LEA13 and LEA30 Are Involved in Tolerance to Water Stress and Stomata Density in Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
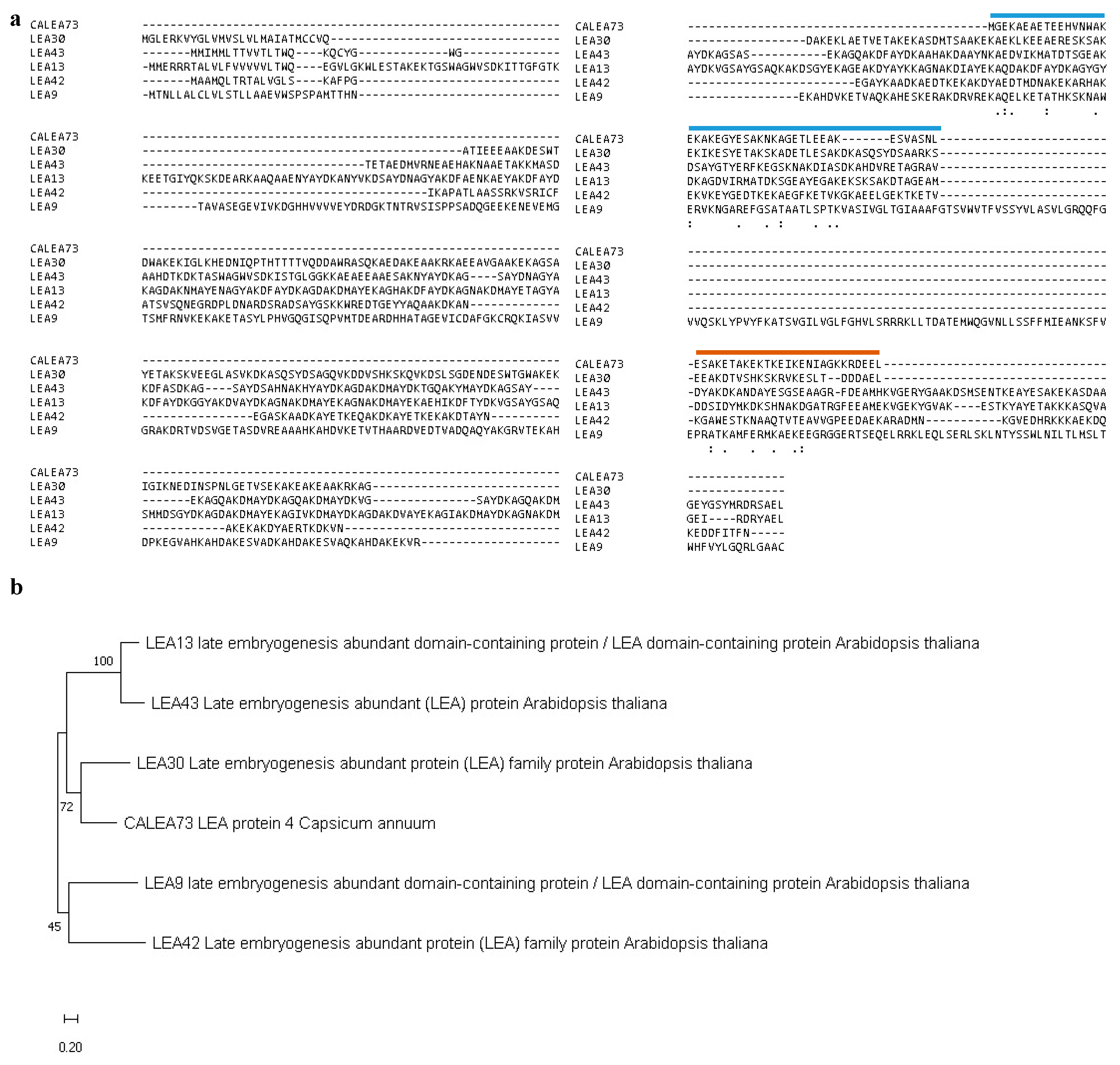
2.1. Identification of Genes Related to CaLEA73
2.2. LEA13 and LEA30 Are Involved in the Tolerance to Water Deficit Stress
2.3. Expression and Functional Analyses of LEA13 and LEA30 under Water Stress Conditions
2.4. Transpiration and Stomatal Density Are Higher in lea13 and lea30
2.5. Stomata Developmental Genes Expression Is Affected in the 35S:CaLEA73 Background
2.6. Differential Gene Expression in lea13
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Sequence Alignments
4.2. Promoter cis-Element Analysis
4.3. Drought Tolerance Experiments and Determination of Water Content
4.4. Gene Expression Analysis by qRT-PCR
4.5. Analysis of Stomatal Development
4.6. Transpiration and Stomatal Conductance
4.7. Statistical Analysis
4.8. Microarray Analysis in lea13 Genetic Background
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Srivastava, R.; Quilichini, T.D.; Dong, H.; Bao, Y.; Horner, H.T.; Howell, S.H. IRE1, a component of the unfolded protein response signaling pathway, protects pollen development in Arabidopsis from heat stress. Plant J. 2016, 88, 93–204. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.S.; Lee, J.H.; Kim, J.J.; Kim, C.H.; Jun, S.S.; Hong, Y.N. Molecular and functional characterization of CaLEA6, the gene for a hydrophobic LEA protein from Capsicum annuum. Gene 2004, 344, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Shinozaki, K.; Shinozaki, K. Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 2011, 52, 1569–1582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Battaglia, M.; Olvera-Carrillo, Y.; Garciarrubio, A.; Campos, F.; Covarrubias, A.A. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 2008, 148, 6–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wakeel, A.; Wang, L.; Xu, M. SPEECHLESS and MUTE mediate feedback Regulation of Signal Transduction during Stomatal Development. Plants 2021, 10, 432. [Google Scholar] [CrossRef]
- Lampard, G.R.; Lukowitz, W.; Ellis, B.E.; Bergmann, D.C. Novel and Expanded Roles for MAPK Signaling in Arabidopsis Stomatal Cell Fate Revealed by Cell Type–Specific Manipulations. Plant Cell 2009, 21, 3506–3517. [Google Scholar] [CrossRef] [Green Version]
- Qi, X.; Torii, K.U. Hormonal and environmental signals guiding stomatal development. BMC Biol. 2018, 16, 21. [Google Scholar] [CrossRef] [Green Version]
- Hong-Bo, S.; Zong-Suo, L.; Ming-An, S. LEA proteins in higher plants: Structure, function, gene expression and regulation. Colloid Surf. B 2005, 45, 131–135. [Google Scholar] [CrossRef]
- Magwanga, R.O.; Lu, P.; Kirungu, J.N.; Lu, H.; Wang, X.; Cai, X.; Zhou, Z.; Zhang, Z.; Salih, H.; Wang, K.; et al. Characterization of the late embryogenesis abundant (LEA) proteins family and their role in drought stress tolerance in upland cotton. BMC Genet. 2018, 15, 19. [Google Scholar] [CrossRef] [Green Version]
- Jia, H.; Wang, X.; Shi, Y.; Wu, X.; Wang, Y.; Liu, J.; Fang, Z.; Li, C.; Dong, K. Overexpression of Medicago sativa LEA4-4 can improve the salt, drought, and oxidation resistance of transgenic Arabidopsis. PLoS ONE 2020, 15, e0234085. [Google Scholar] [CrossRef]
- Cortez-Baheza, E.; Cruz-Fernández, F.; Hernández-Álvarez, M.I.; Peraza-Luna, F.; Aguado-Santacruz, G.A.; Serratos-Arévalo, J.C.; Ponce, P. A new Lea gene is induced during osmopriming of Capsicum annuum L. seeds. Int. J. Bot. 2008, 4, 77–84. [Google Scholar] [CrossRef]
- Acosta-García, G.; Chapa-Oliver, A.M.; Millán-Almaraz, J.R.; Guevara-González, R.G.; Cortez-Baheza, E.; Rangel-Cano, R.M.; Ramírez-Pimentel, J.G.; Cruz-Hernández, A.; Guevara-Olvera, L.; Aguilera-Bibian, J.E.; et al. CaLEA 73 gene from Capsicum annuum L. enhances drought and osmotic tolerance modulating transpiration rate in transgenic Arabidopsis thaliana. Can. J. Plant Sci. 2015, 95, 227–235. [Google Scholar] [CrossRef]
- Bies-Ethève, N.; Gaubier-Comella, P.; Debures, A.; Lasserre, E.; Jobet, E.; Raynal, M.; Cooke, R.; Delseny, M. Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Mol. Biol. 2008, 67, 107–124. [Google Scholar] [CrossRef]
- Hundertmark, M.; Hincha, D.K. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom. 2008, 9, 118. [Google Scholar] [CrossRef] [Green Version]
- Candat, A.; Paszkiewicz, G.; Neveu, M.; Gautier, R.; Logan, D.C.; Avelange-Macherel, M.H.; Macherela, D. The ubiquitous distribution of Late Embryogenesis Abundant proteins across cell compartments in Arabidopsis offers tailored protection against abiotic stress. Plant Cell 2014, 26, 3148–3166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, K.C.; Lin, W.C.; Cheng, W.H. Salt hypersensitive mutant 9, a nucleolar APUM23 protein, is essential for salt sensitivity in association with the ABA signaling pathway in Arabidopsis. BMC Plant Biol. 2018, 18, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Zhang, F.; Yu, Y.; Feng, L.; Jia, J.; Liu, B.; Li, B.; Guo, H.; Zhai, J.A. Comprehensive Online Database for Exploring ∼20,000 Public Arabidopsis RNA-Seq Libraries. Mol. Plant 2020, 13, 1231–1233. [Google Scholar] [CrossRef]
- Cuevas-Velazquez, C.L.; Saab-Rincón, G.; Reyes, J.L.; Covarrubias, A.A. The unstructured N-terminal region of Arabidopsis group 4 Late Embryogenesis Abundant Proteins (LEA) is required for folding and for chaperone-like activity under water deficit. J. Biol. Chem. 2016, 291, 10893–10903. [Google Scholar] [CrossRef] [Green Version]
- Bray, E.A. Plant responses to water deficit. Trends Plant Sci. 1997, 2, 48–54. [Google Scholar] [CrossRef]
- Yongkun Chen, Y.; Li, C.; Zhang, B.; Yi, J.; Yang, Y.; Kong, C.; Lei, C.; Gong, M. The Role of the Late Embryogenesis-Abundant (LEA) Protein Family in Development and the Abiotic Stress Response: A Comprehensive Expression Analysis of Potato (Solanum Tuberosum). Genes 2019, 10, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nir, I.; Moshelion, M.; Weiss, D. The Arabidopsis GIBBERELLIN METHYL TRANSFERASE 1 suppresses gibberellin activity, reduces whole-plant transpiration and promotes drought tolerance in transgenic tomato. Plant Cell Environ. 2014, 37, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Yoo, C.Y.; Hasegawa, P.M.; Mickelbart, M.V. Regulation of stomatal density by the GTL1 transcription factor for improving water use efficiency. Plant Signal. Behav. 2011, 6, 1069–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zoulias, N.; Harrison, E.L.; Casson, S.A.; Gray, J.E. Molecular control of stomatal development. Biochem. J. 2018, 475, 441–454. [Google Scholar] [CrossRef] [Green Version]
- Xiao, B.; Huang, Y.; Tang, N.; Xiong, L. Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor. Appl. Genet. 2007, 115, 35–46. [Google Scholar] [CrossRef]
- Dang, N.X.; Popova, A.V.; Hundertmark, M.; Hincha, D.K. Functional characterization of selected LEA proteins from Arabidopsis thaliana in yeast and in vitro. Planta 2014, 240, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Tomoki Shibuya, T.; Itai, R.; Maeda, M.; Kitashiba, H.; Isuzugawa, K.; Kato, K.; Kanayama, Y. Characterization of PcLEA14, a Group 5 Late Embryogenesis Abundant Protein Gene from Pear (Pyrus communis). Plants 2020, 9, 1138. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Duan, X.; Wang, B.; Hong, B.; Ho, T.H.D.; Wu, R. Expression of a Late Embryogenesis Abundant protein gene, HVA7, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol. 1996, 110, 249–257. [Google Scholar] [CrossRef] [Green Version]
- Salleh, F.M.; Evans, K.; Goodall, B.; Machin, H.; Mowla, S.B.; Mur, L.A.J.; Runions, J.; Theodoulou, F.L.; Foyer, C.H.; Rogers, H.J. A novel function for a redox-related LEA protein (SAG21/AtLEA5) in root development and biotic stress responses. Plant Cell Environ. 2012, 35, 418–429. [Google Scholar] [CrossRef] [Green Version]
- Muniz-Pedrosa, A.; Santos-Martins, C.P.; Pereira-Gonçalves, L.; Cardoso-Costa, M.G. Late Embryogenesis Abundant (LEA) Constitutes a Large and Diverse Family of Proteins Involved in Development and Abiotic Stress Responses in Sweet Orange (Citrus sinensis L. Osb.). PLoS ONE 2015, 10, e0145785. [Google Scholar] [CrossRef] [Green Version]
- Daszkowska-Golec, A.; Szarejko, I. Open or close the gate—Stomata action under the control of phytohormones in drought stress conditions. Front. Plant Sci. 2013, 4, 138. [Google Scholar] [CrossRef] [Green Version]
- Brodribb, T.J.; Sussmilch, F.; McAdam, S.A.M. From reproduction to production, stomata are the master regulators. Plant J. 2020, 101, 756–767. [Google Scholar] [CrossRef]
- Yang, X.H.; Li, X.G.; Li, B.L.; Zhang, D.Q. Genome-wide transcriptional profiling reveals molecular signatures of secondary xylem differentiation in Populus tomentosa. Genet. Mol. Res. 2014, 13, 9489–9504. [Google Scholar] [CrossRef]
- Richard, O.; Paquet, N.; Haudecoeur, E.; Charrier, B. Organization and Expression of the GSK3/Shaggy Kinase gene family in the moss Physcomitrella patens suggest early gene multiplication in land plants and an ancestral response to osmotic stress. J. Mol. Evol. 2005, 61, 99–113. [Google Scholar] [CrossRef]
- Chen, C.; Shu, J.; Li, C.; Thapa, R.K.; Nguyen, V.; Yu, K.; Yuan, Z.C.; Kohalmi, S.E.; Liu, J.; Marsolais, F.; et al. RNA polymerase II-independent recruitment of SPT6L at transcription start sites in Arabidopsis. Nucleic Acids Res. 2019, 47, 6714–6725. [Google Scholar] [CrossRef]
- Roxrud, I.; Lid, S.E.; Fletcher, J.C.; Schmidt, E.D.L.; Opsahl-Sorteberg, H.G. GASA4, One of the 14-Member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol. 2007, 48, 471–483. [Google Scholar] [CrossRef]
- Saibo, N.J.M.; Vriezen, W.H.; Beemster, G.T.S.; Van Der Straeten, D. Growth and stomata development of Arabidopsis hypocotyls are controlled by gibberellins and modulated by ethylene and auxins. Plant J. 2003, 33, 989–1000. [Google Scholar] [CrossRef] [PubMed]
- Richardson, L.G.; Torii, K.U. Take a deep breath: Peptide signalling in stomatal patterning and differentiation. J. Exp. Bot. 2013, 64, 5243–5251. [Google Scholar] [CrossRef] [Green Version]
- Han, S.K.; Torii, K.U. Lineage-specific stem cells, signals and asymmetries during stomatal development. Development 2016, 143, 1259–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.J.; Lee, J.H.; Kim, W.; Jung, H.S.; Huijser, P.; Ahn, J.H. The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 Module Regulates Ambient Temperature-Responsive Flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiol. 2012, 159, 461–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Wang, Y.; Lv, B.; Li, J.; Luo, L.; Lu, S.; Zhang, X.; Ma, H.; Ming, F. The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol. 2014, 55, 604–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pillitteri, L.J.; Dong, J. Stomatal development in Arabidopsis. Arab. Book 2013, 11, e0162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohashi-Ito, K.; Bergmann, D.C. Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 2006, 18, 2493–2505. [Google Scholar] [CrossRef] [Green Version]
- Horstman, A.; Willemsen, V.; Boutilier, K.; Heidstra, R. AINTEGUMENTA-LIKE proteins: Hubs in a plethora of networks. Trends Plant Sci. 2014, 19, 146–157. [Google Scholar] [CrossRef]
- Lai, L.B.; Nadeau, J.A.; Lucas, J.; Lee, E.K.; Nakagawa, T.; Zhao, L.; Geisler, M.; Sack, F.D. The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. Plant Cell 2005, 17, 2754–2767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Z.; Li, D.; Wang, L.; Sack, F.D.; Grotewold, E. Role of the stomatal development regulators FLP/MYB88 in abiotic stress responses. Plant J. 2010, 64, 731–739. [Google Scholar] [CrossRef] [PubMed]
- Wurzinger, B.; Mair, A.; Pfister, B.; Teige, M. Cross-talk of calcium-dependent protein kinase and MAP kinase signaling. Plant Signal. Behav. 2011, 6, 8–12. [Google Scholar] [CrossRef]
- Miwa, H.; Kinoshita, A.; Fukuda, H.; Sawa, S. Plant meristems: CLAVATA3/ESR-related signaling in the shoot apical meristem and the root apical meristem. J. Plant Res. 2009, 122, 31–39. [Google Scholar] [CrossRef]
- Lau, O.S.; Davies, K.A.; Chang, J.; Adrian, J.; Rowe, M.H.; Ballenger, C.E.; Bergmann, D.C. Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science 2014, 345, 1605–1609. [Google Scholar] [CrossRef] [Green Version]
- Kanaoka, M.M.; Pillitteri, L.J.; Fujii, H.; Yoshida, Y.; Bogenschutz, N.L.; Takabayashi, J.; Zhu, J.K.; Torii, K.U. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 2008, 20, 1775–1785. [Google Scholar] [CrossRef] [Green Version]
- Danzer, J.; Mellott, E.; Bui, A.Q.; Le, B.H.; Martin, P.; Hashimoto, M.; Perez-Lesher, J.; Chen, M.; Pelletier, J.M.; Somers, D.A.; et al. Down-regulating the expression of 53 soybean transcription factor genes uncovers a Role for SPEECHLESS in Initiating Stomatal Cell Lineages during Embryo Development. Plant Physiol. 2015, 168, 1025–1035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacAlister, C.A.; Ohashi-Ito, K.; Bergmann, D.C. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 2007, 445, 537. [Google Scholar] [CrossRef]
- MacAlister, C.A.; Bergmann, D.C. Sequence and function of basic helix–loop–helix proteins required for stomatal development in Arabidopsis are deeply conserved in land plants. Evol. Dev. 2011, 13, 182–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weimer, A.K.; Matos, J.L.; Sharma, N.; Patell, F.; Murray, J.A.; Dewitte, W.; Bergmann, D.C. Lineage and stage-specific expressed CYCD7; 1 coordinates the single symmetric division that creates stomatal guard cells. Development 2018, 145, dev160671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torii, K.U. Mix-and-match: Ligand-receptor pairs in stomatal development and beyond. Trends Plant Sci. 2012, 17, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, A.; Torii, K.U. Shouting out loud: Signaling modules in the regulation of stomatal development. Plant Physiol. 2020, 185, 765–780. [Google Scholar] [CrossRef]
- Dimmic, M.W.; Rest, J.S.; Mindell, D.P. rtREV: An amino acid substitution matrix for inference of retrovirus and reverse transcriptase phylogeny. J. Mol. Evol. 2002, 55, 65–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, W.T. The Arabidopsis RING E3 Ubiquitin Ligase AtAIRP3/LOG2 Participates in Positive Regulation of High-Salt and Drought Stress Responses. Plant Physiol. 2013, 162, 1733–1749. [Google Scholar] [CrossRef] [Green Version]
- Yu, S.G.; Cho, N.H.; Kim, J.H.; Oh, T.R.; Kim, W.T. Suppression of DRR1 results in the accumulation of insoluble ubiquitinated proteins, which impairs drought stress tolerance. J. Integr. Plant Biol. 2020, 63, 431–437. [Google Scholar] [CrossRef]








| AGI Code | Zscore | Annotation | Description |
|---|---|---|---|
| AT1G65440 | 5.243761 | GLOBAL TRANSCRIPTION FACTOR GROUP B, global transcription factor group B1 | Transcription elongation factor SPT6-like protein |
| At1g75590 | 4.204775 | F10A5.20, F10A5_20 | SAUR-like auxin-responsive protein |
| At1g17240 | 4.004409 | AtRLP2, F20D23.6, F20D23_6, receptor-like protein 2 | Receptor-like protein 2; Encodes a CLAVATA2 (CLV2)-related gene. |
| At4g15040 | 3.362095 | At4g15040 | Subtilisin-like serine endopeptidase family protein. |
| At2g01420 | 3.242944 | ARABIDOPSIS PIN-FORMED 4, AUXIN TRANSPORTER SPLICE VARIANT B, PIN-FORMED 4 | Auxin efflux carrier component 4. |
| At3g05840 | 3.101374 | ASKGAMMA, F10A16.14, F10A16_14 | Shaggy-related protein kinase gamma; encodes a SHAGGY-like kinase involved in meristem organization. |
| At4g23270 | 3.003471 | F21P8.160, F21P8_160, cysteine-rich RLK (RECEPTOR-like protein kinase) 19 | Cysteine-rich receptor-like protein kinase 19 |
| At3g62670 | 2.782405 | ARR20, MEE41, maternal effect embryo arrest 41, response regulator ARR20 | Putative two-component response regulator ARR20; response regulator 20 |
| At1g76520 | 2.556337 | F14G6.12, F14G6_12 | Auxin efflux carrier family protein. |
| At1g65480 | 2.536566 | F5I14.3, F5I14_3, FLOWERING LOCUS T | FLOWERING LOCUS T protein; |
| At2g39540 | 2.342906 | F12L6.20, F12L6_20 | Gibberellin-regulated protein |
| At5g10310 | 2.315854 | F18D22.80, F18D22_80 | Epidermal patterning factor-like protein |
| AGI Code | Zscore | Annotation | Description |
|---|---|---|---|
| AT4G35580 | −6.65111 | F8D20.90, F8D20_90, NAC transcription factor-like 9 | NAC transcription factor-like 9. |
| At5g65510 | −4.88908 | AT5G65510, AINTEGUMENTA-like 7, PLETHORA 7, PLT7 | AINTEGUMENTA-like 7 protein; Encodes one of three PLETHORA transcription factors required to maintain high levels of PIN1 expression. |
| At3g61900 | −4.14811 | AT3G61900 | SAUR-like auxin-responsive protein. |
| At2g06020 | −3.48404 | At2g06020 | Myb family transcription factor; Homeodomain-like superfamily protein. |
| At5g26950 | −3.45455 | AT5G26950, AGAMOUS-like 93, F2P16.17, F2P16_17 | Agamous-like MADS-box protein AGL93. |
| At3g20640 | −3.19807 | AT3G20640 | Transcription factor bHLH123. |
| At5g40330 | −3.18796 | transcription factor MYB23 | MYB DOMAIN PROTEIN 23, myb domain protein 23; Encodes a MYB gene that, when overexpressed ectopically, can induce ectopic trichome formation. |
| At1g26945 | −3.06999 | AT1G26945, KIDARI | Basic helix-loop-helix protein KIDARI; basic helix-loop-helix (bHLH) DNA-binding superfamily protein KIDARI (KDR). |
| At3g18040 | −3.05605 | AT3G18040, MAP kinase 9 | Mitogen-activated protein kinase 9. |
| At1g01260 | −3.02189 | AT1G01260, F6F3.7, F6F3_7 | Transcription factor bHLH13. |
| At3g25905 | −2.89363 | CLAVATA3/ESR-RELATED 27, CLE27 | Protein CLAVATA3/ESR-related 27; Member of a large family of putative ligands homologous to the Clavata3 gene. |
| At5g58890 | −2.82425 | AGAMOUS-like 82, K19M22.9, K19M22_9 | Protein AGAMOUS-LIKE 82. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Cordova, A.; Ramírez-Medina, H.; Silva-Martinez, G.-A.; González-Cruz, L.; Bernardino-Nicanor, A.; Huanca-Mamani, W.; Montero-Tavera, V.; Tovar-Aguilar, A.; Ramírez-Pimentel, J.-G.; Durán-Figueroa, N.-V.; et al. LEA13 and LEA30 Are Involved in Tolerance to Water Stress and Stomata Density in Arabidopsis thaliana. Plants 2021, 10, 1694. https://doi.org/10.3390/plants10081694
López-Cordova A, Ramírez-Medina H, Silva-Martinez G-A, González-Cruz L, Bernardino-Nicanor A, Huanca-Mamani W, Montero-Tavera V, Tovar-Aguilar A, Ramírez-Pimentel J-G, Durán-Figueroa N-V, et al. LEA13 and LEA30 Are Involved in Tolerance to Water Stress and Stomata Density in Arabidopsis thaliana. Plants. 2021; 10(8):1694. https://doi.org/10.3390/plants10081694
Chicago/Turabian StyleLópez-Cordova, Abigael, Humberto Ramírez-Medina, Guillermo-Antonio Silva-Martinez, Leopoldo González-Cruz, Aurea Bernardino-Nicanor, Wilson Huanca-Mamani, Víctor Montero-Tavera, Andrea Tovar-Aguilar, Juan-Gabriel Ramírez-Pimentel, Noé-Valentín Durán-Figueroa, and et al. 2021. "LEA13 and LEA30 Are Involved in Tolerance to Water Stress and Stomata Density in Arabidopsis thaliana" Plants 10, no. 8: 1694. https://doi.org/10.3390/plants10081694







