15.97 | 0.166 | ns |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mondo, J.M.; Agre, P.A.; Asiedu, R.; Akoroda, M.O.; Asfaw, A. Genome-Wide Association Studies for Sex Determination and Cross-Compatibility in Water Yam (Dioscorea alata L.). Plants 2021, 10, 1412. https://doi.org/10.3390/plants10071412
Mondo JM, Agre PA, Asiedu R, Akoroda MO, Asfaw A. Genome-Wide Association Studies for Sex Determination and Cross-Compatibility in Water Yam (Dioscorea alata L.). Plants. 2021; 10(7):1412. https://doi.org/10.3390/plants10071412
Chicago/Turabian StyleMondo, Jean M., Paterne A. Agre, Robert Asiedu, Malachy O. Akoroda, and Asrat Asfaw. 2021. "Genome-Wide Association Studies for Sex Determination and Cross-Compatibility in Water Yam (Dioscorea alata L.)" Plants 10, no. 7: 1412. https://doi.org/10.3390/plants10071412
APA StyleMondo, J. M., Agre, P. A., Asiedu, R., Akoroda, M. O., & Asfaw, A. (2021). Genome-Wide Association Studies for Sex Determination and Cross-Compatibility in Water Yam (Dioscorea alata L.). Plants, 10(7), 1412. https://doi.org/10.3390/plants10071412







