Dissecting the Genetic Basis of Flowering Time and Height Related-Traits Using Two Doubled Haploid Populations in Maize
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genetic Materials
2.2. Phenotypic Evaluation and Statistical Analysis
2.3. Linkage Map and QTL Analysis
3. Results
3.1. Phenotypic Variation
3.2. Identification of QTLs for Six Traits
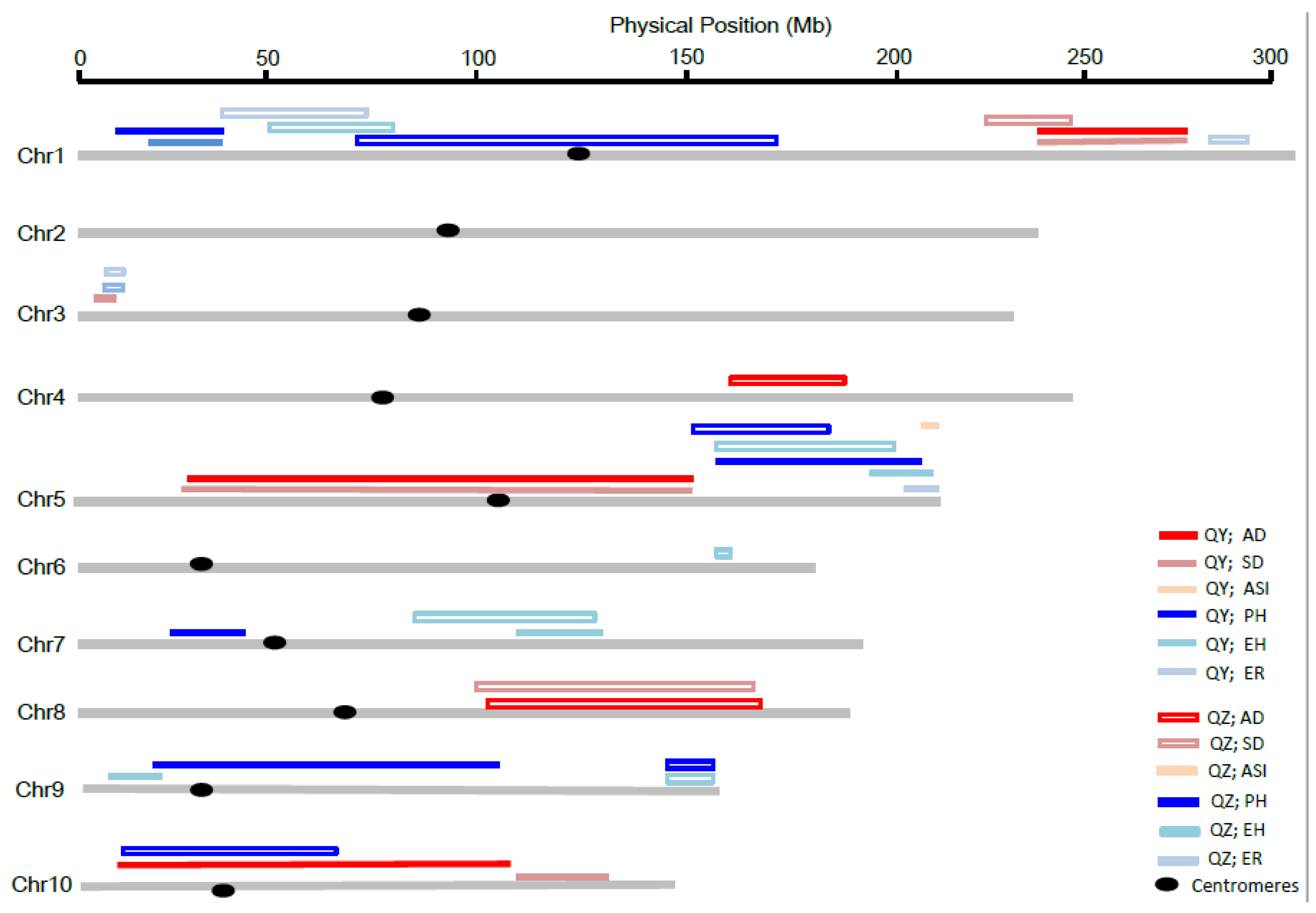
3.3. Co-Localization of QTLs for Different Traits between the Two DH Populations
4. Discussion
4.1. Relationship between Flowering Time and Height-Related Traits
4.2. Potential Utilization of the Present QTLs
4.3. Comparison with QTLs Identified in Previous Studies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Irish, E.; Nelson, T.M. Identification of multiple stages in the conversion of maize meristems from vegetative to floral development. Development 1991, 112, 891–898. [Google Scholar] [CrossRef]
- Hallauer, A.R.; Carena, M.J. Adaptation of tropical maize germplasm to temperate environments. Euphytica 2014, 196, 1–11. [Google Scholar] [CrossRef]
- Bendix, C.; Marshall, C.M.; Harmon, F.G. Circadian Clock Genes Universally Control Key Agricultural Traits. Mol. Plant 2015, 8, 1135–1152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buckler, E.S.; Holland, J.B.; Bradbury, P.J.; Acharya, C.B.; Brown, P.J.; Browne, C.; Ersoz, E.; Flint-Garcia, S.; Garcia, A.; Glaubitz, J.C.; et al. The Genetic Architecture of Maize Flowering Time. Science 2009, 325, 714–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ducrocq, S.; Giauffret, C.; Madur, D.; Combes, V.; Dumas, F.; Jouanne, S.; Coubriche, D.; Jamin, P.; Moreau, L.; Charcosset, A. Fine Mapping and Haplotype Structure Analysis of a Major Flowering Time Quantitative Trait Locus on Maize Chromosome 10. Genetics 2009, 183, 1555–1563. [Google Scholar] [CrossRef] [Green Version]
- Vega, S.H.; Sauer, M.; Orkwiszewski, J.A.; Poethig, R.S. The early phase change Gene in Maize. Plant Cell 2002, 14, 133–147. [Google Scholar] [CrossRef] [Green Version]
- Camus-Kulandaivelu, L.; Veyrieras, J.B.; Madur, D.; Combes, V.; Fourmann, M.; Barraud, S.; Dubreuil, P.; Gouesnard, B.; Manicacci, D.; Charcosset, A. Maize adaptation to temperate climate: Relationship between population structure and polymorphism in the Dwarf8 gene. Genetics 2006, 172, 2449–2463. [Google Scholar] [CrossRef] [Green Version]
- Bomblies, K.; Doebley, J.F. Pleiotropic Effects of the Duplicate Maize FLORICAULA/LEAFY Genes zfl1 and zfl2 on Traits Under Selection During Maize Domestication. Genetics 2006, 172, 519–531. [Google Scholar] [CrossRef] [Green Version]
- Miller, T.A.; Muslin, E.H.; Dorweiler, J.E. A maize CONSTANS-like gene, conz1, exhibits distinct diurnal expression patterns in varied photoperiods. Planta 2008, 227, 1377–1388. [Google Scholar] [CrossRef]
- Coles, N.D.; McMullen, M.D.; Balint-Kurti, P.J.; Pratt, R.C.; Holland, J.B. Genetic Control of Photoperiod Sensitivity in Maize Revealed by Joint Multiple Population Analysis. Genetics 2010, 184, 799–812. [Google Scholar] [CrossRef] [Green Version]
- Meng, X.; Muszynski, M.G.; Danilevskaya, O.N. The FT-Like ZCN8 Gene Functions as a Floral Activator and Is Involved in Photoperiod Sensitivity in Maize. Plant Cell 2011, 23, 942–960. [Google Scholar] [CrossRef] [Green Version]
- Durand, E.; Bouchet, S.; Bertin, P.; Ressayre, A.; Jamin, P.; Charcosset, A.; Dillmann, C.; Tenaillon, M.I. Flowering Time in Maize: Linkage and Epistasis at a Major Effect Locus. Genetics 2012, 190, 1547–1562. [Google Scholar] [CrossRef] [Green Version]
- Romay, M.C.; Millard, M.J.; Glaubitz, J.C.; Peiffer, J.A.; Swarts, K.L.; Casstevens, T.M.; Elshire, R.J.; Acharya, C.B.; Mitchell, S.E.; Flint-Garcia, S.A.; et al. Comprehensive genotyping of the USA national maize inbred seed bank. Genome Biol. 2013, 14, R55. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Li, C.; Bradbury, P.J.; Liu, X.; Lu, F.; Romay, C.M.; Glaubitz, J.C.; Wu, X.; Peng, B.; Shi, Y.; et al. Identification of genetic variants associated with maize flowering time using an extremely large multi-genetic background population. Plant J. 2016, 86, 391–402. [Google Scholar] [CrossRef]
- Hung, H.-Y.; Shannon, L.M.; Tian, F.; Bradbury, P.J.; Chen, C.; Flint-Garcia, S.A.; McMullen, M.D.; Ware, D.; Buckler, E.S.; Doebley, J.F.; et al. ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc. Natl. Acad. Sci. USA 2012, 109, E1913–E1921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Li, Z.; Li, W.; Ku, L.; Wang, C.; Ye, J.; Li, K.; Yang, N.; Li, Y.; Zhong, T.; et al. CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc. Natl. Acad. Sci. USA 2013, 110, 16969–16974. [Google Scholar] [CrossRef] [Green Version]
- Jin, M.; Liu, X.; Jia, W.; Liu, H.; Li, W.; Peng, Y.; Du, Y.; Wang, Y.; Yin, Y.; Zhang, X.; et al. ZmCOL3, a CCT gene represses flowering in maize by interfering with the circadian clock and activating expression of ZmCCT. J. Integr. Plant Biol. 2018, 60, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Colasanti, J.; Yuan, Z.; Sundaresan, V. The indeterminate Gene Encodes a Zinc Finger Protein and Regulates a Leaf-Generated Signal Required for the Transition to Flowering in Maize. Cell 1998, 93, 593–603. [Google Scholar] [CrossRef] [Green Version]
- Muszynski, M.G.; Dam, T.; Li, B.; Shirbroun, D.M.; Hou, Z.; Bruggemann, E.; Archibald, R.; Ananiev, E.V.; Danilevskaya, O.N. Delayed flowering1 Encodes a Basic Leucine Zipper Protein That Mediates Floral Inductive Signals at the Shoot Apex in Maize. Plant Physiol. 2006, 142, 1523–1536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, S.; Luo, Y.; Zhang, Z.; Xu, M.; Wang, W.; Zhao, Y.; Zhang, L.; Fan, Y.; Wang, L. ZmSOC1, a MADS-Box Transcription Factor from Zea mays, Promotes Flowering in Arabidopsis. Int. J. Mol. Sci. 2014, 15, 19987–20003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvi, S.; Sponza, G.; Morgante, M.; Tomes, D.; Niu, X.; Fengler, K.A.; Meeley, R.; Ananiev, E.V.; Svitashev, S.; Bruggemann, E.; et al. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc. Natl. Acad. Sci. USA 2007, 104, 11376–11381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alter, P.; Bircheneder, S.; Zhou, L.Z.; Schlüter, U.; Gahrtz, M.; Sonnewald, U.; Dresselhaus, T. Flowering Time-Regulated Genes in Maize Include the Transcription Factor ZmMADS1. Plant Physiol. 2016, 172, 389–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berke, T.G.; Rocheford, T.R. Quantitative Trait Loci for Flowering, Plant and Ear Height, and Kernel Traits in Maize. Crop Sci. 1995, 35, 1542–1549. [Google Scholar] [CrossRef]
- Peng, J.; Richards, D.E.; Hartley, N.M.; Murphy, G.P.; Devos, K.M.; Flintham, J.E.; Beales, J.; Fish, L.J.; Worland, A.J.; Pelica, F.; et al. Green revolution’ genes encode mutant gibberellin response modulators. Nature 1999, 400, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Y.; Wang, Y.; Liu, Z.; Liu, C.; Peng, B.; Tan, W.; Wang, D.; Shi, Y.; Sun, B.; et al. Stability of QTL across environments and QTL-by-environment interactions for plant and ear height in maize. Agric. Sci. China 2010, 9, 1400–1412. [Google Scholar] [CrossRef]
- Cai, H.; Chu, Q.; Gu, R.; Yuan, L.; Liu, J.; Zhang, X.; Chen, F.; Mi, G.; Zhang, F. Identification of QTLs for plant height, ear height and grain yield in maize (Zea mays L.) in response to nitrogen and phosphorus supply. Plant Breed. 2012, 131, 502–510. [Google Scholar] [CrossRef]
- Vanous, A.; Gardner, C.; Blanco, M.; Martin-Schwarze, A.; Lipka, A.E.; Flint-Garcia, S.; Bohn, M.; Edwards, J.; Lubberstedt, T. Association Mapping of Flowering and Height Traits in Germplasm Enhancement of Maize Doubled Haploid (GEM-DH) Lines. Plant Genome 2018, 11, 170083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Guo, X.; Wang, A.; Liu, P.; Wu, W.; Zhao, Q.; Zhao, M.; Zhu, Y.; Chen, Z. Quantitative trait loci mapping of plant architecture-related traits using the high-throughput genotyping by sequencing method. Euphytica 2019, 215, 212. [Google Scholar] [CrossRef]
- Peiffer, J.A.; Romay, M.C.; Gore, M.A.; Flint-Garcia, S.A.; Zhang, Z.; Millard, M.J.; Gardner, C.A.; McMullen, M.D.; Holland, J.B.; Bradbury, P.J.; et al. The Genetic Architecture Of Maize Height. Genetics 2014, 196, 1337–1356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Zhou, Z.; Ding, J.; Wu, Y.; Zhou, B.; Wang, R.; Ma, J.; Wang, S.; Zhang, X.; Xia, Z.; et al. Combined Linkage and Association Mapping Reveals QTL and Candidate Genes for Plant and Ear Height in Maize. Front. Plant Sci. 2016, 15, 833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Zhang, C.; Zhou, Y.; Hao, Z.; Wang, Z.; Zeng, X.; Di, H.; Li, M.; Zhang, D.; Yong, H.; et al. Genetic dissection of maize plant architecture with an ultra-high density bin map based on recombinant inbred lines. BMC Genom. 2016, 3, 178. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Wang, B.; Dong, X.; Liu, H.; Ren, L.; Chen, J.; Hauck, A.; Song, W.; Lai, J. An ultra-high density bin-map for rapid QTL mapping for tassel and ear architecture in a large F₂ maize population. BMC Genom. 2014, 15, 433. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Zhang, C.; Lu, X.; Wang, L.; Hao, Z.; Li, M.; Zhang, D.; Yong, H.; Zhu, H.; Weng, J.; et al. Dissecting the Genetic Basis Underlying Combining Ability of Plant Height Related Traits in Maize. Front. Plant Sci. 2018, 9, 1117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winkler, R.G.; Helentjaris, T. The maize Dwarf3 gene encodes a cytochrome P450-mediated early step in Gibberellin biosynthesis. Plant Cell 1995, 7, 1307–1317. [Google Scholar] [PubMed] [Green Version]
- Teng, F.; Zhai, L.; Liu, R.; Bai, W.; Wang, L.; Huo, D.; Tao, Y.; Zheng, Y.; Zhang, Z. ZmGA3ox2, a candidate gene for a major QTL, qPH3.1, for plant height in maize. Plant J. 2013, 73, 405–416. [Google Scholar] [CrossRef]
- Hartwig, T.; Chuck, G.S.; Fujioka, S.; Klempien, A.; Weizbauer, R.; Potluri, D.P.; Choe, S.; Johal, G.S.; Schulz, B. Brassinosteroid control of sex determination in maize. Proc. Natl. Acad. Sci. USA 2011, 108, 19814–19819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Multani, D.S.; Briggs, S.P.; Chamberlin, M.A.; Blakeslee, J.J.; Murphy, A.S.; Johal, G.S. Loss of an MDR Transporter in Compact Stalks of Maize br2 and Sorghum dw3 Mutants. Science 2003, 302, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Lawit, S.J.; Wych, H.M.; Xu, D.; Kundu, S.; Tomes, D.T. Maize DELLA Proteins dwarf plant8 and dwarf plant9 as Modulators of Plant Development. Plant Cell Physiol. 2010, 51, 1854–1868. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Ge, F.; Qiang, Z.; Zhu, L.; Zhang, S.; Chen, L.; Wang, X.; Li, J.; Fu, Y. Maize ZmRPH1 encodes a microtubule-associated protein that controls plant and ear height. Plant Biotechnol. J. 2020, 18, 1345–1347. [Google Scholar] [CrossRef] [Green Version]
- Du, L.; Yu, F.; Zhang, H.; Wang, B.; Ma, K.; Yu, C.; Xin, W.; Huang, X.; Liu, Y.; Liu, K. Genetic mapping of quantitative trait loci and a major locus for resistance to grey leaf spot in maize. Theor. Appl. Genet. 2020, 133, 2521–2533. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014; Available online: http://www.R-project.org/ (accessed on 10 July 2014).
- Knapp, S.J.; Stroup, W.W.; Ross, W.M. Exact Confidence Intervals for Heritability on a Progeny Mean Basis. Crop Sci. 1985, 25, 192–194. [Google Scholar] [CrossRef]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67. [Google Scholar] [CrossRef]
- Wang, S.; Basten, C.; Zeng, Z. Windows QTL Cartographer 2.5; Department of Statistics, North Carolina State University: Raleigh, NC, USA, 2012. [Google Scholar]
- McCouch, S.; Cho, Y.; Yano, M.; Paul, E.; Blinstrub, M.; Morishima, H.; Kinoshita, T. Report on QTL nomenclature. Rice Genet. Newsl. 1997, 14, 11–13. [Google Scholar]
- Wei, X.; Xu, J.; Guo, H.; Jiang, L.; Chen, S.; Yu, C.; Zhou, Z.; Hu, P.; Zhai, H.; Wan, J. DTH8 Suppresses Flowering in Rice, Influencing Plant Height and Yield Potential Simultaneously. Plant Physiol. 2010, 153, 1747–1758. [Google Scholar] [CrossRef] [Green Version]
- Weng, X.; Wang, L.; Wang, J.; Hu, Y.; Du, H.; Xu, C.; Xing, Y.; Li, X.; Xiao, J.; Zhang, Q. Grain Number, Plant Height, and Heading Date7 Is a Central Regulator of Growth, Development, and Stress Response. Plant Physiol. 2014, 164, 735–747. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Chen, X.; Xie, K.; Xing, Q.; Wu, Y.; Li, J.; Du, C.; Sun, Z.; Guo, Z. Dlf1, a WRKY Transcription Factor, Is Involved in the Control of Flowering Time and Plant Height in Rice. PLoS ONE 2014, 9, e102529. [Google Scholar] [CrossRef] [Green Version]
- Thornsberry, J.M.; Goodman, M.M.; Doebley, J.; Kresovich, S.; Nielsen, D.; Buckler, E.S. Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 2001, 28, 286–289. [Google Scholar] [CrossRef]
- Wang, J.; Yang, J.; Jia, Q.; Zhu, J.; Shang, Y.; Hua, W.; Zhou, M. A New QTL for Plant Height in Barley (Hordeum vulgare L.) Showing No Negative Effects on Grain Yield. PLoS ONE 2014, 9, e90144. [Google Scholar] [CrossRef]
- Yan, W.H.; Wang, P.; Chen, H.X.; Zhou, H.J.; Li, Q.P.; Wang, C.R.; Ding, Z.H.; Zhang, Y.S.; Yu, S.B.; Xing, Y.Z.; et al. A Major QTL, Ghd8, Plays Pleiotropic Roles in Regulating Grain Productivity, Plant Height, and Heading Date in Rice. Mol. Plant 2011, 4, 319–330. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Wang, K.; Guo, L.; Zhu, Y.J.; Fan, Y.Y.; Cheng, S.H.; Zhuang, J.Y. Pleiotropism of the Photoperiod-Insensitive Allele of Hd1 on Heading Date, Plant Height and Yield Traits in Rice. PLoS ONE 2012, 7, e52538. [Google Scholar] [CrossRef] [Green Version]
- Beavis, W.D.; Grant, D.; Albertsen, M.; Fincher, R. Quantitative trait loci for plant height in four maize populations and their associations with qualitative genetic loci. Theor. Appl. Genet. 1991, 83, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Duvick, D.N. Genetic progress in yield of United States maize. Maydica 2007, 50, 193–202. [Google Scholar]
- Weng, J.; Xie, C.; Hao, Z.; Wang, J.; Liu, C.; Li, M.; Zhang, D.; Bai, L.; Zhang, S.; Li, X. Genome-Wide Association Study Identifies Candidate Genes That Affect Plant Height in Chinese Elite Maize (Zea mays L.) Inbred Lines. PLoS ONE 2011, 6, e29229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farkhari, M.; Krivanek, A.; Xu, Y.; Rong, T.; Naghavi, M.R.; Samadi, B.Y.; Lu, Y. Root-lodging resistance in maize as an example for high-throughput genetic mapping via single nucleotide polymorphism-based selective genotyping. Plant Breed. 2012, 132, 90–98. [Google Scholar] [CrossRef]
- Tadesse, E.B.; Olsen, M.; Das, B.; Gowda, M.; Labuschagne, M. Genetic Dissection of Grain Yield and Agronomic Traits in Maize under Optimum and Low-Nitrogen Stressed Environments. Int. J. Mol. Sci. 2020, 21, 543. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Traits | Q1 | Ye478 | Zheng58 | Population QY | Population QZ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean ± SD a | Range b | Mean ± SD | Skew. c | Kurt. d | Range | Mean ± SD | Skew. | Kurt. | |||
| AD (days) | 89.1 ± 3.2 | 80.0 ± 2.1 | 81.9 ± 2.1 | 74.0–100.0 | 87.8 ± 4.6 | −0.07 | −0.17 | 72.0–99.0 | 84.3 ± 4.7 | 0.13 | −0.47 |
| SD (days) | 94.0 ± 1.8 | 83.5 ± 1.8 | 85.2 ± 1.6 | 77.0–105.0 | 91.3 ± 4.7 | −0.09 | 0.02 | 75.0–105.0 | 88.5 ± 5.3 | 0.05 | −0.19 |
| ASI (days) | 4.9 ± 1.9 | 3.5 ± 0.4 | 3.4 ± 1.4 | 0–12.0 | 3.5 ± 1.9 | 0.78 | 0.66 | −2.0–16.0 | 4.3 ± 2.3 | 0.84 | 1.60 |
| PH (cm) | 270.1 ± 8.9 | 174.5 ± 10.7 | 158.6 ± 9.7 | 123.0–310.0 | 206.7 ± 28.2 | 0.11 | 0.43 | 140.0–305.0 | 208.1 ± 28.4 | 0.16 | −0.10 |
| EH (cm) | 120.3 ± 3.2 | 60.5 ± 3.8 | 50.6 ± 4.3 | 37.0–143.7 | 75.3 ± 18.1 | 0.32 | 0.12 | 34.3–157.0 | 71.0 ± 19.3 | 0.32 | 0.16 |
| ER (%) | 44.6 ± 2.6 | 34.2 ± 4.1 | 31.7 ± 3.2 | 19.8–57.9 | 36.3 ± 5.3 | 0.02 | 0.29 | 18.6–54.9 | 35.8 ± 6.3 | −0.13 | −0.17 |
| Pop. | QTL Name | Chr. | Peak (cM) | LOD | Add. | R2 (%) | 2-LOD Interval (cM) | Range (Mb) | Trait |
|---|---|---|---|---|---|---|---|---|---|
| QY | qPH_Y1a | 1 | 40.2 | 4.2 | −6.7 | 8.8 | 36.6–50.4 | 15.6–33.4 | PH |
| qEH_Y1a | 1 | 44.3 | 3.2 | −3.4 | 7.0 | 41.3–49.6 | 22.6–33.0 | EH | |
| qAD_Y1a | 1 | 173.9 | 6.5 | −1.0 | 12.5 | 165.7–185.4 | 234.3–277.0 | AD | |
| qSD_Y1a | 1 | 173.9 | 5.3 | −1.0 | 10.7 | 162.4–184.3 | 232.2–272.1 | SD | |
| qSD_Y3a | 3 | 35.3 | 3.5 | −0.8 | 6.8 | 28.7–36.1 | 4.9–6.5 | SD | |
| qSD_Y5a | 5 | 62.3 | 8.0 | −2.0 | 16.7 | 54.1–71.3 | 20.3–165.9 | SD | |
| qAD_Y5a | 5 | 66.5 | 4.3 | −0.8 | 8.3 | 54.1–71.1 | 20.3–165.9 | AD | |
| qPH_Y5a | 5 | 91.9 | 7.2 | −9.0 | 15.3 | 74.6–96.0 | 165.9–203.8 | PH | |
| qEH_Y5a | 5 | 102.5 | 5.6 | −4.8 | 13.3 | 91.4–112.9 | 195.8–214.0 | EH | |
| qER_Y5a | 5 | 117.3 | 3.0 | −1.0 | 7.8 | 106.6–121.4 | 207.7–217.3 | ER | |
| qASI_Y5a | 5 | 130.4 | 4.1 | 1.2 | 11.7 | 128.8–140.3 | 218.9–221.1 | ASI | |
| qPH_Y7a | 7 | 44.3 | 3.1 | −5.8 | 6.5 | 42.6–50.0 | 24.7–45.7 | PH | |
| qEH_Y7a | 7 | 70.7 | 3.3 | −3.5 | 7.7 | 63.2–80.6 | 119.5–134.7 | EH | |
| qEH_Y9a | 9 | 37.7 | 5.1 | −4.5 | 12.0 | 26.5–48.4 | 9.4–21.6 | EH | |
| qPH_Y9a | 9 | 52.0 | 6.0 | −8.4 | 14.0 | 43.7–65.6 | 16.8–108.9 | PH | |
| qAD_Y10a | 10 | 34.5 | 4.3 | −0.8 | 8.2 | 31.7–44.9 | 14.4–129.5 | AD | |
| qSD_Y10a | 10 | 56.0 | 6.3 | −1.1 | 13.5 | 45.1–59.9 | 129.5–142.3 | SD | |
| QZ | qER_Z1a | 1 | 90.0 | 7.9 | −1.7 | 13.8 | 73.8–96.0 | 35.5–71.2 | ER |
| qEH_Z1a | 1 | 94.2 | 6.6 | −4.6 | 11.2 | 83.4–107.2 | 51.4–85.1 | EH | |
| qPH_Z1a | 1 | 109.0 | 5.0 | −5.9 | 7.7 | 99.7–125.5 | 72.9–172.2 | PH | |
| qSD_Z1a | 1 | 189.6 | 5.0 | −0.9 | 10.0 | 176–197.0 | 221.1–240.4 | SD | |
| qER_Z1a | 1 | 239.6 | 3.0 | −1.0 | 4.8 | 235.3–250.5 | 279.6–285.5 | ER | |
| qEH_Z3a | 3 | 60.6 | 4.1 | −3.5 | 6.6 | 49.6–65.5 | 6.7–11.9 | EH | |
| qER_Z3a | 3 | 60.6 | 12.0 | −2.1 | 22.0 | 55.6–63.7 | 8.5–10.5 | ER | |
| qAD_Z4a | 4 | 99.8 | 6.4 | −0.8 | 11.3 | 83.4–104.9 | 158.2–181.5 | AD | |
| qPH_Z5a | 5 | 111.7 | 7.5 | −7.4 | 12.4 | 99.3–123.7 | 147.4–175.0 | PH | |
| qEH_Z5a | 5 | 128.0 | 6.8 | −4.7 | 11.6 | 116.7–138.5 | 168.6–199.3 | EH | |
| qEH_Z6a | 6 | 153.7 | 3.0 | 2.9 | 4.4 | 146.5–160.7 | 166.8–169.7 | EH | |
| qEH_Z7a | 7 | 68.0 | 3.2 | 3.1 | 5.1 | 56.8–78.7 | 87.4–126.7 | EH | |
| qSD_Z8a | 8 | 85.5 | 3.8 | −0.7 | 7.3 | 79.4–96.5 | 110.5–156.3 | SD | |
| qAD_Z8a | 8 | 97.8 | 3.2 | −0.6 | 5.6 | 80.9–99.3 | 112.4–163.8 | AD | |
| qPH_Z9a | 9 | 97.9 | 8.9 | −9.2 | 14.7 | 90.0–108.8 | 147.7–155.6 | PH | |
| qEH_Z9a | 9 | 97.9 | 4.2 | −4.0 | 6.8 | 89.7–105.9 | 147.7–155.2 | EH | |
| qPH_Z10a | 10 | 29.0 | 3.1 | −4.8 | 4.8 | 23.5–30.3 | 17.4–69.0 | PH |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, L.; Zhang, H.; Xin, W.; Ma, K.; Du, D.; Yu, C.; Liu, Y. Dissecting the Genetic Basis of Flowering Time and Height Related-Traits Using Two Doubled Haploid Populations in Maize. Plants 2021, 10, 1585. https://doi.org/10.3390/plants10081585
Du L, Zhang H, Xin W, Ma K, Du D, Yu C, Liu Y. Dissecting the Genetic Basis of Flowering Time and Height Related-Traits Using Two Doubled Haploid Populations in Maize. Plants. 2021; 10(8):1585. https://doi.org/10.3390/plants10081585
Chicago/Turabian StyleDu, Lei, Hao Zhang, Wangsen Xin, Kejun Ma, Dengxiang Du, Changping Yu, and Yongzhong Liu. 2021. "Dissecting the Genetic Basis of Flowering Time and Height Related-Traits Using Two Doubled Haploid Populations in Maize" Plants 10, no. 8: 1585. https://doi.org/10.3390/plants10081585
APA StyleDu, L., Zhang, H., Xin, W., Ma, K., Du, D., Yu, C., & Liu, Y. (2021). Dissecting the Genetic Basis of Flowering Time and Height Related-Traits Using Two Doubled Haploid Populations in Maize. Plants, 10(8), 1585. https://doi.org/10.3390/plants10081585





