The Coordinated Upregulated Expression of Genes Involved in MEP, Chlorophyll, Carotenoid and Tocopherol Pathways, Mirrored the Corresponding Metabolite Contents in Rice Leaves during De-Etiolation
Abstract
:1. Introduction
2. Results
2.1. Tocochromanol Levels in De-Etiolated Rice Leaves
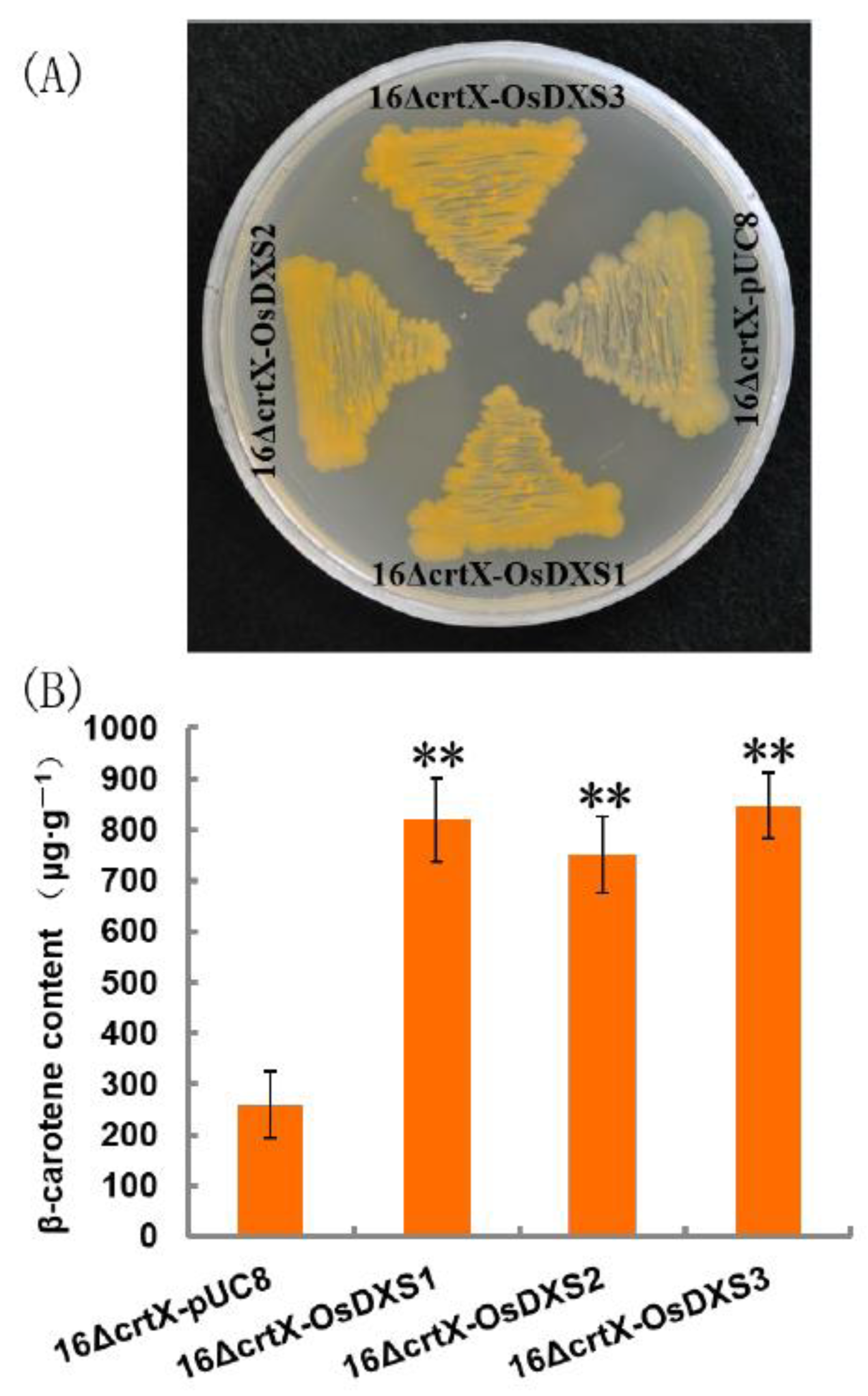
2.2. Functional Analysis of OsDXS Gene Family
2.3. Expression Analysis of Genes Involved in MEP and MVA Pathways
2.4. Expression Analysis of Genes Involved in Carotenoid, Tocopherol, and Chlorophyll Biosynthetic Pathways
2.5. In Silico Promoter Analysis of Light-Responsive Cis-Regulatory Elements in the Promoter Regions of Light-Upregulated Genes
3. Discussion
3.1. The Coordinated Expression Levels of the MEP and MVA Pathway Genes and Carotenoid, Tocopherol and Chlorophyll Biosynthetic Pathway Genes Mirrored the Levels of the Corresponding Metabolite Changes during De-Etiolation
3.2. The Underpinning Mechanistic Basis of Coordinated Light-Responsive Gene Expression during the De-Etiolation Process
4. Materials and Methods
4.1. Plant Material
4.2. Tocochromanols Analysis
4.3. Rice DXS cDNA Cloning and Functional Complementation Analysis
4.4. Gene Expression Analysis with Quantitative Real-Time PCR (qRT-PCR)
4.5. Retrieval of Promoter Regions and Analysis of Cis-Regulatory Elements
4.6. Data and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, P.; Chen, H.; Liang, Y.; Shen, S. Proteomic analysis of de-etiolated rice seedlings upon exposure to light. Proteomics 2007, 7, 2459–2468. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, H. Coordinated shoot and root responses to light signaling in Arabidopsis. Plant Commun. 2020, 1, 100026. [Google Scholar] [CrossRef]
- Jiao, Y.; Lau, O.S.; Deng, X.W. Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 2007, 8, 217–230. [Google Scholar] [CrossRef]
- Jung, K.H.; Lee, J.; Dardick, C.; Seo, Y.S.; Cao, P.; Canlas, P.; Phetsom, J.; Xu, X.; Ouyang, S.; An, K.; et al. Identification and functional analysis of light-responsive unique genes and gene family members in rice. PLoS Genet. 2008, 4, e1000164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chory, J.; Chatterjee, M.; Cook, R.K.; Elich, T.; Fankhauser, C.; Li, J.; Nagpal, P.; Neff, M.; Pepper, A.; Poole, D.; et al. From seed germination to flowering, light controls plant development via the pigment phytochrome. Proc. Natl. Acad. Sci. USA 1996, 93, 12066–12071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clouse, S.D. Integration of light and brassinosteroid signals in etiolated seedling growth. Trends Plant Sci. 2001, 6, 443–445. [Google Scholar] [CrossRef]
- Li, Y.; Swaminathan, K.; Hudson, M.E. Rapid, organ-specific transcriptional responses to light regulate photomorphogenic development in dicot seedlings. Plant Physiol. 2011, 156, 2124–2140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vranova, E.; Coman, D.; Gruissem, W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 2013, 64, 665–700. [Google Scholar] [CrossRef] [PubMed]
- Ghassemian, M.; Lutes, J.; Tepperman, J.M.; Chang, H.S.; Zhu, T.; Wang, X.; Quail, P.H.; Lange, B.M. Integrative analysis of transcript and metabolite profiling data sets to evaluate the regulation of biochemical pathways during photomorphogenesis. Arch. Biochem. Biophys. 2006, 448, 45–59. [Google Scholar] [CrossRef]
- Jin, X.; Baysal, C.; Gao, L.; Medina, V.; Drapal, M.; Ni, X.; Sheng, Y.; Shi, L.; Capell, T.; Fraser, P.D.; et al. The subcellular localization of two isopentenyl diphosphate isomerases in rice suggests a role for the endoplasmic reticulum in isoprenoid biosynthesis. Plant Cell Rep. 2020, 39, 119–133. [Google Scholar] [CrossRef]
- Kim, B.R.; Kim, S.U.; Chang, Y.J. Differential expression of three 1-deoxy-D: -xylulose-5-phosphate synthase genes in rice. Biotechnol. Lett. 2005, 27, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Okada, A.; Shimizu, T.; Okada, K.; Kuzuyama, T.; Koga, J.; Shibuya, N.; Nojiri, H.; Yamane, H. Elicitor induced activation of the methylerythritol phosphate pathway toward phytoalexins biosynthesis in rice. Plant Mol. Biol. 2007, 65, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Kajiwara, S.; Fraser, P.D.; Kondo, K.; Misawa, N. Expression of an exogenous isopentenyl diphosphate isomerase gene enhances isoprenoid biosynthesis in Escherichia coli. Biochem. J. 1997, 324, 421–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, R.; Wang, Y.; Wang, P.; Li, C.; Xiao, F.; Chen, N.; Li, N.; Li, C.; Sun, C.; Li, L.; et al. A single nucleotide mutation of IspF gene involved in the MEP pathway for isoprenoid biosynthesis causes yellow-green leaf phenotype in rice. Plant Mol. Biol. 2018, 96, 5–16. [Google Scholar] [CrossRef]
- Gallagher, C.E.; Matthews, P.D.; Li, F.; Wurtzel, E.T. Gene duplication in the carotenoid biosynthetic pathway preceded evolution of the grasses. Plant Physiol. 2004, 135, 1776–1783. [Google Scholar] [CrossRef] [Green Version]
- Fang, J.; Chai, C.; Qian, Q.; Li, C.; Tang, J.; Sun, L.; Huang, Z.; Guo, X.; Sun, C.; Liu, M.; et al. Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J. 2008, 54, 177–189. [Google Scholar] [CrossRef] [Green Version]
- Chaudhary, N.; Khurana, P. Vitamin E biosynthesis genes in rice: Molecular characterization, expression profiling and comparative phylogenetic analysis. Plant Sci. 2009, 177, 479–491. [Google Scholar] [CrossRef]
- Lee, S.; Kim, J.H.; Yoo, E.S.; Lee, C.H.; Hirochika, H.; An, G. Differential regulation of chlorophyll a oxygenase genes in rice. Plant Mol. Biol. 2005, 57, 805–818. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, X.; He, B.; Diao, L.; Sheng, S.; Wang, J.; Guo, X.; Su, N.; Wang, L.; Jiang, L.; et al. A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol. 2007, 145, 29–40. [Google Scholar] [CrossRef] [Green Version]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto, K.; Matsumoto, T.; Okada, A.; Komiyama, K.; Chujo, T.; Yoshikawa, H.; Nojiri, H.; Yamane, H.; Okada, K. Identification of target genes of the bZIP transcription factor OsTGAP1, whose overexpression causes elicitor-induced hyperaccumulation of diterpenoid phytoalexins in rice cells. PLoS ONE 2014, 9, e105823. [Google Scholar] [CrossRef] [Green Version]
- Von Lintig, J.; Welsch, R.; Bonk, M.; Giuliano, G.; Batschauer, A.; Kleinig, H. Light-dependent regulation of carotenoid biosynthesis occurs at the level of phytoene synthase expression and is mediated by phytochrome in Sinapis alba and Arabidopsis thaliana seedlings. Plant J. 1997, 12, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Welsch, R.; Beyer, P.; Hugueney, P.; Kleinig, H.; von Lintig, J. Regulation and activation of phytoene synthase, a key enzyme in carotenoid biosynthesis, during photomorphogenesis. Planta 2000, 211, 846–854. [Google Scholar] [CrossRef]
- Welsch, R.; Wust, F.; Bar, C.; Al-Babili, S.; Beyer, P. A third phytoene synthase is devoted to abiotic stress-induced abscisic acid formation in rice and defines functional diversification of phytoene synthase genes. Plant Physiol. 2008, 147, 367–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaudhary, N.; Nijhawan, A.; Khurana, J.P.; Khurana, P. Carotenoid biosynthesis genes in rice: Structural analysis, genome-wide expression profiling and phylogenetic analysis. Mol. Genet. Genom. 2010, 283, 13–33. [Google Scholar] [CrossRef] [PubMed]
- Collakova, E.; DellaPenna, D. Homogentisate phytyltransferase activity is limiting for tocopherol biosynthesis in Arabidopsis. Plant Physiol. 2003, 131, 632–642. [Google Scholar] [CrossRef] [Green Version]
- Terzaghi, W.B.; Cashmore, A.R. Light-regulated transcription. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1995, 46, 445–474. [Google Scholar] [CrossRef]
- Enfissi, E.M.; Barneche, F.; Ahmed, I.; Lichtle, C.; Gerrish, C.; McQuinn, R.P.; Giovannoni, J.J.; Lopez-Juez, E.; Bowler, C.; Bramley, P.M.; et al. Integrative transcript and metabolite analysis of nutritionally enhanced DE-ETIOLATED1 downregulated tomato fruit. Plant Cell 2010, 22, 1190–1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almeida, J.; Perez-Fons, L.; Fraser, P.D. A transcriptomic, metabolomic and cellular approach to the physiological adaptation of tomato fruit to high temperature. Plant Cell Environ. 2020. [Google Scholar] [CrossRef]
- Misawa, N.; Satomi, Y.; Kondo, K.; Yokoyama, A.; Kajiwara, S.; Saito, T.; Ohtani, T.; Miki, W. Structure and functional analysis of a marine bacterial carotenoid biosynthesis gene cluster and astaxanthin biosynthetic pathway proposed at the gene level. J. Bacteriol. 1995, 177, 6575–6584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baysal, C.; He, W.; Drapal, M.; Villorbina, G.; Medina, V.; Capell, T.; Khush, G.S.; Zhu, C.; Fraser, P.D.; Christou, P. Inactivation of rice starch branching enzyme IIb triggers broad and unexpected changes in metabolism by transcriptional reprogramming. Proc. Natl. Acad. Sci. USA 2020, 117, 26503–26512. [Google Scholar] [CrossRef] [PubMed]





| Gene Name | Cis-Acting Regulatory Elements Involved in Light Responsiveness | GenBank Accession Numbers |
|---|---|---|
| OsDXS1 | G-box (CACGTC, -102); IBOXCORE (I box; GATAA, -422, -485, -510); Box 4 (ATTAAT, -501, -748); GT1-motif (GGTTAA, -990) | XM_015785019 (mRNA); AP014961 (gDNA, genomic DNA) |
| OsDXR | G-box (CACGTT, -52); IBOXCORE (GATAA, -200, -233); Box 4 (ATTAAT, -664); MRE (AACCTAA, -852) | AK099702 (mRNA); AP014957 (gDNA) |
| OsMCT | G-box (TAAACGTG, -35); Box 4 (ATTAAT, -54); Sp1 (GGGCGG, -819, -952) | XM_015771410 (mRNA); AP003346 (gDNA) |
| OsCMK | IBOXCORE (GATAA, -552) | XM_015766647 (mRNA); AP003221 (gDNA) |
| OsMDS | IBOXCORE (GATAA, -384, -564, -667); GT1-motif (GGTTAA, -857) | NC_029257 (mRNA); AP005823 (gDNA) |
| OsHDS | G-box (CACGAC, -46); IBOXCORE (GATAA, -980) | XM_015770238 (mRNA); AP004124 (gDNA) |
| OsHDR1 | IBOXCORE (GATAA, -17) | XM_015777111 (mRNA); AC103550 (gDNA) |
| OsIPPI1 | G-box (CACGTG, -143); LTR (CCGAAA, -278); IBOXCORE (GATAA, -898); G-box (CACGAC, -974) | XM_015791312 (mRNA); AP014963 (gDNA) |
| OsPORB | Box II (CCACGTGGC, -55); AE-box (AGAAACAA, -143); IBOXCORE (GATAA, -156); G-box (CGCGTC, -514); G-box (GCCACGTGGA, -557); G-box (CACGTT, -566); GATA-motif (GATAGGG, -628) | XM_015759459 (mRNA); AC068923 (gDNA) |
| OsCHLG | GATA-motif (GATAGGA, -189); TCT-motif (TCTTAC, -297); IBOXCORE (GATAA, -978) | XM_015782164 (mRNA); AC136221 (gDNA) |
| OsCAO1 | LTR (CCGAAA, -841, -810); Box 4 (ATTAAT, -749, -697); G-box (CACGTC, -519, -8); Box II (CCACGTGGC, -489); TCT-motif (TCTTAC, -358); IBOXCORE (I-box; GATAA, -70) | XM_015758600 (mRNA); AP014966 (gDNA) |
| OsPSY1 | TCCC-motif (TCTCCCT, -24); G-box (CACGTC, -30); IBOXCORE (GATAA, -70, -926); Box 4 (ATTAAT, -464, -590, -749); AE-box (AGAAACAA, -877) | XM_015787027 (mRNA); AP005750 (gDNA) |
| OsPSY2 | G-box (CACGTG, -86, -92); Box 4 (ATTAAT, -183, -464, -630); IBOXCORE (GATAA, -940) | AK073290 (mRNA); AL831803 (gDNA) |
| OsPDS | G-box (TACGTG, -173); IBOXCORE (GATAA, -341, -379); G-box (CACGAC, -864) | XM_015777615 (mRNA); AC079633 (gDNA) |
| OsZDS | IBOXCORE (GATAA, -66, -538, -569); TCCC-motif (TCTCCCT, -314); Box 4 (ATTAAT, -902) | XM_015791038 (mRNA); AP004273 (gDNA) |
| OsLYCB | Box 4 (ATTAAT, -171, -569); IBOXCORE (GATAA, -634); GT1-motif (GGTTAA, -738); G-box (CACGTT, -769) | XM_015771748 (mRNA); AP005849 (gDNA) |
| OsLYCE | Sp1 (GGGCGG, -115, -815); GT1-motif (GGTTAA, -288); G-box (CACGTC, -376) | XM_015766712 (mRNA); AP003332 (gDNA) |
| OsGGR | MRE (AACCTAA, -22); IBOXCORE (GATAA, -348, -457); GT1-motif (GGTTAA, -612); GT1-motif (GGTTAAT, -836) | XM_015771626 (mRNA); AP004114 (gDNA) |
| OsVTE1 | G-box (CCACGTAA, -74); Sp1 (GGGCGG, -156); G-box (CACGTG, -184); IBOXCORE (GATAA, -498, -573) | XM_015770843 (mRNA); AP005536 (gDNA) |
| OsVTE2 | Box 4 (ATTAAT, -275); MRE (AACCTAA, -916) | XM_015789025 (mRNA); AP005760 (gDNA) |
| OsVTE4 | Box 4 (ATTAAT, -87); IBOXCORE (GATAA, -746, -809) | XM_015767600 (mRNA); AP003994 (gDNA) |
| OsVTE5 | AE-box (AGAAACAA, -817); IBOXCORE (GATAA, -846) | XM_015769509 (mRNA); AP003252 (gDNA) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, X.; Baysal, C.; Drapal, M.; Sheng, Y.; Huang, X.; He, W.; Shi, L.; Capell, T.; Fraser, P.D.; Christou, P.; et al. The Coordinated Upregulated Expression of Genes Involved in MEP, Chlorophyll, Carotenoid and Tocopherol Pathways, Mirrored the Corresponding Metabolite Contents in Rice Leaves during De-Etiolation. Plants 2021, 10, 1456. https://doi.org/10.3390/plants10071456
Jin X, Baysal C, Drapal M, Sheng Y, Huang X, He W, Shi L, Capell T, Fraser PD, Christou P, et al. The Coordinated Upregulated Expression of Genes Involved in MEP, Chlorophyll, Carotenoid and Tocopherol Pathways, Mirrored the Corresponding Metabolite Contents in Rice Leaves during De-Etiolation. Plants. 2021; 10(7):1456. https://doi.org/10.3390/plants10071456
Chicago/Turabian StyleJin, Xin, Can Baysal, Margit Drapal, Yanmin Sheng, Xin Huang, Wenshu He, Lianxuan Shi, Teresa Capell, Paul D. Fraser, Paul Christou, and et al. 2021. "The Coordinated Upregulated Expression of Genes Involved in MEP, Chlorophyll, Carotenoid and Tocopherol Pathways, Mirrored the Corresponding Metabolite Contents in Rice Leaves during De-Etiolation" Plants 10, no. 7: 1456. https://doi.org/10.3390/plants10071456
APA StyleJin, X., Baysal, C., Drapal, M., Sheng, Y., Huang, X., He, W., Shi, L., Capell, T., Fraser, P. D., Christou, P., & Zhu, C. (2021). The Coordinated Upregulated Expression of Genes Involved in MEP, Chlorophyll, Carotenoid and Tocopherol Pathways, Mirrored the Corresponding Metabolite Contents in Rice Leaves during De-Etiolation. Plants, 10(7), 1456. https://doi.org/10.3390/plants10071456






