Isolation and Characterization of Pseudomonas chlororaphis Strain ST9; Rhizomicrobiota and in Planta Studies
Abstract
:1. Introduction
2. Results
2.1. Identification and Characterization of P. chlororaphis ST9
2.2. P. chlororaphis ST9 Root Colonization Ability, Persistence and Effect on the Rhizosphere Microbiota
2.3. Plant Gene Expression and Phenotypic Analysis of P. chlororaphis Inoculated Plants
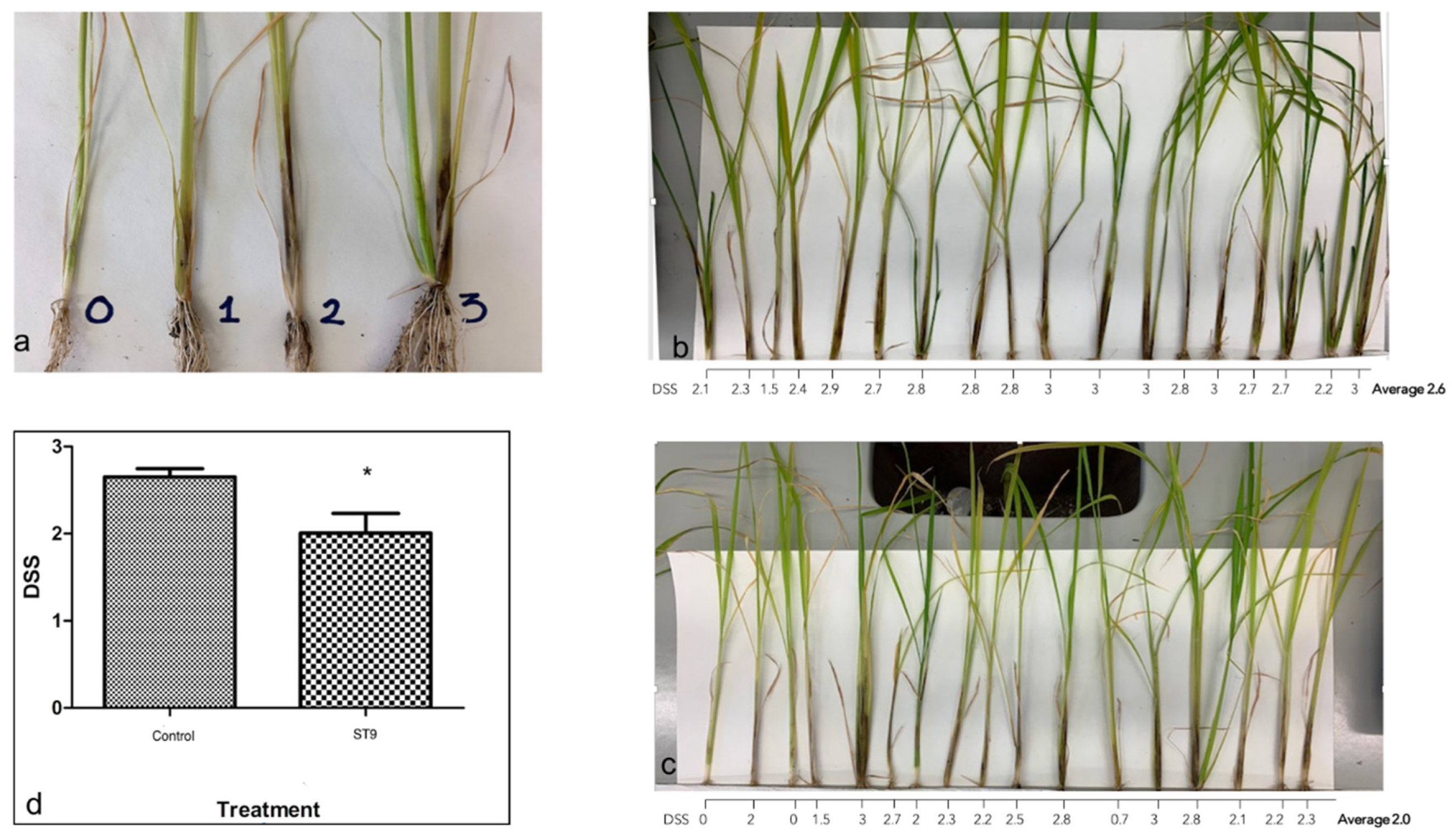
3. Discussion
4. Material and Methods
4.1. Strain Isolation, Growth and Identification
4.2. In Vitro Phenotypic Characterization
4.3. Generation of a Rifampicin-Resistant Spontaneous Mutants
4.4. Bacterial Genomic DNA Extraction and Sequencing
4.5. P. chlororaphis ST9 Taxonomy
4.6. Plant Inoculation
4.7. Greenhouse Experiments
4.8. Colonization Counts
4.9. In Vivo Biocontrol Activity of P. chlororaphis ST9 against Dickeya Zeae
4.10. Bacterial Genomic DNA Extraction, 16S rRNA Gene Amplicon Library Preparation and Sequencing
4.11. Microbiome Sequence Analysis
4.12. RNA Extraction and cDNA Conversion
4.13. Primer Selection
4.14. Gene Expression Analysis
4.15. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hayat, R.; Ali, S.; Amara, U.; Khalid, R.; Ahmed, I. Soil beneficial bacteria and their role in plant growth promotion: A review. Ann. Microbiol. 2010, 60, 579–598. [Google Scholar] [CrossRef]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant growth-promoting rhizobacteria: Context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef] [Green Version]
- Berendsen, R.L.; Pieterse, C.M.; Bakker, P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Van Themaat, E.V.L.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khatoon, Z.; Huang, S.; Rafique, M.; Fakhar, A.; Kamran, M.A.; Santoyo, G. Unlocking the potential of plant growth-promoting rhizobacteria on soil health and the sustainability of agricultural systems. J. Environ. Manag. 2020, 273, 111118. [Google Scholar] [CrossRef] [PubMed]
- Bender, S.F.; Wagg, C.; van der Heijden, M.G. An underground revolution: Biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol. Evol. 2016, 31, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Pii, Y.; Mimmo, T.; Tomasi, N.; Terzano, R.; Cesco, S.; Crecchio, C. Microbial interactions in the rhizosphere: Beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. A review. Biol. Fertil. Soils 2015, 51, 403–415. [Google Scholar] [CrossRef]
- Spaepen, S.; Bossuyt, S.; Engelen, K.; Marchal, K.; Vanderleyden, J. Phenotypical and molecular responses of Arabidopsis thaliana roots as a result of inoculation with the auxin-producing bacterium Azospirillum brasilense. New Phytol. 2014, 201, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Glick, B.R.; Pasternak, J. Plant-microbial interaction under gnotobiotic conditions: A scanning electron microscope study. Curr. Microbiol. 1991, 23, 111–114. [Google Scholar] [CrossRef]
- Ruzzi, M.; Aroca, R. Plant growth-promoting rhizobacteria act as biostimulants in horticulture. Sci. Hortic. 2015, 196, 124–134. [Google Scholar] [CrossRef]
- Mehnaz, S. An overview of globally available bioformulations. Bioformulations Sustain. Agric. 2016, 267–281. [Google Scholar] [CrossRef]
- Hazra, K.; Swain, D.; Bohra, A.; Singh, S.; Kumar, N.; Nath, C. Organic rice: Potential production strategies, challenges and prospects. Org. Agric. 2018, 8, 39–56. [Google Scholar] [CrossRef]
- Oleńska, E.; Małek, W.; Wójcik, M.; Swiecicka, I.; Thijs, S.; Vangronsveld, J. Beneficial features of plant growth-promoting rhizobacteria for improving plant growth and health in challenging conditions: A methodical review. Sci. Total Environ. 2020, 140682. [Google Scholar] [CrossRef] [PubMed]
- Biessy, A.; Novinscak, A.; Blom, J.; Léger, G.; Thomashow, L.S.; Cazorla, F.M.; Josic, D.; Filion, M. Diversity of phytobeneficial traits revealed by whole-genome analysis of worldwide-isolated phenazine-producing Pseudomonas Spp. Environ. Microbiol. 2019, 21, 437–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burr, S.E.; Gobeli, S.; Kuhnert, P.; Goldschmidt-Clermont, E.; Frey, J. Pseudomonas chlororaphis subsp. piscium subsp. nov., isolated from freshwater fish. Int. J. Syst. Evol. Microbiol. 2010, 60, 2753–2757. [Google Scholar] [CrossRef] [Green Version]
- Peix, A.; Valverde, A.; Rivas, R.; Igual, J.M.; Ramirez-Bahena, M.-H.; Mateos, P.F.; Santa-Regina, I.; Rodriguez-Barrueco, C.; Martínez-Molina, E.; Velázquez, E. Reclassification of Pseudomonas aurantiaca as a synonym of Pseudomonas chlororaphis and proposal of three subspecies, P. chlororaphis subsp. chlororaphis subsp. nov., P. chlororaphis subsp. aureofaciens subsp. nov., comb. nov. and P. chlororaphis subsp. aurantiaca subsp. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2007, 57, 1286–1290. [Google Scholar]
- Anderson, A.J.; Kim, Y.C. Insights into plant-beneficial traits of probiotic Pseudomonas chlororaphis isolates. J. Med. Microbiol. 2020, 69, 361–371. [Google Scholar] [CrossRef]
- Arrebola, E.; Tienda, S.; Vida, C.; De Vicente, A.; Cazorla, F.M. Fitness features involved in the biocontrol interaction of Pseudomonas chlororaphis with host plants: The case study of PcPCL1606. Front. Microbiol. 2019, 10, 719. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.F.; Xu, Y.J.; Ma, Z.Q.; Deng, Z.S.; Shan, C.J.; Wei, G.H. Colonization and plant growth promoting characterization of endophytic Pseudomonas chlororaphis strain Zong1 isolated from Sophora alopecuroides root nodules. Braz. J. Microbiol. 2013, 44, 629–637. [Google Scholar] [CrossRef] [Green Version]
- Arrebola, E.; Cazorla, F.M. Aer receptors influence the Pseudomonas chlororaphis PCL1606 lifestyle. Front. Microbiol. 2020, 11, 1560. [Google Scholar] [CrossRef]
- Calderón, C.E.; Pérez-García, A.; de Vicente, A.; Cazorla, F.M. The dar genes of Pseudomonas chlororaphis PCL1606 are crucial for biocontrol activity via production of the antifungal compound 2-hexyl, 5-propyl resorcinol. Mol. Plant Microbe Interact. 2013, 26, 554–565. [Google Scholar] [CrossRef] [Green Version]
- Ahemad, M. Phosphate-solubilizing bacteria-assisted phytoremediation of metalliferous soils: A review. 3 Biotech 2015, 5, 111–121. [Google Scholar] [CrossRef] [Green Version]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.R.; Yang, K.Y.; Cho, B.H.; Han, T.H.; Kim, I.S.; Lee, M.C.; Anderson, A.J.; Kim, Y.C. Production of indole-3-acetic acid in the plant-beneficial strain Pseudomonas chlororaphis O6 is negatively regulated by the global sensor kinase GacS. Curr. Microbiol. 2006, 52, 473–476. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.A.; Kim, J.S.; Park, J.Y.; Han, S.H.; Dimkpa, C.; Anderson, A.J.; Kim, Y.C. The RpoS sigma factor negatively regulates production of IAA and siderophore in a biocontrol rhizobacterium, Pseudomonas chlororaphis O6. Plant Pathol. J. 2013, 29, 323. [Google Scholar] [CrossRef] [Green Version]
- Chin-A-Woeng, T.F.; Bloemberg, G.V.; van der Bij, A.J.; van der Drift, K.M.; Schripsema, J.; Kroon, B.; Scheffer, R.J.; Keel, C.; Bakker, P.A.; Tichy, H.-V. Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. Mol. Plant Microbe Interact. 1998, 11, 1069–1077. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Gao, Q.; Hamada, M.S.; Dawood, D.H.; Zheng, J.; Chen, Y.; Ma, Z. Potential of Pseudomonas chlororaphis subsp. aurantiaca strain Pcho10 as a biocontrol agent against Fusarium graminearum. Phytopathology 2014, 104, 1289–1297. [Google Scholar] [CrossRef] [Green Version]
- Spence, C.A.; Raman, V.; Donofrio, N.M.; Bais, H.P. Global gene expression in rice blast pathogen Magnaporthe oryzae treated with a natural rice soil isolate. Planta 2014, 239, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; He, Y.; Jiang, H.; Peng, H.; Huang, X.; Zhang, X.; Thomashow, L.S.; Xu, Y. Characterization of a phenazine-producing strain Pseudomonas chlororaphis GP72 with broad-spectrum antifungal activity from green pepper rhizosphere. Curr. Microbiol. 2007, 54, 302–306. [Google Scholar] [CrossRef]
- Tagele, S.B.; Lee, H.G.; Kim, S.W.; Lee, Y.S. Phenazine and 1-undecene producing Pseudomonas chlororaphis subsp. aurantiaca strain KNU17Pc1 for growth promotion and disease suppression in Korean maize cultivars. J. Microbiol. Biotechnol. 2019, 29, 66–78. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Khabbaz, S.; Wang, A.; Li, H.; Abbasi, P. Detection and characterization of broad-spectrum antipathogen activity of novel rhizobacterial isolates and suppression of Fusarium crown and root rot disease of tomato. J. Appl. Microbiol. 2015, 118, 685–703. [Google Scholar] [CrossRef]
- Nam, H.S.; Anderson, A.J.; Kim, Y.C. Biocontrol efficacy of formulated Pseudomonas chlororaphis O6 against plant diseases and root-knot nematodes. Plant Pathol. J. 2018, 34, 241. [Google Scholar] [CrossRef] [PubMed]
- Hilario, E.; Buckley, T.R.; Young, J.M. Improved resolution on the phylogenetic relationships among Pseudomonas by the combined analysis of atp D, car A, rec A and 16S rDNA. Antonie Van Leeuwenhoek 2004, 86, 51–64. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Liu, K.; Hu, H.; Wang, W.; Zhang, X. Genetic engineering of Pseudomonas chlororaphis GP72 for the enhanced production of 2-hydroxyphenazine. Microb. Cell Factories 2016, 15, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorosky, R.J.; Yu, J.M.; Pierson, L.S.; Pierson, E.A. Pseudomonas chlororaphis produces two distinct R-tailocins that contribute to bacterial competition in biofilms and on roots. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nandi, M.; Selin, C.; Brassinga, A.K.C.; Belmonte, M.F.; Fernando, W.D.; Loewen, P.C.; De Kievit, T.R. Pyrrolnitrin and hydrogen cyanide production by Pseudomonas chlororaphis strain PA23 exhibits nematicidal and repellent activity against Caenorhabditis elegans. PLoS ONE 2015, 10, e0123184. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Van Pee, K.-H.; Naismith, J.H. The ternary complex of PrnB (the second enzyme in the pyrrolnitrin biosynthesis pathway), tryptophan, and cyanide yields new mechanistic insights into the indolamine dioxygenase superfamily. J. Biol. Chem. 2010, 285, 21126–21133. [Google Scholar] [CrossRef] [Green Version]
- Cazorla, F.M.; Duckett, S.B.; Bergström, E.T.; Noreen, S.; Odijk, R.; Lugtenberg, B.J.; Thomas-Oates, J.E.; Bloemberg, G.V. Biocontrol of avocado dematophora root rot by antagonistic Pseudomonas fluorescens PCL1606 correlates with the production of 2-hexyl 5-propyl resorcinol. Mol. Plant Microbe Interact. 2006, 19, 418–428. [Google Scholar] [CrossRef] [Green Version]
- Shah, N.; Gislason, A.S.; Becker, M.; Belmonte, M.F.; Fernando, W.D.; de Kievit, T.R. Investigation of the quorum-sensing regulon of the biocontrol bacterium Pseudomonas chlororaphis strain PA23. PLoS ONE 2020, 15, e0226232. [Google Scholar]
- Cho, S.-T.; Chang, H.-H.; Egamberdieva, D.; Kamilova, F.; Lugtenberg, B.; Kuo, C.-H. Genome analysis of Pseudomonas fluorescens PCL1751: A rhizobacterium that controls root diseases and alleviates salt stress for its plant host. PLoS ONE 2015, 10, e0140231. [Google Scholar] [CrossRef] [PubMed]
- Flury, P.; Aellen, N.; Ruffner, B.; Péchy-Tarr, M.; Fataar, S.; Metla, Z.; Dominguez-Ferreras, A.; Bloemberg, G.; Frey, J.; Goesmann, A. Insect pathogenicity in plant-beneficial pseudomonads: Phylogenetic distribution and comparative genomics. ISME J. 2016, 10, 2527–2542. [Google Scholar] [CrossRef] [PubMed]
- Lund, B.; Brocklehurst, T.; Wyatt, G. Characterization of Strains of Clostridium puniceum sp. no v., a Pink-pigmented, Pectolytic Bacterium. Microbiology 1981, 122, 17–26. [Google Scholar] [CrossRef] [Green Version]
- García-González, T.; Sáenz-Hidalgo, H.K.; Silva-Rojas, H.V.; Morales-Nieto, C.; Vancheva, T.; Koebnik, R.; Ávila-Quezada, G.D. Enterobacter cloacae, an emerging plant-pathogenic bacterium affecting chili pepper seedlings. Plant Pathol. J. 2018, 34, 1. [Google Scholar] [CrossRef] [PubMed]
- Haack, F.S.; Poehlein, A.; Kröger, C.; Voigt, C.A.; Piepenbring, M.; Bode, H.B.; Daniel, R.; Schäfer, W.; Streit, W.R. Molecular keys to the Janthinobacterium and Duganella spp. interaction with the plant pathogen Fusarium graminearum. Front. Microbiol. 2016, 7, 1668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolton, M.; Erlacher, A.; Berg, G.; Cytryn, E. The Flavobacterium genus in the plant holobiont: Ecological, physiological, and applicative insights. In Microbial Models: From Environmental to Industrial Sustainability; Springer: Berlin/Heidelberg, Germany, 2016; pp. 189–207. [Google Scholar]
- Berg, G. Plant–microbe interactions promoting plant growth and health: Perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 2009, 84, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C.M.; Soares, H.M.; Soares, E.V. Promising bacterial genera for agricultural practices: An insight on plant growth-promoting properties and microbial safety aspects. Sci. Total Environ. 2019, 682, 779–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greetatorn, T.; Hashimoto, S.; Sarapat, S.; Tittabutr, P.; Boonkerd, N.; Uchiumi, T.; Teaumroong, N. Empowering rice seedling growth by endophytic Bradyrhizobium sp. SUTN 9-2. Lett. Appl. Microbiol. 2019, 68, 258–266. [Google Scholar] [CrossRef]
- Radhapriya, P.; Ramachandran, A.; Palani, P. Indigenous plant growth-promoting bacteria enhance plant growth, biomass, and nutrient uptake in degraded forest plants. 3 Biotech 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Sansinenea, E. Bacillus spp.: As plant growth-promoting bacteria. In Secondary Metabolites of Plant Growth Promoting Rhizomicroorganisms; Springer: Berlin/Heidelberg, Germany, 2019; pp. 225–237. [Google Scholar] [CrossRef]
- Souza, R.d.; Ambrosini, A.; Passaglia, L.M. Plant growth-promoting bacteria as inoculants in agricultural soils. Genet. Mol. Biol. 2015, 38, 401–419. [Google Scholar] [CrossRef]
- Vargas, L.; de Carvalho, T.L.G.; Ferreira, P.C.G.; Baldani, V.L.D.; Baldani, J.I.; Hemerly, A.S. Early responses of rice (Oryza sativa L.) seedlings to inoculation with beneficial diazotrophic bacteria are dependent on plant and bacterial genotypes. Plant Soil 2012, 356, 127–137. [Google Scholar] [CrossRef]
- Song, Y.; Wang, L.; Xiong, L. Comprehensive expression profiling analysis of OsIAA gene family in developmental processes and in response to phytohormone and stress treatments. Planta 2009, 229, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Hong, J.; Kim, S. First report of leaf blight caused by Pantoea agglomerans on rice in Korea. Plant Dis. 2010, 94, 1372. [Google Scholar] [CrossRef] [PubMed]
- Brusamarello-Santos, L.; Pacheco, F.; Aljanabi, S.; Monteiro, R.; Cruz, L.; Baura, V.; Pedrosa, F.; Souza, E.; Wassem, R. Differential gene expression of rice roots inoculated with the diazotroph Herbaspirillum seropedicae. Plant Soil 2012, 356, 113–125. [Google Scholar] [CrossRef]
- Chaudhary, D.; Narula, N.; Sindhu, S.; Behl, R. Plant growth stimulation of wheat (Triticum aestivum L.) by inoculation of salinity tolerant Azotobacter strains. Physiol. Mol. Biol. Plants 2013, 19, 515–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solanki, M.; Garg, F.C. The use of lacZ marker in enumeration of Azotobacter chroococcum in carrier based inoculants. Braz. J. Microbiol. 2014, 45, 595–601. [Google Scholar] [CrossRef] [Green Version]
- Mosquito, S.; Bertani, I.; Licastro, D.; Compant, S.; Myers, M.P.; Hinarejos, E.; Levy, A.; Venturi, V. In planta colonization and role of T6SS in two rice Kosakonia endophytes. Mol. Plant Microbe Interact. 2020, 33, 349–363. [Google Scholar] [CrossRef]
- Andreozzi, A.; Prieto, P.; Mercado-Blanco, J.; Monaco, S.; Zampieri, E.; Romano, S.; Valè, G.; Defez, R.; Bianco, C. Efficient colonization of the endophytes Herbaspirillum huttiense RCA24 and Enterobacter cloacae RCA25 influences the physiological parameters of Oryza sativa L. cv. Baldo rice. Environ. Microbiol. 2019, 21, 3489–3504. [Google Scholar] [CrossRef]
- Ambreetha, S.; Chinnadurai, C.; Marimuthu, P.; Balachandar, D. Plant-associated Bacillus modulates the expression of auxin-responsive genes of rice and modifies the root architecture. Rhizosphere 2018, 5, 57–66. [Google Scholar] [CrossRef]
- Smeltzer, M.S.; Hart, M.E.; Iandolo, J.J. Quantitative spectrophotometric assay for staphylococcal lipase. Appl. Environ. Microbiol. 1992, 58, 2815–2819. [Google Scholar] [CrossRef] [Green Version]
- Huber, B.; Riedel, K.; Hentzer, M.; Heydorn, A.; Gotschlich, A.; Givskov, M.; Molin, S.; Eberl, L. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 2001, 147, 2517–2528. [Google Scholar] [CrossRef] [Green Version]
- Zlosnik, J.E.; Hird, T.J.; Fraenkel, M.C.; Moreira, L.M.; Henry, D.A.; Speert, D.P. Differential mucoid exopolysaccharide production by members of the Burkholderia cepacia complex. J. Clin. Microbiol. 2008, 46, 1470–1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bric, J.M.; Bostock, R.M.; Silverstone, S.E. Rapid in situ assay for indoleacetic acid production by bacteria immobilized on a nitrocellulose membrane. Appl. Environ. Microbiol. 1991, 57, 535–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penrose, D.M.; Glick, B.R. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plant. 2003, 118, 10–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Steindler, L.; Venturi, V. Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. FEMS Microbiol. Lett. 2007, 266, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Köhler, T.; Curty, L.K.; Barja, F.; Van Delden, C.; Pechère, J.-C. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 2000, 182, 5990–5996. [Google Scholar] [CrossRef] [Green Version]
- Better, M.; Lewis, B.; Corbin, D.; Ditta, G.; Helinski, D.R. Structural relationships among Rhizobium meliloti symbiotic promoters. Cell 1983, 35, 479–485. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Lemoine, F.; Correia, D.; Lefort, V.; Doppelt-Azeroual, O.; Mareuil, F.; Cohen-Boulakia, S.; Gascuel, O. NGPhylogeny. fr: New generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019, 47, W260–W265. [Google Scholar] [CrossRef] [Green Version]
- Tremblay, N.; Wang, Z.; Cerovic, Z.G. Sensing crop nitrogen status with fluorescence indicators. A review. Agron. Sustain. Dev. 2012, 32, 451–464. [Google Scholar] [CrossRef] [Green Version]
- Goulas, Y.; Cerovic, Z.G.; Cartelat, A.; Moya, I. Dualex: A new instrument for field measurements of epidermal ultraviolet absorbance by chlorophyll fluorescence. Appl. Opt. 2004, 43, 4488–4496. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’hara, R.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Package ‘vegan’. Community Ecol. Package Version 2013, 2, 1–295. [Google Scholar]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language And Environment For Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- Anderson, J.A.; Staley, J.; Challender, M.; Heuton, J. Safety of Pseudomonas chlororaphis as a gene source for genetically modified crops. Transgenic Res. 2018, 27, 103–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosas, S.B. Pseudomonas chlororaphis subsp. aurantiaca SR1: Isolated from rhizosphere and its return as inoculant. A review. Int. Biol. Rev. 2017, 1. [Google Scholar] [CrossRef]
- Peng, H.; Zhang, P.; Bilal, M.; Wang, W.; Hu, H.; Zhang, X. Enhanced biosynthesis of phenazine-1-carboxamide by engineered Pseudomonas chlororaphis HT66. Microb. Cell Factories 2018, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]






| System/Compound | Target/Beneficial Effect | References | ST9 Genes |
|---|---|---|---|
| Antagonism | |||
| Phenazine 1-carboxylic acid (PCA) | Antifungal redox-active antibiotic | [26] | 7 genes cluster From fig|286.2086.peg.5005 to fig|286.2086.peg.5011 phzABCDEFG |
| 2-hydroxy phenazine | Fungistatic and bacteriostatic | [35] | fig|286.2086.peg.5012; phzO |
| Tailocine R | Antibacterial Persistence within the rhizosphere microbiome | [36] | 40 genes cluster From fig|286.2086.peg.1042 to fig|286.2086.peg.1082 |
| Pirrolnitrin | Antifungal compound | [37,38] | 4 genes cluster From fig|286.2086.peg.3071 to fig|286.2086.peg.3074; prnA, prnB, prnC, prnD |
| 2-hexyl, 5-propyl resorcinol (HRP) | Antifungal compound | [21,39] | 5 genes cluster From fig|286.2086.peg.4110 to fig|286.2086.peg.4114; darA, darB, darC, darR, darS |
| Hydrogen cyanide | Metalloenzymes inhibitor and antifungal | [37] | 2 genes cluster fig|286.2086.peg.2368 and fig|286.2086.peg.2369 |
| Signal Interferences | |||
| Acyl-homoserine lactone acylase | Quorum quenching | fig|286.2086.peg.2769 | |
| N-acyl-L-homoserine lactone synthetase | Quorum sensing | [40] | 3 systems fig|286.2086.peg.5003 and fig|286.2086.peg.5004; phzI, phzR fig|286.2086.peg.4930 and fig|286.2086.peg.4931; aurI, aurR fig|286.2086.peg.2434 and fig|286.2086.peg.2435; luxI, luxR |
| Niche Colonization | |||
| poly-beta-1,6-N-acetyl-D-glucosamine | Biofilm adhesin | [41] | 4 genes cluster From fig|286.2086.peg.20 to fig|286.2086.peg.24 pgaA, pgaB, pgaC, pgaD |
| Motility and chemotaxis | Flagella | [41] | 6 genes cluster From fig|286.2086.peg.1427 to fig|286.2086.peg.1433 flgF, flgG, flgH, flgI, flgK, flgL 36 gene cluster From fig|286.2086.peg.1451 to fig|286.2086.peg.1486 9 gene cluster From fig|286.2086.peg.4398 to fig|286.2086.peg.4406 |
| Plant Protection | |||
| Chitinase | insecticidal | [42] | 5 genes fig|286.2086.peg.1868 and fig|286.2086.peg.1869 fig|286.2086.peg.3224 and fig|286.2086.peg.3225 fig|286.2086.peg.1868 4987 chitin binding protein and chitinases |
| Proteases | insecticidal | [42] | fig|286.2086.peg.3194 aprX/prtA and other related genes |
| Lipases | insecticidal | [42] | fig|286.2086.peg.3118 phospholipase C and others related genes |
| rebB | insecticidal | [42] | 2 genes fig|286.2086.peg.35 and fig|286.2086.peg.36 rebB-like protein (refractile inclusion bodies) |
| Beneficial Activities to Plant | |||
| Phosphate solubilization | P solubilization enzyme | fig|286.2086.peg.702; alkD | |
| Achromobactine | Fe chelation and competition | 13 genes cluster From fig|286.2086.peg.3101 to fig|286.2086.peg.3113 Achromobactin synthesis, receptor, transport | |
| In Vitro PGP Activity | In Vitro Antagonistic Activity | In Vivo Activity | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protease | Lipase | IAA Production | P Solubilization | ACC Deaminase | EPS Production | Motility | Dickeya zeae | Pseudomonas fuscovaginae | Aspergillus nidulans | Fusarium graminearum | Magnaporthe oryzae | Emergence 4 dpi | Coleoptile Length 10 dpi | Dry Weight Biomass 12 dpi | Bacterial Foot Rot Disease Control | |
| P. chlororaphis ST9 | + | + | - | + | - | - | + | + | + | + | + | + | - | + * | - | + ** |
| Gene Name | Putative Function | Reference |
|---|---|---|
| OsERS1 | Ethylene response sensor 1 | [53] |
| OsERS2 | Ethylene response sensor 2 | [53] |
| OsETR2 | Ethylene responsive 2 | [53] |
| OsETR3 | Ethylene responsive 3 | [53] |
| OsIAA1 | Auxin-responsive protein IAA1-like | [54] |
| OsIAA4 | Auxin-responsive protein IAA4 | [54] |
| OsIAA11 | Auxin-responsive protein IAA11 | [54] |
| OsIAA13 | Auxin-responsive protein IAA13-like | [54] |
| OsIAA14 | Auxin-responsive protein IAA14-like | [54] |
| OsACT1 | Actin 1 | [55] |
| OsARF2 | Similar to auxin response factor 2 | [56] |
| OsERF2 | Similar to ethylene response factor 2 | [56] |
| OsERF3 | Similar to ethylene response binding factor 3 | [56] |
| OsISAP1 | Multiple stress-responsive zinc-finger protein | [56] |
| Osmetallo-tionein | Metallothionein-like protein type 1 | [56] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bertani, I.; Zampieri, E.; Bez, C.; Volante, A.; Venturi, V.; Monaco, S. Isolation and Characterization of Pseudomonas chlororaphis Strain ST9; Rhizomicrobiota and in Planta Studies. Plants 2021, 10, 1466. https://doi.org/10.3390/plants10071466
Bertani I, Zampieri E, Bez C, Volante A, Venturi V, Monaco S. Isolation and Characterization of Pseudomonas chlororaphis Strain ST9; Rhizomicrobiota and in Planta Studies. Plants. 2021; 10(7):1466. https://doi.org/10.3390/plants10071466
Chicago/Turabian StyleBertani, Iris, Elisa Zampieri, Cristina Bez, Andrea Volante, Vittorio Venturi, and Stefano Monaco. 2021. "Isolation and Characterization of Pseudomonas chlororaphis Strain ST9; Rhizomicrobiota and in Planta Studies" Plants 10, no. 7: 1466. https://doi.org/10.3390/plants10071466
APA StyleBertani, I., Zampieri, E., Bez, C., Volante, A., Venturi, V., & Monaco, S. (2021). Isolation and Characterization of Pseudomonas chlororaphis Strain ST9; Rhizomicrobiota and in Planta Studies. Plants, 10(7), 1466. https://doi.org/10.3390/plants10071466







