Current and Future Pathotyping Platforms for Plasmodiophora brassicae in Canada
Abstract
1. Introduction
1.1. Clubroot Disease
1.2. Emergence of ‘New’ Virulent Pathotypes
1.3. Need for Rapid Pathotyping
2. Current Diagnostic Methods
2.1. Phenotypic Approaches
2.2. Microscopy
2.3. Molecular Approaches
| Year | Technique | Primer Sequences (5′ to 3′) | ||
|---|---|---|---|---|
| Ito et al. [43] | 1999 | Nested PCR | Outer | PBTZS-2: CCGAATTCGCGTCAGCGTGA a |
| Inner | PBTZS-3: CCACGTCGATCACGTTGCAAT PBTZS-4: GCTGGCGTTGATGTACTGGAA | |||
| Faggian et al. [44] | 1999 | Nested PCR | Outer | PbITS1: ACTTGCATCGATTACGTCCC PbITS2: GGCATTCTCGAGGGTATCAA |
| Inner | PbITS6: CAACGAGTCAGCTTGAATGC PbITS7: TGTTTCGGCTAGGATGGTTC | |||
| Wallenhammar & Arwidsson [45] | 2001 | Nested PCR | Outer | PBAW-10: CCCCGGGGATCACGATAAATAACA PBAW-11: GGAAGGCCGCCCAGGACTACC PBAW-12: GCCGGCCAGCATCTCCAT PBAW-13: CCCCAGGGTTCACAGCGTTCAA |
| Inner | PBTZS-3 [43] PBTZS-4 [43] | |||
| Cao et al. [46] | 2007 | PCR | TC1F: GTGGTCGAACTTCATTAAATTTGGGCTCTT TC1R: TTCACCTACGGAACGTATATGTGCATGTGA | |
| Sundelin et al. [47] | 2010 | qPCR | Pb4-1: TACCATACCCAGGGCGATT PbITS6 [44] | |
| Rennie et al. [48] | 2011 | qPCR | DC1F: CCTAGCGCTGCATCCCATAT DC1R: CGGCTAGGATGGTTCGAAAA | |
| Wallenhammar et al. [49] | 2012 | TaqMan qPCR | PbF: AAACAACGAGTCAGCTTGAATGC PbR: TTCGCGCACAAGCAC TTG (Probe) PbP: CGCGCCATGCGACACTGTTAAATT | |
| Cao et al. [51] | 2014 | TaqMan qPCR | TC1F: GTGGTCGAACTTCATTAAATTTGGGCTCTT RTPbR1a: TCAGCACCGTTTCCGGCTGCTAAGGC (Probe) TCPb1: AAGAAGGAGAAGTCGTAACAAGGTTTC | |
| Deora et al. [50] | 2015 | TaqMan qPCR | PBGFPuv3F: CCTAGCGCTGCATCCCATATCGATGGCCCTGTCCTTTTAC PBGFPuv3R: CGGCTAGGATGG TTCGAAAGTGTAATCCCAGCAGCAGTTA (Probe) GFP1: ACCATTACCTGTCGACACAATCTGCCCT | |
| Al-Daoud et al. [52] | 2016 | PMA-PCR | PBGFPuv3F [50] PBGFPuv3R [50] (Probe) GFP1 [50] | |
| Wen et al. [53] | 2020 | ddPCR | DC1F: CCT AGC GCT GCA TCC CAT AT DC1mR: CGGCTAGGATGGTTCGAAA (Probe) PB1: /56-FAM/CCA TGTGAA/ZEN/CCG GTGACGTGCG/3IABkFQ/ | |
3. Rapid Molecular Pathotyping
3.1. Amplicon Length Distinction
3.2. SNP-Based Distinction
3.3. RNase-H Dependent PCR (rhPCR)
3.4. Single Base Extension (SBE)
3.5. Can Metabarcoding Be Used in Clubroot?
3.6. General Limitations of Molecular Approaches
4. Future Perspectives
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dixon, G.R. The Occurrence and Economic Impact of Plasmodiophora brassicae and Clubroot Disease. J. Plant Growth Regul. 2009, 28, 194–202. [Google Scholar] [CrossRef]
- Canola Industry in Canada, from Farm to Global Markets. Available online: https://www.canolacouncil.org/about-canola/industry/ (accessed on 12 April 2021).
- Strelkov, S.E.; Hwang, S.F. Clubroot in the Canadian Canola Crop: 10 Years into the Outbreak. Can. J. Plant Pathol. 2014, 36, 27–36. [Google Scholar] [CrossRef]
- Pageau, D.; Lajeunesse, J.; Lafond, J. Impact of clubroot [Plasmodiophora brassicae] on the yield and quality of canola. Can. J. Plant Pathol. 2006, 28, 137–143. [Google Scholar] [CrossRef]
- Tewari, J.P.; Strelkov, S.E.; Orchard, D.; Hartman, M.; Lange, R.M.; Turkington, T.K. Identification of Clubroot of Crucifers on Canola (Brassica napus) in Alberta. Can. J. Plant Pathol. 2005, 27, 143–144. [Google Scholar] [CrossRef]
- Howard, R.J.; Strelkov, S.E.; Harding, M.W. Clubroot of Cruciferous Crops—New Perspectives on an Old Disease. Can. J. Plant Pathol. 2010, 32, 43–57. [Google Scholar] [CrossRef]
- Strelkov, S.E.; Hwang, S.F.; Howard, R.J.; Hartman, M.; Turkington, T.K. Progress towards the Sustainable Management of Clubroot (Plasmodiophora brassicae) of Canola on the Canadian Prairies. Prairie Soils Crops J. 2011, 4, 114–121. [Google Scholar]
- Strelkov, S.E.; Manolii, V.P.; Harding, M.W.; Daniels, G.C.; Nuffer, P.; Hwang, S.F. The Occurrence and Spread of Clubroot on Canola in Alberta in 2019. Canadian Plant Disease Survey 2020 Vol. 100: Disease Highlights 2019. Can. J. Plant Pathol. 2020, 42, 117–120. [Google Scholar] [CrossRef]
- Strelkov, S.E.; Manolii, V.P.; Aigu, Y.; Harding, M.W.; Hwang, S.F.; Daniels, G.C. The Occurrence and Spread of Clubroot on Canola in Alberta in 2020. Canadian Plant Disease Survey 2021 Vol. 101: Disease Highlights 2020. Can. J. Plant Pathol. 2021, 43. in press. [Google Scholar]
- Al-Daoud, F.; Moran, M.; Gossen, B.D.; McDonald, M.R. First Report of Clubroot (Plasmodiophora brassicae) on Canola in Ontario. Can. J. Plant Pathol. 2018, 40, 96–99. [Google Scholar] [CrossRef]
- Chapara, V.; Kalwar, N.; Lubenow, L.; Chirumamilla, A. Prevalence of Clubroot on Canola in North Dakota. J. Agron. Agric. Sci. 2019, 2, 8. [Google Scholar] [CrossRef]
- Canola Encyclopedia. Clubroot Disease. Available online: https://www.canolacouncil.org/canola-encyclopedia/diseases/clubroot/ (accessed on 12 April 2021).
- Peng, G.; Lahlali, R.; Hwang, S.F.; Pageau, D.; Hynes, R.K.; McDonald, M.R.; Gossen, B.D.; Strelkov, S.E. Crop Rotation, Cultivar Resistance, and Fungicides/Biofungicides for Managing Clubroot (Plasmodiophora brassicae) on Canola. Can. J. Plant Pathol. 2014, 36, 99–112. [Google Scholar] [CrossRef]
- Donald, C.; Porter, I. Integrated Control of Clubroot. J. Plant Growth Regul. 2009, 28, 289. [Google Scholar] [CrossRef]
- Rahman, H.; Peng, G.; Yu, F.; Falk, K.C.; Kulkarni, M.; Selvaraj, G. Genetics and Breeding for Clubroot Resistance in Canadian Spring Canola (Brassica napus L.). Can. J. Plant Pathol. 2014, 36, 122–134. [Google Scholar] [CrossRef]
- Fredua-Agyeman, R.; Hwang, S.F.; Strelkov, S.E.; Zhou, Q.; Feindel, D. Potential Loss of Clubroot Resistance Genes from Donor Parent Brassica rapa subsp. rapifera (ECD 04) during Doubled Haploid Production. Plant Pathol. 2018, 67, 892–901. [Google Scholar] [CrossRef]
- LeBoldus, J.M.; Manolii, V.P.; Turkington, T.K.; Strelkov, S.E. Adaptation to Brassica Host Genotypes by a Single-Spore Isolate and Population of Plasmodiophora brassicae (Clubroot). Plant Dis. 2012, 96, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Strelkov, S.E.; Hwang, S.F.; Manolii, V.P.; Cao, T.; Feindel, D. Emergence of New Virulence Phenotypes of Plasmodiophora brassicae on Canola (Brassica napus) in Alberta, Canada. Eur. J. Plant Pathol. 2016, 145, 517–529. [Google Scholar] [CrossRef]
- Strelkov, S.E.; Manolii, V.P.; Harding, M.W.; Hwang, S.F.; Poscente, N.; Lisowski, S.L.I.; Pugh, C.A.; Burke, D.A. The Occurrence of Clubroot on Canola in Alberta in 2013. Can. Plant Dis. Surv. 2014, 94, 158–161. [Google Scholar]
- Strelkov, S.E.; Hwang, S.F.; Manolii, V.P.; Cao, T.; Fredua-Agyeman, R.; Harding, M.W.; Peng, G.; Gossen, B.D.; McDonald, M.R.; Feindel, D. Virulence and Pathotype Classification of Plasmodiophora brassicae Populations Collected from Clubroot Resistant Canola (Brassica napus) in Canada. Can. J. Plant Pathol. 2018, 40, 284–298. [Google Scholar] [CrossRef]
- Hollman, K.B.; Hwang, S.F.; Manolii, V.P.; Strelkov, S.E. Pathotypes of Plasmodiophora brassicae Collected from Clubroot Resistant Canola (Brassica napus L.) Cultivars in Western Canada in 2017–2018. Can. J. Plant Pathol. 2021. [Google Scholar] [CrossRef]
- Strelkov, S.E.; Hwang, S.F.; Manolii, V.P.; Turnbull, G.D.; Fredua-Agyeman, R.; Hollman, K.B.; Kaus, S. Characterization of Clubroot (Plasmodiophora brassicae) from Canola (Brassica napus) in the Peace Country of Alberta, Canada. Can. J. Plant Pathol. 2021, 43, 155–161. [Google Scholar] [CrossRef]
- Askarian, H.; Akhavan, A.; Manolii, V.P.; Cao, T.; Hwang, S.F.; Strelkov, S.E. Virulence Spectrum of Single-Spore and Field Isolates of Plasmodiophora brassicae Able to Overcome Resistance in Canola (Brassica napus). Plant Dis. 2020, 105, 43–52. [Google Scholar] [CrossRef]
- Samuel, G.; Garrett, S.D. The Infected Root-Hair Count for Estimating the Activity of Plasmodiophora brassicae Woron. in the Soil. Ann. Appl. Biol. 1945, 32, 96–101. [Google Scholar] [CrossRef]
- Macfarlane, I. Factors Affecting the Survival of Plasmodio-Phora Brassicae Wor. in the Soil and Its Assessment by a Host Test. Ann. Appl. Biol. 1952, 39, 239–256. [Google Scholar] [CrossRef]
- Colhoun, J. A Technique for Examining Soil for the Presence of Plasmodiophora brassicae Woron. Ann. Appl. Biol. 1957, 45, 559–565. [Google Scholar] [CrossRef]
- Melville, S.C.; Hawken, R.H. Soil Testing for Club Root in Devon and Cornwall. Plant Pathol. 1967, 16, 145–147. [Google Scholar] [CrossRef]
- Faggian, R.; Strelkov, S.E. Detection and Measurement of Plasmodiophora brassicae. J. Plant Growth Regul. 2009, 28, 282–288. [Google Scholar] [CrossRef]
- Williams, P.H. A system for the determination of races of Plasmodiophora brassicae that infect Cabbage and Rutabaga. Phytopathology 1966, 56, 624–626. [Google Scholar]
- Buczacki, S.T.; Toxopeus, H.; Mattusch, P.; Johnston, T.D.; Dixon, G.R.; Hobolth, L.A. Study of Physiologic Specialization in Plasmodiophora brassicae: Proposals for Attempted Rationalization through an International Approach. Trans. Br. Mycol. Soc. 1975, 65, 295–303. [Google Scholar] [CrossRef]
- Somé, A.; Manzanares, M.J.; Laurens, F.; Baron, F.; Thomas, G.; Rouxel, F. Variation for Virulence on Brassica napus L. amongst Plasmodiophora brassicae Collections from France and Derived Single-Spore Isolates. Plant Pathol. 1996, 45, 432–439. [Google Scholar] [CrossRef]
- Pang, W.; Liang, Y.; Zhan, Z.; Li, X.; Piao, Z. Development of a Sinitic Clubroot Differential Set for the Pathotype Classification of Plasmodiophora brassicae. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Buczacki, S.T.; Moxham, S.E. A Triple Stain for Differentiating Resin-Embedded Sections of Plasmodiophora brassicae in Host Tissues under the Light Microscope. Trans. Br. Mycol. Soc. UK 1979, 72, 311. [Google Scholar] [CrossRef]
- Buczacki, S.T.; Moxham, S.E. Structure of the Resting Spore Wall of Plasmodiophora brassicae Revealed by Electron Microscopy and Chemical Digestion. Trans. Br. Mycol. Soc. 1983, 81, 221–231. [Google Scholar] [CrossRef]
- Sharma, K.; Gossen, B.D.; McDonald, M.R. Effect of Temperature on Cortical Infection by Plasmodiophora brassicae and Clubroot Severity. Phytopathology 2011, 101, 1424–1432. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.S.; Rahman, M.H.; Deyholos, M.K.; Basu, U.; Kav, N.N.V. Differential Expression of miRNAs in Brassica napus Root Following Infection with Plasmodiophora brassicae. PLoS ONE 2014, 9, e86648. [Google Scholar] [CrossRef] [PubMed]
- Deora, A.; Gossen, B.D.; McDonald, M.R. Cytology of Infection, Development and Expression of Resistance to Plasmodiophora brassicae in Canola. Ann. Appl. Biol. 2013, 163, 56–71. [Google Scholar] [CrossRef]
- Tanaka, S.; Kochi, S.; Kunita, H.; Ito, S.; Kameya-Iwaki, M. Biological Mode of Action of the Fungicide, Flusulfamide, Against Plasmodiophora brassicae (Clubroot). Eur. J. Plant Pathol. 1999, 105, 577–584. [Google Scholar] [CrossRef]
- Harding, M.W.; Hill, T.B.; Yang, Y.; Daniels, G.C.; Hwang, S.F.; Strelkov, S.E.; Howard, R.J.; Feng, J. An Improved Evans Blue Staining Method for Consistent, Accurate Assessment of Plasmodiophora brassicae Resting Spore Viability. Plant Dis. 2019, 103, 2330–2336. [Google Scholar] [CrossRef]
- White, J.G.; Buczacki, S.T. Observations on Suppression of Clubroot by Artificial or Natural Heating of Soil. Trans. Br. Mycol. Soc. 1979, 73, 271–275. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamaguchi, T. A Method for Assessing the Pathogenic Activity of Resting Spores of Plasmodiophora brassicae by Fluorescence Microscopy. Jpn. J. Phytopathol. 1988, 54, 466–475. [Google Scholar] [CrossRef]
- Naiki, T.; Dixon, G.R.; Ikegami, H. Quantitative Estimation of Spore Germination of Plasmodiophora brassicae. Trans. Br. Mycol. Soc. 1987, 89, 569–572. [Google Scholar] [CrossRef]
- Ito, S.; Maehara, T.; Maruno, E.; Tanaka, S.; Kameya-Iwaki, M.; Kishi, F. Development of a PCR-Based Assay for the Detection of Plasmodiophora brassicae in Soil. J. Phytopathol. 1999, 147, 83–88. [Google Scholar] [CrossRef]
- Faggian, R.; Bulman, S.R.; Lawrie, A.C.; Porter, I.J. Specific Polymerase Chain Reaction Primers for the Detection of Plasmodiophora brassicae in Soil and Water. Phytopathology 1999, 89, 392–397. [Google Scholar] [CrossRef]
- Wallenhammar, A.-C.; Arwidsson, O. Detection of Plasmodiophora brassicae By PCR in Naturally Infested Soils. Eur. J. Plant Pathol. 2001, 107, 313–321. [Google Scholar] [CrossRef]
- Cao, T.; Tewari, J.P.; Strelkov, S.E. Molecular Detection of Plasmodiophora brassicae, Causal Agent of Clubroot of Crucifers, in Plant and Soil. Plant Dis. 2007, 91, 80–87. [Google Scholar] [CrossRef]
- Sundelin, T.; Christensen, C.B.; Larsen, J.; Møller, K.; Lübeck, M.; Bødker, L.; Jensen, B. In Planta Quantification of Plasmodiophora brassicae Using Signature Fatty Acids and Real-Time PCR. Plant Dis. 2010, 94, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Rennie, D.C.; Manolii, V.P.; Cao, T.; Hwang, S.F.; Howard, R.J.; Strelkov, S.E. Direct Evidence of Surface Infestation of Seeds and Tubers by Plasmodiophora brassicae and Quantification of Spore Loads. Plant Pathol. 2011, 60, 811–819. [Google Scholar] [CrossRef]
- Wallenhammar, A.-C.; Almquist, C.; Söderström, M.; Jonsson, A. In-Field Distribution of Plasmodiophora brassicae Measured Using Quantitative Real-Time PCR. Plant Pathol. 2012, 61, 16–28. [Google Scholar] [CrossRef]
- Deora, A.; Gossen, B.D.; Amirsadeghi, S.; McDonald, M.R. A Multiplex QPCR Assay for Detection and Quantification of Plasmodiophora brassicae in Soil. Plant Dis. 2015, 99, 1002–1009. [Google Scholar] [CrossRef] [PubMed]
- Cao, T.; Rennie, D.C.; Manolii, V.P.; Hwang, S.F.; Falak, I.; Strelkov, S.E. Quantifying Resistance to Plasmodiophora brassicae in Brassica Hosts. Plant Pathol. 2014, 63, 715–726. [Google Scholar] [CrossRef]
- Al-Daoud, F.; Gossen, B.D.; Robson, J.; McDonald, M.R. Propidium Monoazide Improves Quantification of Resting Spores of Plasmodiophora brassicae with qPCR. Plant Dis. 2016, 101, 442–447. [Google Scholar] [CrossRef]
- Wen, R.; Lee, J.; Chu, M.; Tonu, N.; Dumonceaux, T.; Gossen, B.D.; Yu, F.; Peng, G. Quantification of Plasmodiophora brassicae Resting Spores in Soils Using Droplet Digital PCR (ddPCR). Plant Dis. 2020, 104, 1188–1194. [Google Scholar] [CrossRef]
- Manzanares-Dauleux, M.J.; Barret, P.; Thomas, G. Development of a Pathotype Specific SCAR Marker in Plasmodiophora brassicae. Eur. J. Plant Pathol. 2000, 106, 781–787. [Google Scholar] [CrossRef]
- Zhang, H.; Feng, J.; Manolii, V.P.; Strelkov, S.E.; Hwang, S.F. Characterization of a Gene Identified in Pathotype 5 of the Clubroot Pathogen Plasmodiophora brassicae. Phytopathology 2015, 105, 764–770. [Google Scholar] [CrossRef] [PubMed]
- Holtz, M.D.; Hwang, S.F.; Strelkov, S.E. Genotyping of Plasmodiophora brassicae Reveals the Presence of Distinct Populations. BMC Genom. 2018, 19, 254. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Hwang, S.F.; Strelkov, S.E.; Fredua-Agyeman, R.; Manolii, V.P. A Molecular Marker for the Specific Detection of New Pathotype 5-like Strains of Plasmodiophora brassicae in Canola. Plant Pathol. 2018, 67, 1582–1588. [Google Scholar] [CrossRef]
- Erdogan, O.; Nemli, S.; Oncu, T.; Tanyolac, B. Genetic Variation among Pathotypes of Verticillium dahliae Kleb. from Cotton in Western Turkey Revealed by AFLP. Can. J. Plant Pathol. 2013, 35, 354–362. [Google Scholar] [CrossRef]
- Chagné, D.; Batley, J.; Edwards, D.; Forster, J.W. Single Nucleotide Polymorphism Genotyping in Plants. In Association Mapping in Plants; Oraguzie, N.C., Rikkerink, E.H.A., Gardiner, S.E., De Silva, H.N., Eds.; Springer: New York, NY, USA, 2007; pp. 77–94. ISBN 978-0-387-36011-9. [Google Scholar]
- Udupa, S.M.; Weigand, F.; Saxena, M.C.; Kahl, G. Genotyping with RAPD and Microsatellite Markers Resolves Pathotype Diversity in the Ascochyta Blight Pathogen of Chickpea. Theor. Appl. Genet. 1998, 97, 299–307. [Google Scholar] [CrossRef]
- Mercado-Blanco, J.; Rodríguez-Jurado, D.; Pérez-Artés, E.; Jiménez-Díaz, R.M. Detection of the Defoliating Pathotype of in Infected Olive Plants by Nested PCR. Eur. J. Plant Pathol. 2002, 108, 1–13. [Google Scholar] [CrossRef]
- Holtz, M.D.; Hwang, S.F.; Manolii, V.P.; Strelkov, I.S.; Strelkov, S.E. Development of Molecular Markers to Identify Distinct Populations of Plasmodiophora brassicae. Eur. J. Plant Pathol. 2021, 159, 637–654. [Google Scholar] [CrossRef]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-Throughput Droplet Digital PCR System for Absolute Quantitation of DNA Copy Number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef] [PubMed]
- Dobosy, J.R.; Rose, S.D.; Beltz, K.R.; Rupp, S.M.; Powers, K.M.; Behlke, M.A.; Walder, J.A. RNase H-Dependent PCR (rhPCR): Improved Specificity and Single Nucleotide Polymorphism Detection Using Blocked Cleavable Primers. BMC Biotechnol. 2011, 11, 80. [Google Scholar] [CrossRef] [PubMed]
- Zuzak, K.; Yang, Y.; Kimmel, N.; Harding, M.W.; Feindel, D.; Feng, J. Identification of Native and Invasive Subspecies of Common Reed (Phragmites australis) in Alberta, Canada, by RNase-H-Dependent PCR. Botany 2017. [Google Scholar] [CrossRef]
- McAllister, C.H.; Fortier, C.E.; St Onge, K.R.; Sacchi, B.M.; Nawrot, M.J.; Locke, T.; Cooke, J.E.K. A Novel Application of RNase H2-Dependent Quantitative PCR for Detection and Quantification of Grosmannia clavigera, a Mountain Pine Beetle Fungal Symbiont, in Environmental Samples. Tree Physiol. 2018, 38, 485–501. [Google Scholar] [CrossRef] [PubMed]
- Labbé, G.; Rankin, M.A.; Robertson, J.; Moffat, J.; Giang, E.; Lee, L.K.; Ziebell, K.; MacKinnon, J.; Laing, C.R.; Parmley, E.J.; et al. Targeting Discriminatory SNPs in Salmonella enterica serovar Heidelberg Genomes Using RNase H2-Dependent PCR. J. Microbiol. Methods 2019, 157, 81–87. [Google Scholar] [CrossRef]
- Rodgers, T.W.; Olson, J.R.; Mock, K.E. Use of RNase H-Dependent PCR for Discrimination and Detection of Closely Related Species from Environmental DNA. Methods Ecol. Evol. 2019, 10, 1091–1096. [Google Scholar] [CrossRef]
- Yang, Y.; Zuzak, K.; Harding, M.W.; Strelkov, S.E.; Hwang, S.F.; Feindel, D.; Feng, J. DNA Sequence Dimorphisms in Populations of the Clubroot Pathogen Plasmodiophora brassicae. Plant Dis. 2018, 102, 1703–1707. [Google Scholar] [CrossRef]
- Fu, H.; Yang, Y.; Mishra, V.; Zhou, Q.; Zuzak, K.; Feindel, D.; Harding, M.W.; Feng, J. Most Plasmodiophora brassicae Populations in Single Canola Root Galls from Alberta Fields Are Mixtures of Multiple Strains. Plant Dis. 2019, 104, 116–120. [Google Scholar] [CrossRef]
- SNaPshot Multiplex System for SNP Genotyping—CD Genomics. Available online: https://www.cd-genomics.com/SNaPshot.html (accessed on 12 April 2021).
- Rolland, M.; Glais, L.; Kerlan, C.; Jacquot, E. A Multiple Single Nucleotide Polymorphisms Interrogation Assay for Reliable Potato Virus Y Group and Variant Characterization. J. Virol. Methods 2008, 147, 108–117. [Google Scholar] [CrossRef]
- Balme-Sinibaldi, V.; Tribodet, M.; Croizat, F.; Lefeuvre, P.; Kerlan, C.; Jacquot, E. Improvement of Potato Virus Y (PVY) Detection and Quantitation Using PVYN- and PVYO-Specific Real-Time RT-PCR Assays. J. Virol. Methods 2006, 134, 261–266. [Google Scholar] [CrossRef]
- Egamberdiev, S.S.; Salahutdinov, I.B.; Abdullaev, A.A.; Ulloa, M.; Saha, S.; Radjapov, F.; Mullaohunov, B.; Mansurov, D.; Jenkins, J.N.; Abdurakhmonov, I.Y. Detection of Fusarium oxysporum f. Sp. vasinfectum Race 3 by Single-Base Extension Method and Allele-Specific Polymerase Chain Reaction. Can. J. Plant Pathol. 2014, 36, 216–223. [Google Scholar] [CrossRef]
- Rahman, M.; Sun, Z.; McVetty, P.B.E.; Li, G. High Throughput Genome-Specific and Gene-Specific Molecular Markers for Erucic Acid Genes in Brassica napus (L.) for Marker-Assisted Selection in Plant Breeding. Theor. Appl. Genet. 2008, 117, 895–904. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Baizan-Edge, A.; MacFarlane, S.; Torrance, L. Viral Diagnostics in Plants Using Next Generation Sequencing: Computational Analysis in Practice. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Al Rwahnih, M.; Daubert, S.; Golino, D.; Islas, C.; Rowhani, A. Comparison of Next-Generation Sequencing Versus Biological Indexing for the Optimal Detection of Viral Pathogens in Grapevine. Phytopathology 2015, 105, 758–763. [Google Scholar] [CrossRef]
- Kreuze, J.F.; Perez, A.; Untiveros, M.; Quispe, D.; Fuentes, S.; Barker, I.; Simon, R. Complete Viral Genome Sequence and Discovery of Novel Viruses by Deep Sequencing of Small RNAs: A Generic Method for Diagnosis, Discovery and Sequencing of Viruses. Virology 2009, 388, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Verdin, E.; Wipf-Scheibel, C.; Gognalons, P.; Aller, F.; Jacquemond, M.; Tepfer, M. Sequencing Viral siRNAs to Identify Previously Undescribed Viruses and Viroids in a Panel of Ornamental Plant Samples Structured as a Matrix of Pools. Virus Res. 2017, 241, 19–28. [Google Scholar] [CrossRef]
- Rott, M.; Xiang, Y.; Boyes, I.; Belton, M.; Saeed, H.; Kesanakurti, P.; Hayes, S.; Lawrence, T.; Birch, C.; Bhagwat, B.; et al. Application of Next Generation Sequencing for Diagnostic Testing of Tree Fruit Viruses and Viroids. Plant Dis. 2017, 101, 1489–1499. [Google Scholar] [CrossRef]
- Taberlet, P.; Coissac, E.; Pompanon, F.; Brochmann, C.; Willerslev, E. Towards Next-Generation Biodiversity Assessment Using DNA Metabarcoding. Mol. Ecol. 2012, 21, 2045–2050. [Google Scholar] [CrossRef]
- Lamb, P.D.; Hunter, E.; Pinnegar, J.K.; Creer, S.; Davies, R.G.; Taylor, M.I. How Quantitative Is Metabarcoding: A Meta-Analytical Approach. Mol. Ecol. 2019, 28, 420–430. [Google Scholar] [CrossRef]
- Choudhary, P.; Singh, B.N.; Chakdar, H.; Saxena, A.K. DNA Barcoding of Phytopathogens for Disease Diagnostics and Bio-Surveillance. World J. Microbiol. Biotechnol. 2021, 37, 54. [Google Scholar] [CrossRef]
- Head, S.R.; Komori, H.K.; LaMere, S.A.; Whisenant, T.; Van Nieuwerburgh, F.; Salomon, D.R.; Ordoukhanian, P. Library Construction for Next-Generation Sequencing: Overviews and Challenges. BioTechniques 2014, 56, 61–77. [Google Scholar] [CrossRef]
- Tedersoo, L.; Drenkhan, R.; Anslan, S.; Morales-Rodriguez, C.; Cleary, M. High-Throughput Identification and Diagnostics of Pathogens and Pests: Overview and Practical Recommendations. Mol. Ecol. Resour. 2019, 19, 47–76. [Google Scholar] [CrossRef]
- Baloğlu, B.; Chen, Z.; Elbrecht, V.; Braukmann, T.; MacDonald, S.; Steinke, D. A Workflow for Accurate Metabarcoding Using Nanopore MinION Sequencing. Methods Ecol. Evol. 2021. [Google Scholar] [CrossRef]
- Wit, P.D.; Pespeni, M.H.; Palumbi, S.R. SNP Genotyping and Population Genomics from Expressed Sequences—Current Advances and Future Possibilities. Mol. Ecol. 2015, 24, 2310–2323. [Google Scholar] [CrossRef] [PubMed]
- Henry, I.M.; Nagalakshmi, U.; Lieberman, M.C.; Ngo, K.J.; Krasileva, K.V.; Vasquez-Gross, H.; Akhunova, A.; Akhunov, E.; Dubcovsky, J.; Tai, T.H.; et al. Efficient Genome-Wide Detection and Cataloging of EMS-Induced Mutations Using Exome Capture and Next-Generation Sequencing. Plant Cell 2014, 26, 1382–1397. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, L.-J.; Brabbs, T.; Akhunov, A.; Jordan, K.; Budak, H.; Richmond, T.; Singh, S.; Catchpole, L.; Akhunov, E.; Hall, A. Integrating Genomic Resources to Present Full Gene and Putative Promoter Capture Probe Sets for Bread Wheat. GigaScience 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Abdelfattah, A.; Malacrinò, A.; Wisniewski, M.; Cacciola, S.O.; Schena, L. Metabarcoding: A Powerful Tool to Investigate Microbial Communities and Shape Future Plant Protection Strategies. Biol. Control 2018, 120, 1–10. [Google Scholar] [CrossRef]
- Nicolaisen, M.; West, J.S.; Sapkota, R.; Canning, G.G.M.; Schoen, C.; Justesen, A.F. Fungal Communities Including Plant Pathogens in Near Surface Air Are Similar across Northwestern Europe. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Banchi, E.; Ametrano, C.G.; Stanković, D.; Verardo, P.; Moretti, O.; Gabrielli, F.; Lazzarin, S.; Borney, M.F.; Tassan, F.; Tretiach, M.; et al. DNA Metabarcoding Uncovers Fungal Diversity of Mixed Airborne Samples in Italy. PLoS ONE 2018, 13, e0194489. [Google Scholar] [CrossRef] [PubMed]
- Cobo-Díaz, J.F.; Baroncelli, R.; Le Floch, G.; Picot, A. A Novel Metabarcoding Approach to Investigate Fusarium Species Composition in Soil and Plant Samples. FEMS Microbiol. Ecol. 2019, 95. [Google Scholar] [CrossRef]
- Legeay, J.; Husson, C.; Cordier, T.; Vacher, C.; Marcais, B.; Buée, M. Comparison and Validation of Oomycetes Metabarcoding Primers for Phytophthora High Throughput Sequencing. J. Plant Pathol. 2019, 101, 743–748. [Google Scholar] [CrossRef]
- Da Lio, D.; Cobo-Díaz, J.F.; Masson, C.; Chalopin, M.; Kebe, D.; Giraud, M.; Verhaeghe, A.; Nodet, P.; Sarrocco, S.; Le Floch, G.; et al. Combined Metabarcoding and Multi-Locus Approach for Genetic Characterization of Colletotrichum Species Associated with Common Walnut (Juglans regia) Anthracnose in France. Sci. Rep. 2018, 8, 10765. [Google Scholar] [CrossRef] [PubMed]
- Geisen, S.; Laros, I.; Vizcaíno, A.; Bonkowski, M.; de Groot, G.A. Not All Are Free-Living: High-Throughput DNA Metabarcoding Reveals a Diverse Community of Protists Parasitizing Soil Metazoa. Mol. Ecol. 2015, 24, 4556–4569. [Google Scholar] [CrossRef] [PubMed]
- Castaño, C.; Berlin, A.; Durling, M.B.; Ihrmark, K.; Lindahl, B.D.; Stenlid, J.; Clemmensen, K.E.; Olson, Å. Optimized Metabarcoding with Pacific Biosciences Enables Semi-Quantitative Analysis of Fungal Communities. New Phytol. 2020, 228. [Google Scholar] [CrossRef] [PubMed]
- RNase H2 Enzyme. Available online: https://www.idtdna.com/pages/products/qpcr-and-pcr/master-mixes-reagents/rnase-h-enzyme (accessed on 25 April 2021).
- rhPCR Primers. Available online: https://www.idtdna.com/pages/products/qpcr-and-pcr/custom-primers/rhpcr-primers (accessed on 26 April 2021).



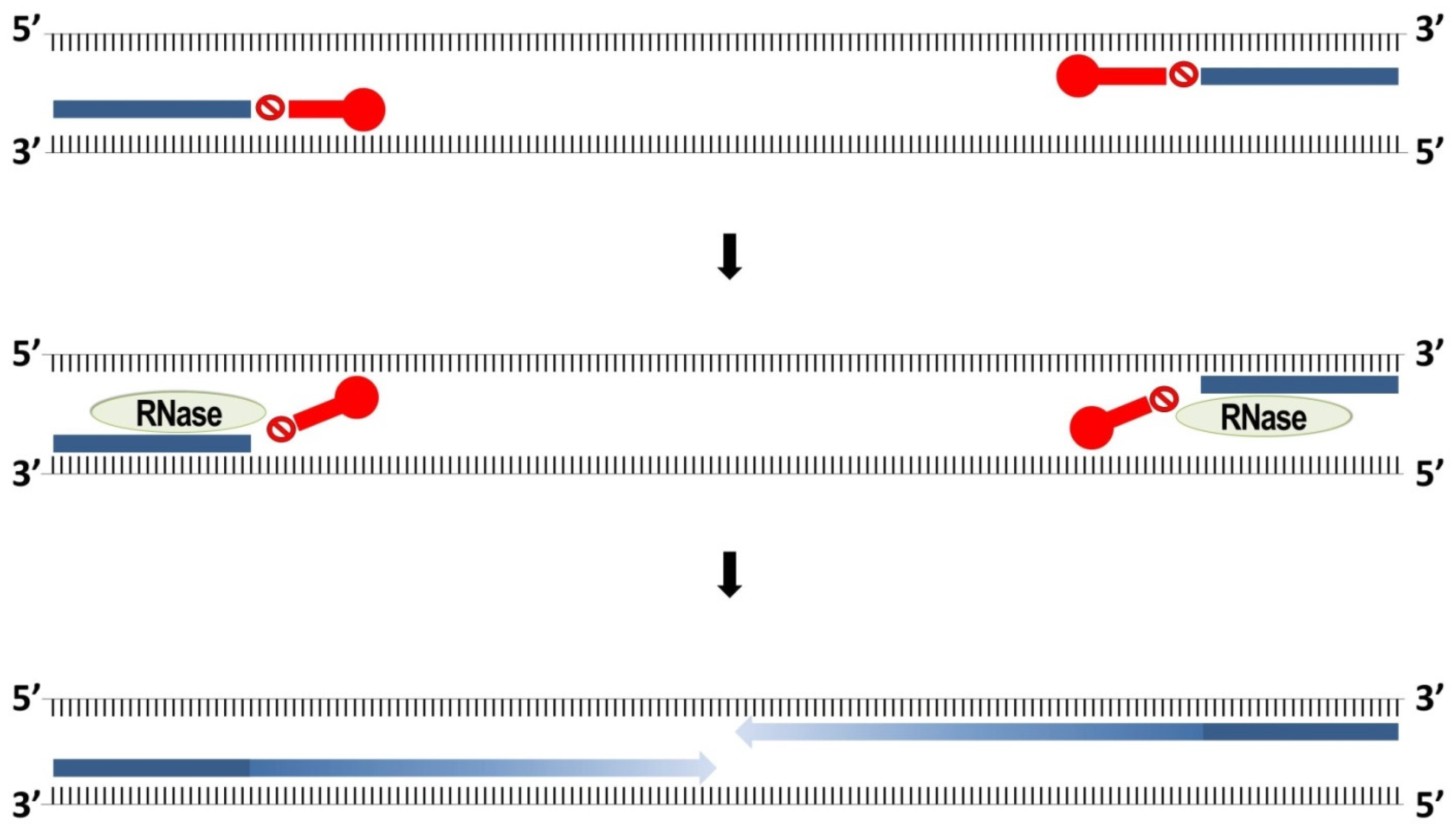
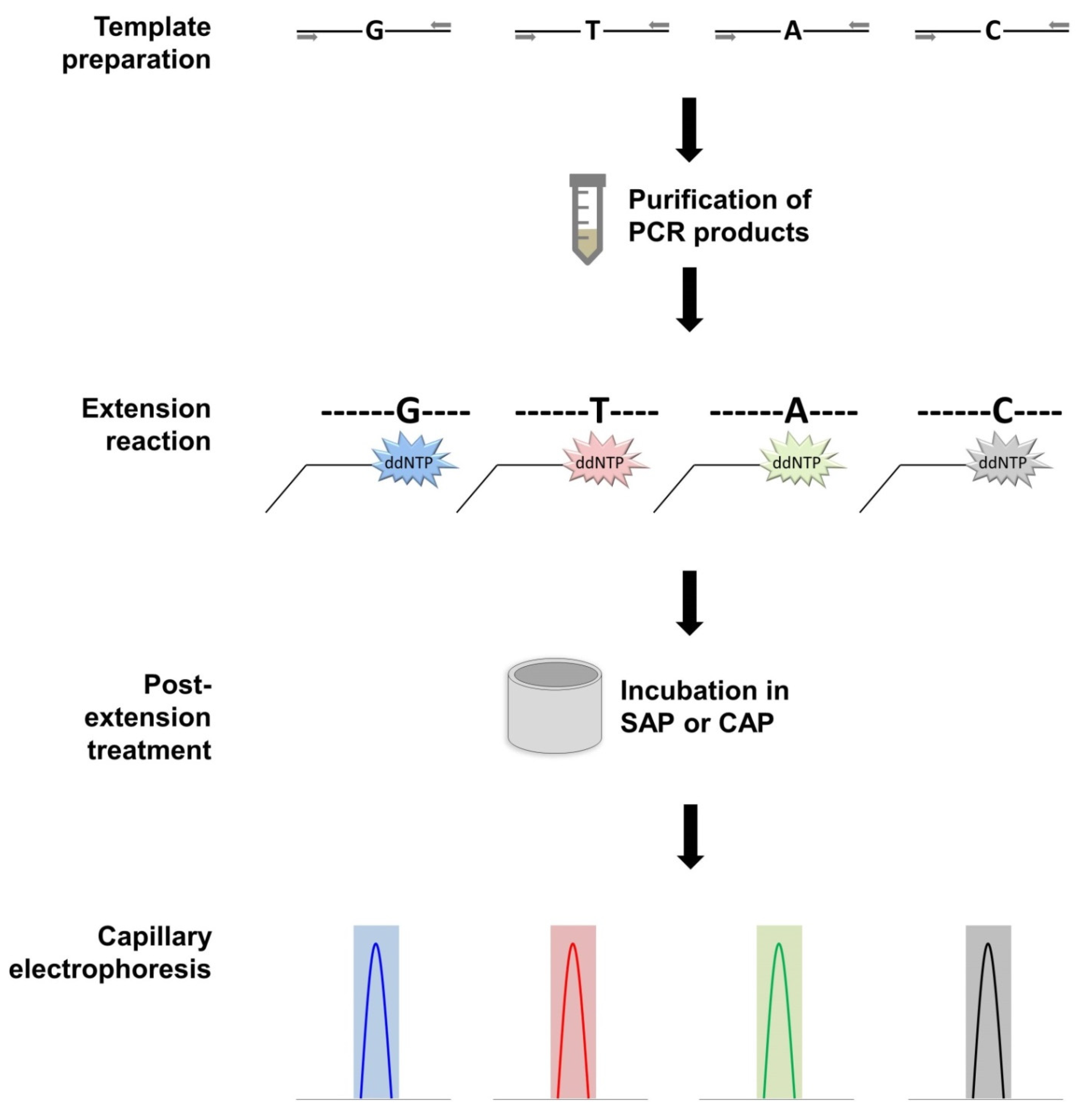

| Technique | Efficiency a | Specificity | Quantitative Potential | Primers Required | Costs | Main Advantages | Main Disadvantages |
|---|---|---|---|---|---|---|---|
| Amplicon length distinction | Low | Low | No b | 1 pair per indel | Low | Conserved primers | Low scalability |
| SNP-based distinction | Low | Low | Yes | 1 or 2 pairs per polymorphic region c | Low | Simple procedure | Low scalability and sensitivity |
| rhPCR | Low | Moderate | Yes | 1 or 2 pairs per polymorphic region c | Moderate d | Simple procedure | Low scalability |
| SBE | Moderate | Moderate | No | 1 pair + 1 SBE primer per SNP e | High | Scalable; can detect any allele | Non-quantitative; lengthy procedure |
| Metabarcoding | High | High | Partially f | 1 pair per barcoding sequence | Very High | High sensitivity and scalability | High costs and expertise required; lengthy procedure |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tso, H.H.; Galindo-González, L.; Strelkov, S.E. Current and Future Pathotyping Platforms for Plasmodiophora brassicae in Canada. Plants 2021, 10, 1446. https://doi.org/10.3390/plants10071446
Tso HH, Galindo-González L, Strelkov SE. Current and Future Pathotyping Platforms for Plasmodiophora brassicae in Canada. Plants. 2021; 10(7):1446. https://doi.org/10.3390/plants10071446
Chicago/Turabian StyleTso, Heather H., Leonardo Galindo-González, and Stephen E. Strelkov. 2021. "Current and Future Pathotyping Platforms for Plasmodiophora brassicae in Canada" Plants 10, no. 7: 1446. https://doi.org/10.3390/plants10071446
APA StyleTso, H. H., Galindo-González, L., & Strelkov, S. E. (2021). Current and Future Pathotyping Platforms for Plasmodiophora brassicae in Canada. Plants, 10(7), 1446. https://doi.org/10.3390/plants10071446






