Identification and Fine-Mapping of Quantitative Trait Loci Controlling Plant Height in Central European Winter Triticale (×Triticosecale Wittmack)
Abstract
1. Introduction
2. Results
3. Discussion
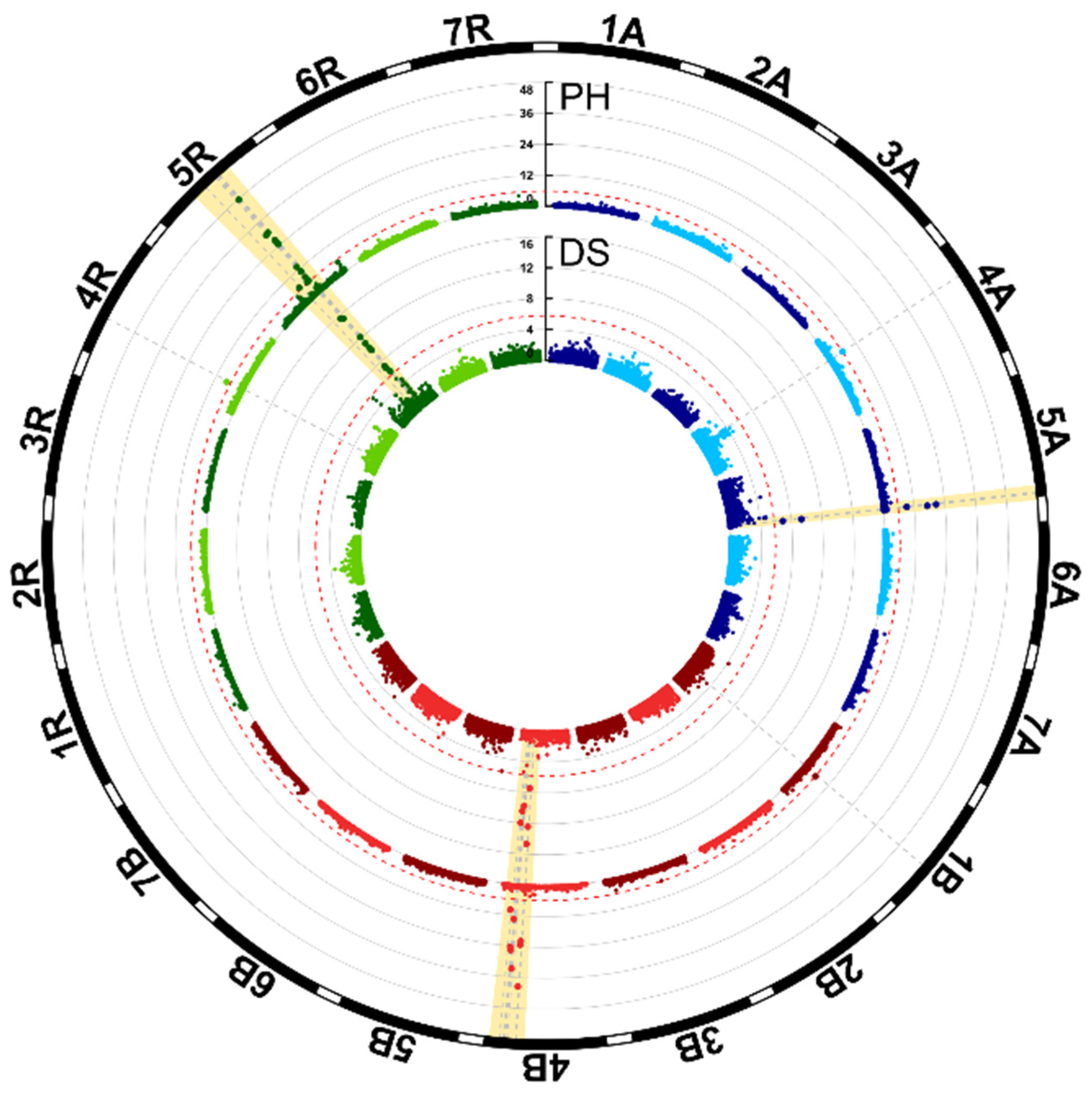
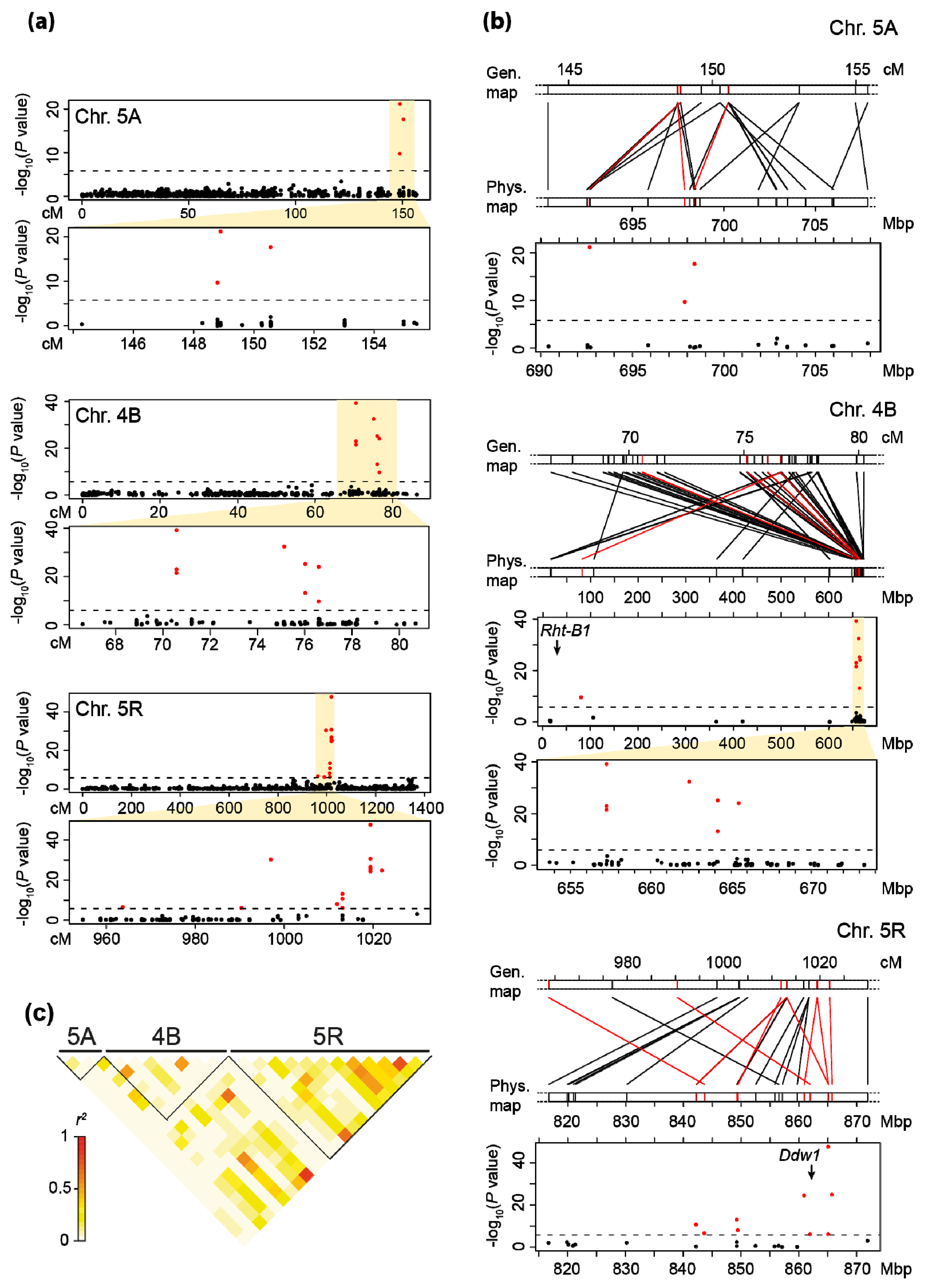
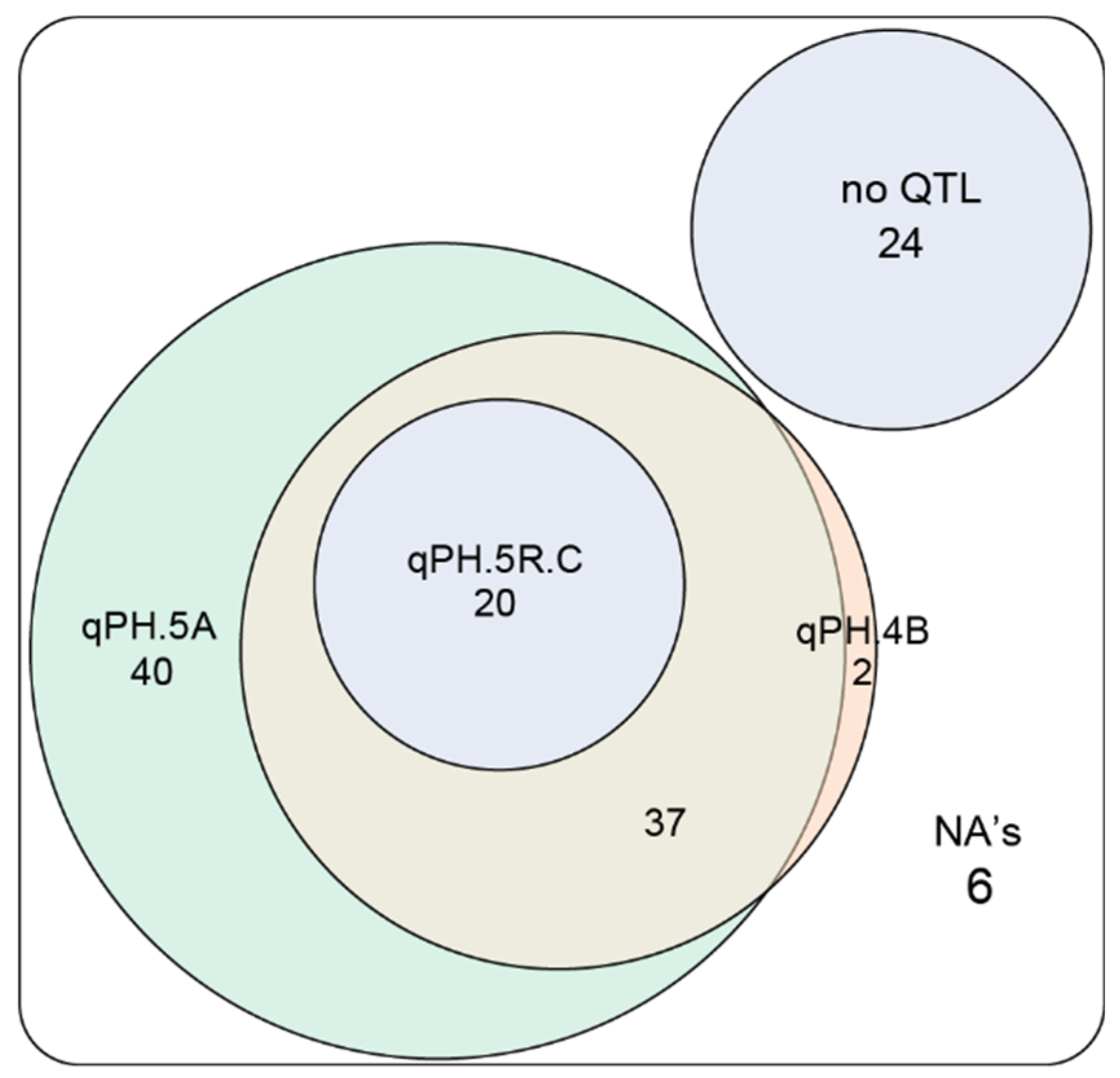
3.1. Genome-Wide Association Mapping and Characterization of QTL for Plant Height
3.2. Long-Term Trends of Plant Height in Triticale
3.3. Conclusions
4. Materials and Methods
4.1. Phenotypic Data
4.2. Statistical Analysis
4.3. Molecular Data Analysis
4.4. Genome-Wide Association Study
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mergoum, M.; Singh, P.K.; Peña, R.J.; Lozano-del Río, A.J.; Cooper, K.V.; Salmon, D.F.; Gómez Macpherson, H. Triticale: A “New” Crop with Old Challenges. In Cereals; Carena, M.J., Ed.; Springer US: New York, NY, USA, 2009; pp. 267–287. ISBN 978-0-387-72294-8. [Google Scholar]
- FAOSTAT. Statistical Databases and Datasets of the Food and Agriculture Organization of the United Nations. Available online: http://www.fao.org/faostat (accessed on 4 May 2021).
- McGoverin, C.M.; Snyders, F.; Muller, N.; Botes, W.; Fox, G.; Manley, M. A review of triticale uses and the effect of growth environment on grain quality. J. Sci. Food Agric. 2011, 91, 1155–1165. [Google Scholar] [CrossRef] [PubMed]
- Myer, A.; Del Lozano Río, A.J. Triticale as animal feed. In Triticale Improvement and Production; Mergoum, M., Gómez Macpherson, H., Eds.; Food and Agriculture Organization Of The United Nations: Rome, Italy, 2004; pp. 49–58. [Google Scholar]
- EU. Directive (EU) 2018/2001 of the European Parliament and of the Council of 11 December 2018 on the Promotion of the Use of Energy from Renewable Sources (recast). Off. J. Eur. Union 2018, 61, 82–209. [Google Scholar]
- EU. Directive 2009/28/EC of the European Parliament and of the Council of 23 April 2009 on the Promotion of the Use of Energy from Renewable Sources and Amending and Subsequently Repealing Directives 2001/77/EC and 2003/30/EC. Off. J. Eur. Union 2009, 52, 16–62. [Google Scholar]
- EU. Directive 2001/77/EC of the European Parliament and of the Council of 27 September 2001 on the Promotion of Electricity Produced from Renewable Energy Sources in the Internal Electricity Market. Off. J. Eur. Union 2001, 44, 33–40. [Google Scholar]
- Weiland, P. Biogas production: Current state and perspectives. Appl. Microbiol. Biotechnol. 2010, 85, 849–860. [Google Scholar] [CrossRef]
- Losert, D.; Maurer, H.P.; Marulanda, J.J.; Würschum, T. Phenotypic and genotypic analyses of diversity and breeding progress in European triticale (×Triticosecale Wittmack). Plant Breed. 2017, 136, 18–27. [Google Scholar] [CrossRef]
- Liu, W.; Leiser, W.L.; Maurer, H.P.; Li, J.; Weissmann, S.; Hahn, V.; Würschum, T. Evaluation of genomic approaches for marker-based improvement of lodging tolerance in triticale. Plant Breed. 2015, 134, 416–422. [Google Scholar] [CrossRef]
- Rajkumara, S. Lodging in cereals—A review. Agric. Rev. 2008, 29, 55–60. [Google Scholar]
- Miedaner, T.; Voss, H.-H. Effect of Dwarfing Rht Genes on Fusarium Head Blight Resistance in Two Sets of Near-Isogenic Lines of Wheat and Check Cultivars. Crop. Sci. 2008, 48, 2115–2122. [Google Scholar] [CrossRef]
- Yan, W.; Li, H.B.; Cai, S.B.; Ma, H.X.; Rebetzke, G.J.; Liu, C.J. Effects of plant height on type I and type II resistance to fusarium head blight in wheat. Plant Pathol. 2011, 60, 506–512. [Google Scholar] [CrossRef]
- Kalih, R.; Maurer, H.P.; Hackauf, B.; Miedaner, T. Effect of a rye dwarfing gene on plant height, heading stage, and Fusarium head blight in triticale (×Triticosecale Wittmack). Theor. Appl. Genet. 2014, 127, 1527–1536. [Google Scholar] [CrossRef]
- Gowda, M.; Hahn, V.; Reif, J.C.; Longin, C.F.H.; Alheit, K.; Maurer, H.P. Potential for simultaneous improvement of grain and biomass yield in Central European winter triticale germplasm. Field Crop. Res. 2011, 121, 153–157. [Google Scholar] [CrossRef]
- Losert, D.; Maurer, H.P.; Weissmann, S.; Würschum, T. Hybrid breeding for biomass yield in winter triticale: I. Hybrid performance, trait correlations and heterosis. Plant Breed. 2016, 135, 560–566. [Google Scholar] [CrossRef]
- Alheit, K.V.; Busemeyer, L.; Liu, W.; Maurer, H.P.; Gowda, M.; Hahn, V.; Weissmann, S.; Ruckelshausen, A.; Reif, J.C.; Würschum, T. Multiple-line cross QTL mapping for biomass yield and plant height in triticale (×Triticosecale Wittmack). Theor. Appl. Genet. 2014, 127, 251–260. [Google Scholar] [CrossRef]
- LTZ. Energiepflanzen für Biogasanlagen. Available online: https://pflanzen.fnr.de/ (accessed on 4 April 2021).
- Longin, C.F.H.; Mühleisen, J.; Maurer, H.P.; Zhang, H.; Gowda, M.; Reif, J.C. Hybrid breeding in autogamous cereals. Theor. Appl. Genet. 2012, 125, 1087–1096. [Google Scholar] [CrossRef] [PubMed]
- Würschum, T.; Langer, S.M.; Longin, C.F.H. Genetic control of plant height in European winter wheat cultivars. Theor. Appl. Genet. 2015, 128, 865–874. [Google Scholar] [CrossRef]
- Börner, A.; Plaschke, J.; Korzun, V.; Worland, A.J. The relationships between the dwarfing genes of wheat and rye. Euphytica 1996, 89, 69–75. [Google Scholar] [CrossRef]
- Griffiths, S.; Simmonds, J.; Leverington, M.; Wang, Y.; Fish, L.; Sayers, L.; Alibert, L.; Orford, S.; Wingen, L.; Snape, J. Meta-QTL analysis of the genetic control of crop height in elite European winter wheat germplasm. Mol. Breed. 2012, 29, 159–171. [Google Scholar] [CrossRef]
- Ellis, M.H.; Rebetzke, G.J.; Chandler, P.; Bonnett, D.; Spielmeyer, W.; Richards, R.A. The effect of different height reducing genes on the early growth of wheat. Funct. Plant Biol. 2004, 31, 583–589. [Google Scholar] [CrossRef]
- Botwright, T.; Rebetzke, G.; Condon, T.; Richards, R. The effect of rht genotype and temperature on coleoptile growth and dry matter partitioning in young wheat seedlings. Funct. Plant Biol. 2001, 28, 417–423. [Google Scholar] [CrossRef]
- Rebetzke, G.J.; Appels, R.; Morrison, A.D.; Richards, R.A.; McDonald, G.; Ellis, M.H.; Spielmeyer, W.; Bonnett, D.G. Quantitative trait loci on chromosome 4B for coleoptile length and early vigour in wheat (Triticum aestivum L.). Aust. J. Agric. Res. 2001, 52, 1221–1224. [Google Scholar] [CrossRef]
- Bai, C.; Liang, Y.; Hawkesford, M.J. Identification of QTLs associated with seedling root traits and their correlation with plant height in wheat. J. Exp. Bot. 2013, 64, 1745–1753. [Google Scholar] [CrossRef]
- Kantarek, Z.; Masojć, P.; Bienias, A.; Milczarski, P. Identification of a novel, dominant dwarfing gene (Ddw4) and its effect on morphological traits of rye. PLoS ONE 2018, 13, e0199335. [Google Scholar] [CrossRef] [PubMed]
- Stojałowski, S.; Myśków, B.; Hanek, M. Phenotypic effect and chromosomal localization of Ddw3, the dominant dwarfing gene in rye (Secale cereale L.). Euphytica 2015, 201, 43–52. [Google Scholar] [CrossRef][Green Version]
- McIntosh, R.A.; Dubcovsky, J.; Rogers, W.J.; Morris, C.; Xia, X.C. Catalogue of Gene Symbols for Wheat: 2017 Supplement. Available online: https://shigen.nig.ac.jp/wheat/komugi/genes/symbolClassList.jsp (accessed on 14 May 2021).
- McIntosh, R.A.; Yamazaki, Y.; Dubcovsky, Y.J.; Rogers, J.; Morris, C.; Apples, R.; Xia, X.C. Catalogue of Gene Symbols for Wheat. Available online: https://shigen.nig.ac.jp/wheat/komugi/genes/symbolClassList.jsp (accessed on 14 May 2021).
- Chernook, A.; Kroupin, P.; Karlov, G.; Soloviev, A.; Korshunova, A.; Rubets, V.; Igonin, V.; Divashuk, M. Effects of Rht-B1b and Ddw1 Dwarfing Genes in Two Connecting Populations of Spring Triticale under Greenhouse Experiment Conditions. Agriculture 2019, 9, 119. [Google Scholar] [CrossRef]
- Würschum, T.; Liu, W.; Busemeyer, L.; Tucker, M.R.; Reif, J.C.; Weissmann, E.A.; Hahn, V.; Ruckelshausen, A.; Maurer, H.P. Mapping dynamic QTL for plant height in triticale. BMC Genet. 2014, 15, 59. [Google Scholar] [CrossRef] [PubMed]
- Braun, E.M.; Tsvetkova, N.; Rotter, B.; Siekmann, D.; Schwefel, K.; Krezdorn, N.; Plieske, J.; Winter, P.; Melz, G.; Voylokov, A.V.; et al. Gene Expression Profiling and Fine Mapping Identifies a Gibberellin 2-Oxidase Gene Co-segregating With the Dominant Dwarfing Gene Ddw1 in Rye (Secale cereale L.). Front. Plant. Sci. 2019, 10, 857. [Google Scholar] [CrossRef]
- Korzun, V.; Börner, A.; Melz, G. RFLP mapping of the dwarfing (Ddw1) and hairy peduncle (Hp) genes on chromosome of rye (Secale cereale L.). Theor. Appl. Genet. 1996, 92, 1073–1077. [Google Scholar] [CrossRef] [PubMed]
- Kroupin, P.; Chernook, A.; Karlov, G.; Soloviev, A.; Divashuk, M. Effect of Dwarfing Gene Ddw1 on Height and Agronomic Traits in Spring Triticale in Greenhouse and Field Experiments in a Non-Black Earth Region of Russia. Plants 2019, 8, 131. [Google Scholar] [CrossRef]
- Sun, L.; Yang, W.; Li, Y.; Shan, Q.; Ye, X.; Wang, D.; Yu, K.; Lu, W.; Xin, P.; Pei, Z.; et al. A wheat dominant dwarfing line with Rht12, which reduces stem cell length and affects gibberellic acid synthesis, is a 5AL terminal deletion line. Plant. J. 2019, 97, 887–900. [Google Scholar] [CrossRef]
- Chen, L.; Phillips, A.L.; Condon, A.G.; Parry, M.A.J.; Hu, Y.-G. GA-responsive dwarfing gene Rht12 affects the developmental and agronomic traits in common bread wheat. PLoS ONE 2013, 8, e62285. [Google Scholar] [CrossRef]
- Worland, A.J.; Sayers, E.J.; Borner, A. The Genetics and Breeding Potential of Rht12, a Dominant Dwarfing Gene in Wheat. Plant. Breed. 1994, 113, 187–196. [Google Scholar] [CrossRef]
- Würschum, T.; Langer, S.M.; Longin, C.F.H.; Tucker, M.R.; Leiser, W.L. A modern Green Revolution gene for reduced height in wheat. Plant. J. 2017, 92, 892–903. [Google Scholar] [CrossRef]
- Flintham, J.E.; Börner, A.; Worland, A.J.; Gale, M.D. Optimizing wheat grain yield: Effects of Rht (gibberellin-insensitive) dwarfing genes. J. Agric. Sci. 1997, 128, 11–25. [Google Scholar] [CrossRef]
- Wilhelm, E.P.; Mackay, I.J.; Saville, R.J.; Korolev, A.V.; Balfourier, F.; Greenland, A.J.; Boulton, M.I.; Powell, W. Haplotype dictionary for the Rht-1 loci in wheat. Theor. Appl. Genet. 2013, 126, 1733–1747. [Google Scholar] [CrossRef] [PubMed]
- Ericsson, K.; Rosenqvist, H.; Nilsson, L.J. Energy crop production costs in the EU. Biomass Bioenergy 2009, 33, 1577–1586. [Google Scholar] [CrossRef]
- Worland, A.J.; Korzun, V.; Röder, M.S.; Ganal, M.W.; Law, C.N. Genetic analysis of the dwarfing gene Rht8 in wheat. Part II. The distribution and adaptive significance of allelic variants at the Rht8 locus of wheat as revealed by microsatellite screening. Theor. Appl. Genet. 1998, 96, 1110–1120. [Google Scholar] [CrossRef]
- Mathews, K.L.; Chapman, S.C.; Trethowan, R.; Singh, R.P.; Crossa, J.; Pfeiffer, W.; Ginkel, M.; DeLacy, I. Global Adaptation of Spring Bread and Durum Wheat Lines Near-Isogenic for Major Reduced Height Genes. Crop. Sci. 2006, 46, 603–613. [Google Scholar] [CrossRef]
- Butler, J.D.; Byrne, P.F.; Mohammadi, V.; Chapman, P.L.; Haley, S.D. Agronomic Performance of Rht Alleles in a Spring Wheat Population across a Range of Moisture Levels. Crop. Sci. 2005, 45, 939–947. [Google Scholar] [CrossRef]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S.; et al. Crop Production under Drought and Heat Stress: Plant Responses and Management Options. Front. Plant. Sci. 2017, 8, 1147. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, S.; Grando, S.; Maatougui, M.; Michael, M.; Slash, M.; Haghparast, R.; Rahmanian, M.; Taheri, A.; Al-Yassin, A.; Benbelkacem, A.; et al. Plant breeding and climate changes. J. Agric. Sci. 2010, 148, 627–637. [Google Scholar] [CrossRef]
- Singh, K.; Khanna-Chopra, R. Physiology and QTL anaylsis of coleoptile length, a trait for drought tolerance in wheat. J. Plant. Biol. 2010, 37, 1–9. [Google Scholar]
- Botwright, T.L.; Rebetzke, G.J.; Condon, A.G.; Richards, R.A. Influence of variety, seed position and seed source on screening for coleoptile length in bread wheat (Triticum aestivum L.). Euphytica 2001, 119, 349–356. [Google Scholar] [CrossRef]
- Neuweiler, J.E.; Maurer, H.P.; Würschum, T. Long-term trends and genetic architecture of seed characteristics, grain yield and correlated agronomic traits in triticale (×Triticosecale Wittmack). Plant. Breed. 2020, 139, 717–729. [Google Scholar] [CrossRef]
- Williams, E.; Piepho, H.P.; Whitaker, D. Augmented p-rep designs. Biom. J. 2011, 53, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Piepho, H.P.; Büchse, A.; Emrich, K. A hitchhiker’s guide to mixed models for randomized experiments. J. Agron. Crop. Sci. 2003, 189, 310–322. [Google Scholar] [CrossRef]
- Butler, D.G.; Cullis, B.R.; Gilmour, A.R.; Gogel, B.J. ASReml-R Reference Manual; The State of Queensland, Department of Primary Industries and Fisherie: Queensland, Australia, 2009.
- Stram, D.O.; Lee, J.W. Variance components testing in the longitudinal mixed effects model. Biometrics 1994, 50, 1171. [Google Scholar] [CrossRef]
- Cullis, B.R.; Smith, A.B.; Coombes, N.E. On the design of early generation variety trials with correlated data. J. Agric. Biol. Environ. Stat. 2006, 11, 381–393. [Google Scholar] [CrossRef]
- Li, H.; Vikram, P.; Singh, R.P.; Kilian, A.; Carling, J.; Song, J.; Burgueno-Ferreira, J.A.; Bhavani, S.; Huerta-Espino, J.; Payne, T.; et al. A high density GBS map of bread wheat and its application for dissecting complex disease resistance traits. BMC Genom. 2015, 16, 216. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh Bi, I.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 2397–2399. [Google Scholar] [CrossRef]
- Money, D.; Gardner, K.; Migicovsky, Z.; Schwaninger, H.; Zhong, G.-Y.; Myles, S. LinkImpute: Fast and Accurate Genotype Imputation for Nonmodel Organisms. G3 Genes Genomes Genet. 2015, 5, 2383–2390. [Google Scholar] [CrossRef] [PubMed]
- Utz, H.F.; Melchinger, A.E.; Schön, C.C. Bias and Sampling Error of the Estimated Proportion of Genotypic Variance Explained by Quantitative Trait Loci Determined from Experimental Data in Maize Using Cross Validation and Validation With Independent Samples. Genetics 2000, 154, 1839–1849. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Walsh, B. Genetics and Analysis of Quantitative Traits, 2nd ed.; Sinauer: Sunderland, MA, USA, 1998; ISBN 978-0878934812. [Google Scholar]
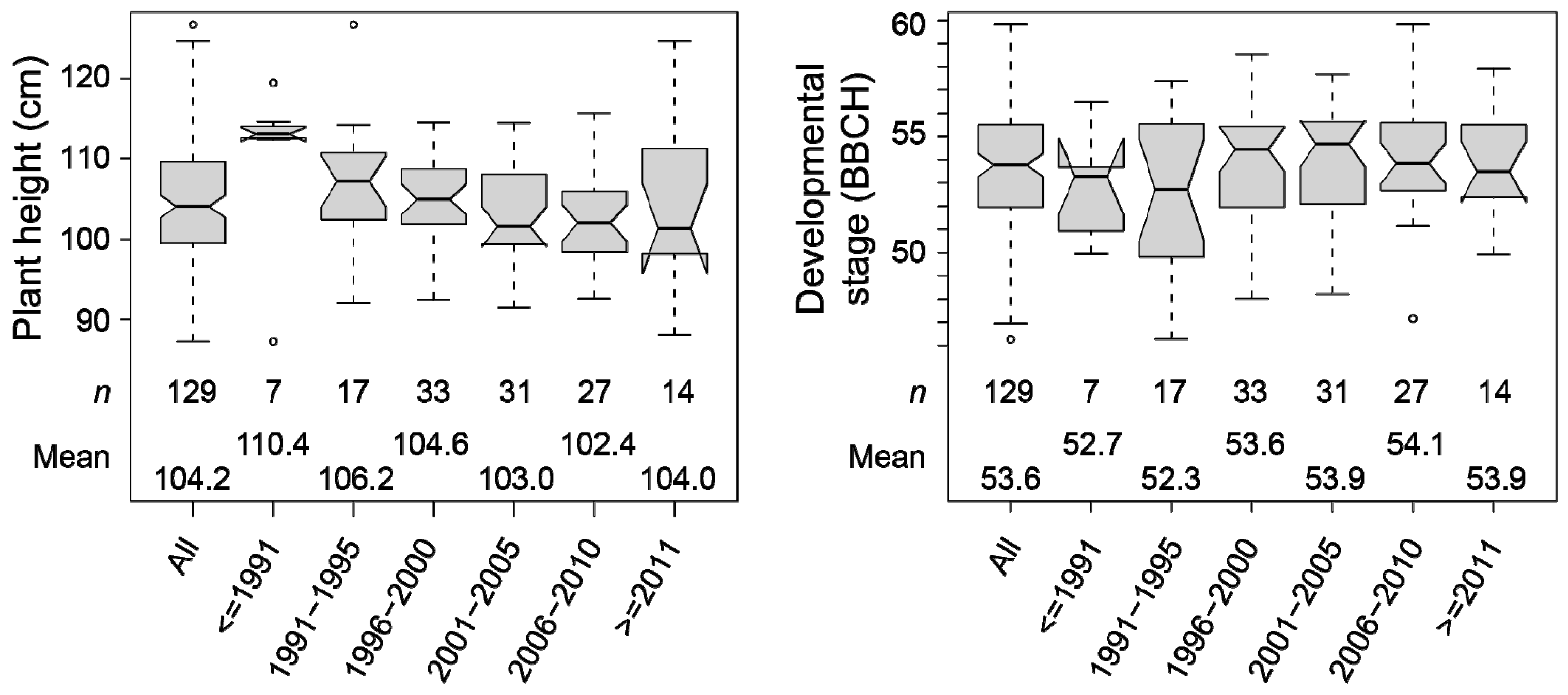


| Plant Height | Developmental Stage | |
|---|---|---|
| Min | 80.2 | 43.0 |
| Mean | 100.8 | 53.4 |
| Max | 126.6 | 59.8 |
| LSD0.05 | 4.0 | 1.3 |
| 77.98 *** | 6.09 *** | |
| 7.38 *** | 1.21 *** | |
| † | 18.32 | 1.27 |
| H2 | 0.81 | 0.80 |
- ***
- Significantly different from zero at the 0.001 probability level.
- †
- Mean residual error variance across environment-trial combinations.
| QTL | Marker | Chr. | Pos. (cm) | p-Value | pG Joint a | pG Single b | α-Effect (Single Fit) | pc |
|---|---|---|---|---|---|---|---|---|
| Plant height (42.16% pG total) | ||||||||
| qPH.5A | D10506872 | 5A | 150.6 | 2.3 × 10−18 | 0.40 | 13.06 | 3.72 | 0.25 |
| qPH.4B | D4371530 d | 4B | 70.6 | 1.2 × 10−23 | 2.34 | 18.97 | −4.02 | 0.36 |
| qPH.4R | D10523748 | 4R | 173.0 | 7.7 × 10−8 | 1.99 | 1.35 | 1.03 | 0.48 |
| qPH.5R.A | D10519777 | 5R | 990.4 | 7.3 × 10−7 | 1.79 | 4.56 | 2.26 | 0.23 |
| qPH.5R.B | D4348428 | 5R | 997.0 | 5.5 × 10−31 | 1.91 | 19.12 | −3.94 | 0.41 |
| qPH.5R.C | S4341499 d | 5R | 1019.4 | 2.0 × 10−48 | 29.38 | 29.38 | 5.30 | 0.63 |
| Developmental stage (29.31% pG total) | ||||||||
| qEC.5A | D10506872 | 5A | 150.6 | 3.3 × 10−10 | 3.26 | 16.38 | 1.15 | 0.25 |
| qEC.4B | D4372007 | 4B | 70.6 | 2.7 × 10−15 | 0.03 | 12.83 | −0.88 | 0.51 |
| qEC.5R.B | D4348428 | 5R | 997.0 | 2.4 × 10−16 | 16.74 | 16.74 | −1.02 | 0.37 |
| qEC.5R.C | S4341499 d | 5R | 1019.4 | 4.1 × 10−16 | 5.35 | 19.43 | 1.19 | 0.63 |
- a
- pG values obtained by a joint fit of all significant markers for the respective trait in a linear model; markers were ordered according to their p-value (lowest first).
- b
- pG values obtained when each significant marker was fitted in a linear model for the respective trait.
- c
- Frequency of trait-increasing allele, i.e., development stages (for earlier genotypes), plant height (for taller genotypes).
- d
- Unmapped marker that was assigned to its most probable position based on its LD with mapped markers.
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trini, J.; Maurer, H.P.; Neuweiler, J.E.; Würschum, T. Identification and Fine-Mapping of Quantitative Trait Loci Controlling Plant Height in Central European Winter Triticale (×Triticosecale Wittmack). Plants 2021, 10, 1592. https://doi.org/10.3390/plants10081592
Trini J, Maurer HP, Neuweiler JE, Würschum T. Identification and Fine-Mapping of Quantitative Trait Loci Controlling Plant Height in Central European Winter Triticale (×Triticosecale Wittmack). Plants. 2021; 10(8):1592. https://doi.org/10.3390/plants10081592
Chicago/Turabian StyleTrini, Johannes, Hans Peter Maurer, Jan Eric Neuweiler, and Tobias Würschum. 2021. "Identification and Fine-Mapping of Quantitative Trait Loci Controlling Plant Height in Central European Winter Triticale (×Triticosecale Wittmack)" Plants 10, no. 8: 1592. https://doi.org/10.3390/plants10081592
APA StyleTrini, J., Maurer, H. P., Neuweiler, J. E., & Würschum, T. (2021). Identification and Fine-Mapping of Quantitative Trait Loci Controlling Plant Height in Central European Winter Triticale (×Triticosecale Wittmack). Plants, 10(8), 1592. https://doi.org/10.3390/plants10081592






