1. Introduction
Wheat is one of the world’s main crops and food sources for human consumption [
1]. Wheat accounts for the largest planting area, and the food security of the population of most countries of the world depends on its yield. One of the main factors affecting wheat yield is fungi diseases: rust, septoria of leaves and ears, and powdery mildew [
2] (
Figure 1).
Leaf rust, powdery mildew, and septoria (pathogens
Puccinia triticina Erikss.,
Blumeria graminis (DC.) Speer,
Zymoseptoria tritici Rob., and
Parastaganospora nodorum Berk.) are cosmopolitan pathogens. They are observed in the phytopathogenic complex on grain crops widely. However, the total damage caused by the causative agents of the above-listed diseases does not exceed 20% and is controlled by the timely use of fungicides [
3]. The consequences of the epiphytoties of yellow and stem rust, leading to grain shortages of more than 30%, are of serious economic importance. In general, grain yield losses from these diseases, depending on the region and season conditions, can vary from 15 to 30% or more [
4,
5]. The most effective way to combat these diseases is their prevention and timely implementation of protective actions [
4,
6,
7]. However, such a strategy is impossible without a timely and correct diagnosis of pathogens. In this case, it is important to identify diseases at the stage of seedlings, since at later stages of plant development, resistance to the pathogen is higher [
8]. In turn, the effectiveness of such diagnostics largely depends on how accurate, labor-intensive, and resource-intensive they are.
Visual assessment of the diseases has been the main diagnostic method throughout the history of wheat cultivation. It allows to identify plaque, pustules, spots, or necrosis [
9,
10]. It requires the training of specialists in phytopathology and a lot of routine work, in addition to the observations (keeping a record book, statistical processing of observations, etc.). In recent decades, molecular, spectral methods, and methods based on the analysis of digital images have appeared and have become widespread [
11,
12]. These approaches differ in the labor intensity, as well as in its cost and accuracy. The methods, using the analysis of digital RGB images, are based on determining changes in the color, texture, and shape of plant organs that arise as a result of changes in their pigment composition under the influence of the vital activity of pathogens [
13,
14,
15,
16,
17]. The advantages of such methods are the low cost of monitoring equipment (it is sufficient to use a digital camera or a mobile phone) and the high monitoring speed. The disadvantages include low sensitivity (in comparison, for example, with spectral methods; see References [
12,
18]).
Recently the technologies for plant-disease monitoring based on digital RGB images have received a powerful impulse due to the improvement of machine learning methods based on the use of neural network algorithms. A feature of deep learning neural networks in comparison with other methods is the multilayer architecture of neurons, in which the next layer uses the output of the previous layer as input data to derive ideas regarding the analyzed objects [
19,
20]. For example, for such an important task as image labeling, some of the most successful methods are convolutional neural networks (CNNs), for which several architecture options are used. Among the first types of CNN architecture were AlexNet [
21] and VGG [
22]. Further development of these approaches made it possible to improve the convergence of the proposed algorithms (ResNet network) [
23], reduce the number of parameters due to deep convolution (MobileNet network) [
24], and improve model training results due to adaptive recalibration of responses across channels (SENet network) [
25]. These advances have expanded the applicability of deep learning neural networks. Moreover, they have also demonstrated exceptional success on complex problems of plant phenotyping [
19,
26,
27,
28,
29,
30].
The deep learning neural networks were implemented efficiently in disease detection for various plant species [
31,
32,
33,
34,
35,
36,
37]. It was demonstrated that deep learning approaches outperform machine learning algorithms, such as support vector machine, random forest, stochastic gradient descent [
38]. The development of the deep learning methods towards better disease recognition in plants included transfer learning approaches [
39], implementing networks of various architectures [
37,
40,
41,
42,
43,
44], working with the limited amount of data [
45], and Bayesian deep learning [
46].
A number of deep learning methods, which have been developed to identify wheat diseases by using digital RGB images, have proved to be effective. In the work by Barbedo [
47], CNN of the GoogLeNet architecture was used to detect lesions in the leaf image, and, on this basis, wheat diseases, such as blast, leaf rust, tan spot, and powdery mildew were identified. In the work by Picon et al. [
48], a method was developed for identifying four types of wheat diseases, taking into account their stage of development (septoria, tan spot, and two types of rust) based on deep CNNs. Lu et al. [
49] have developed an automatic system for diagnosing six types of wheat diseases that recognizes the type of disease and localizes the lesions in the image, obtained in the field conditions.
The increase in the use of deep learning architectures provide significant progress in the diagnosis of wheat diseases based on the digital images. However, there are still gaps to be investigated regarding the use of especially new deep learning architectures in wheat leaf fungal disease detection.
First of all, to build a successful algorithm for disease recognition, a set of a large number of labeled images is required [
50]. The availability of such data in the public domain is the basis for improving modern methods of image recognition for plant phenotyping [
51,
52] and identification of pathogens [
53].
Second, progress in disease recognition depends largely on the choice of neural network architecture [
37]. New and modified deep learning architectures are constantly being introduced to better/transparent plant disease detection [
37]. Improved versions of state-of-the-art models tend to provide high accuracy in disease detection [
31,
54]. Recently, the EfficientNet network architecture was proposed in Reference [
55], and it has shown high efficiency in image labeling. It uses a new activation function called Swish instead of the Rectifier Linear Unit (ReLU) activation function implemented in other CNN models. EfficientNet is a family of CNNs of similar architecture (B0 to B7) which differ from each other in the depth of the layers, their width, and the size of the input image, while maintaining the ratios between these sets of parameters. Thus, as the model number grows, the number of calculated parameters does not increase much. On the ImageNet task, the EfficientNet-B7 model with 66 M parameters achieved an accuracy of 84.3% [
55]. This architecture was used for the fruit recognition and demonstrated higher performance compared to the ConvNet architecture [
29]. Atila et al. demonstrated that EfficientNet architecture outperforms AlexNet, ResNet, VGG16, and Inception V3 networks in the plant-disease recognition task on the PlantVillage dataset [
42]. Zhang et al. [
41] implemented EfficientNet for greenhouse cucumber-disease recognition. They demonstrated that the EfficientNet-B4 model outperforms AlexNet, VGG16, VGG19, Inception V4, ResNet50, ResNet101, SqueeseNet, and DenseNet networks.
Another difficulty is the simultaneous presence of symptoms caused by different diseases [
56]. On the one hand, the problem of multiple classifications is difficult to solve, since it requires a larger volume of images for correct classification, in which various combinations of lesions will be presented in sufficient numbers. On the other hand, the visual manifestations of each of the diseases can be very similar (see
Figure 1), which makes it difficult to make a correct classification.
Finally, one of the important conditions for creating a recognition method is the possibility of using it in field conditions [
57]. This increases the efficiency of disease monitoring and, consequently, the chances of successful plant treatment by fungicides. One of the approaches is using mobile devices for this, both for semi-automatic determination of the degree of plant damage [
58] and fully automatic analysis, including computer vision methods [
59,
60] and deep learning networks [
48,
49,
61,
62].
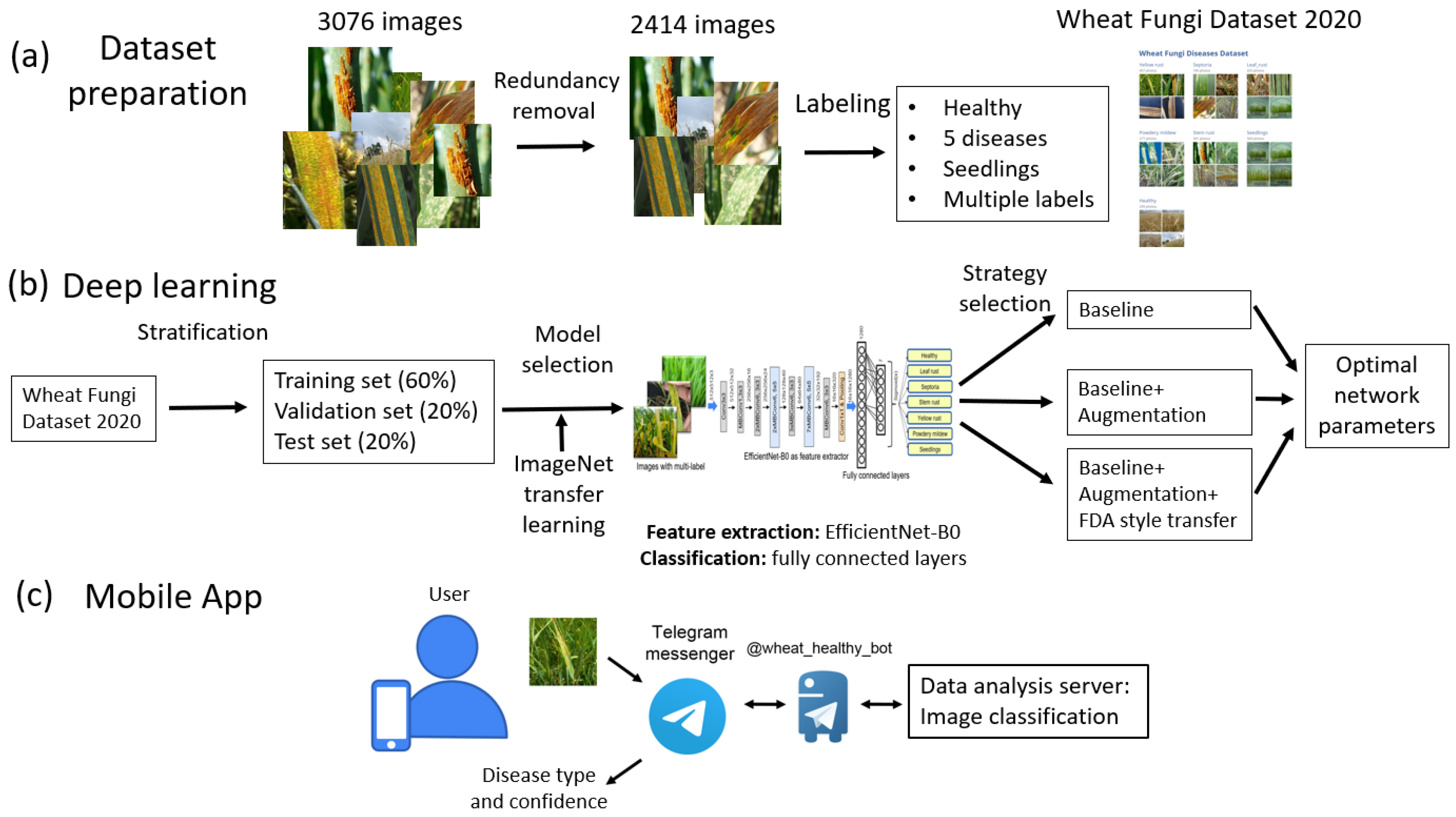
Here we propose a method for the recognition of five fungi diseases of wheat shoots (leaf rust, stem rust, yellow rust, powdery mildew, and septoria), both separately and in combination, with the possibility of identifying the stage of plant development. Our paper makes the following contributions:
The Wheat Fungi Diseases (WFD2020) dataset of 2414 wheat images for which expert labeling was performed by the type of disease. In the process of creating this set, a data-redundancy reduction procedure was applied based on the image hashing algorithm.
The disease-recognition algorithm based on the use of a network with the EfficientNet-B0 architecture with transfer learning from ImageNet dataset and augmentation, including style transfer.
The implementation of the recognition method as a chatbot on the Telegram platform, which allows for the assessment of plants’ symptoms in the field conditions.
4. Discussion
In this paper, a method was proposed for identifying wheat diseases based on digital images obtained in the field conditions and the algorithm based on the EfficientNet-B0 architecture network.
It should be noted that a sample of well-annotated images for training and testing is one of the important conditions for creating a successful method of disease recognition [
50]. Unfortunately, the existing open databases of images of plant diseases contain few images of wheat. For example, there are nine of them in the PDDB database [
34], but they are absent in the well-known PlantVillage dataset [
53]. For the analysis of wheat diseases, in the work by Picon et al. [
48], the sample included 8178 images (according to the imaging protocol, these data contained images of individual leaves, but not the whole plant). In the work by Lu et al. [
49], a sample of Wheat Disease Database 2017 (WDD2017) was collected from 9230 images of not only leaves but also plants in field conditions. Arsenovich et al. [
54] used a dataset that included information on 5596 images of wheat (the authors do not provide details). These three abovementioned sets, however, are not freely available. Therefore, in our work, based on several open sources and our own data, a set of wheat images was formed both in the field and in the laboratory conditions. It is important to note that, in our sample, images from external sources were relabeled by phytopathologists for six types of diseases, both separately and jointly. The WFD2020 data are freely available for downloading.
One of the features of the sample is a special class of images for seedlings. Special attention was paid to this, since the timely diagnosis of the harmful complex in the early periods of the vegetational season is of particular importance. Under favorable weather conditions, diseases can develop rapidly and lead to significant losses of yield. Therefore, to obtain large yields, it is important to monitor the lesion of flag and pre-flag leaves and diagnose the type of pathogen of the disease, which makes it possible to select effective means of protection [
83]. As a result of the research, it appeared that the accuracy of disease prediction is higher for seedlings. However, this may also be related to the dataset peculiarities: the variety of shooting conditions for seedlings is less than when shooting adult plants.
The accuracy of determining diseases by using the developed method for the most optimal learning strategy (EfficientNet-B0_FDA) was 0.942 which is comparable to the results of other authors. For example, Picon et al. [
48] used a network architecture based on ResNet50 to identify lesions of septoria, tan spot, or rust (either leaf or yellow). The accuracy of disease identification was 97%. Lu et al. [
49] tried several options of neural networks for deep learning and multiple instance learning to identify wheat lesions by the six most common diseases. The performance of our approach is comparable to the accuracy of methods for assessing plant diseases not only for wheat but also for other crops. The recognition accuracy ranged from 95 to 97% for plant-disease symptoms in Reference [
54]; on the basis of the PlantVillage dataset and the authors’ data, a two-stage network architecture was developed, which showed an average accuracy of disease recognition of 0.936. In Reference [
60], the problem of simultaneous identification of 14 crop species and 26 diseases was also solved on the basis of the PlantVillage dataset. The network of the ResNet50 architecture was used, which achieved an accuracy of 99.24%.
It is interesting to compare our results with the accuracy obtained for plant-disease recognition from using similar architecture, EfficientNet. Atila et al. [
42] evaluated the performance of all eight topologies of this family on the original PlantVillage dataset and demonstrated that the highest average accuracy in disease recognition was achieved for the B5 architecture (99.91%). However, all models of the family obtained average accuracy very close to each other.
Zhang et al.’s work [
41] is very similar to ours. They predicted the disease symptoms for a single crop, cucumber, on the images taken in a naturally complex greenhouse background. They detected three types of labels (powdery mildew, downy mildew, and healthy leaves) and combination of powdery mildew and downy mildew. Their dataset included 2816 images, which is close to our dataset size. They used different optimization methods and network architectures and obtained the best accuracy for the EfficientNet-B4 network and Ranger optimizer, 96.39% on the test sample. Interestingly, the EfficientNet-B0 architecture demonstrated accuracy of 94.39% on this dataset [
41], which is very close to our results (94.2%).
It should be noted that the difference in the accuracy of our method between the validation and test datasets is small (0.1%), which indicates the overfitting absence for our model. This is not surprising, because we used the EfficientNet architecture with the smallest number of parameters (B0) and the strategy of the dataset stratification.
Our analysis of the matrix of errors showed that their main sources were the incorrect classifications of diseases, for example, rust ones, among themselves. Interestingly, the cross-misclassification between rust diseases and the other two (septoria and powdery mildew) was found to be higher for septoria. This is explainable since the visual symptoms of these diseases can be very similar. In addition, for some images, the prediction of the pathogen type was not reliable enough to perform classification (“no prediction” result).
The possibility of using the technology of identification of crops by pathogens in the field is one of the necessary properties of the system for operational monitoring of diseases. In this direction, methods are being actively developed that integrate the results of prediction by the method of deep learning and the implementation of access to them via smartphones and tablets [
48,
49,
61,
62]. This, however, requires additional effort to build and maintain mobile applications. Here, we took advantage of the possibility of a simple interface through the use of a Telegram messenger bot. Recently, this type of application has become popular and is used for crowdsourcing problems in social research [
84], for predicting real-estate prices [
85], for determining the type of trees [
86], etc. Its advantages are that there is no need to develop and maintain a smartphone graphical interface, and, at the same time, that it allows us to send an image as a request to a server for further processing and displaying the results. At the same time, access to the service is possible wherever there is access to the Internet both from a mobile device and from a desktop PC.
5. Conclusions
The paper proposed a method for recognizing plant diseases on images obtained in field conditions based on the deep machine learning. Fungal diseases of wheat as leaf rust, stem rust, yellow rust, powdery mildew, septoria, and their combinations were recognized. Additionally, the network determines whether the plant is a seedling. The image dataset represents a specially formed and labeled sample of 2414 images, which is freely available.
The algorithm is based on the EfficientNet-B0 neural network architecture. We implemented several techniques to achieve better performance of the network: transfer learning; dataset stratification into training, validation and testing samples; and augmentation, including image style transfer.
Of the three examined learning strategies, the best accuracy was provided by the method using augmentation and transfer of image styles (the accuracy was 0.942). These results are comparable with the performance obtained by other deep learning methods for plant-disease recognition. Our network performance is in good agreement with the results obtained by implementation of the EfficientNet architectures for plant-disease recognition.
The interface to the recognition method was implemented on the basis of the Telegram chatbot, which provides users with convenient access to the program via the Internet, both from a mobile device and using a desktop PC. This allows users to utilize our recognition method for wheat plants in the field conditions by using the images obtained via smartphone camera under real-world circumstances.