Induction of Conjugation and Zygospore Cell Wall Characteristics in the Alpine Spirogyra mirabilis (Zygnematophyceae, Charophyta): Advantage under Climate Change Scenarios?
Abstract
:1. Introduction
2. Results
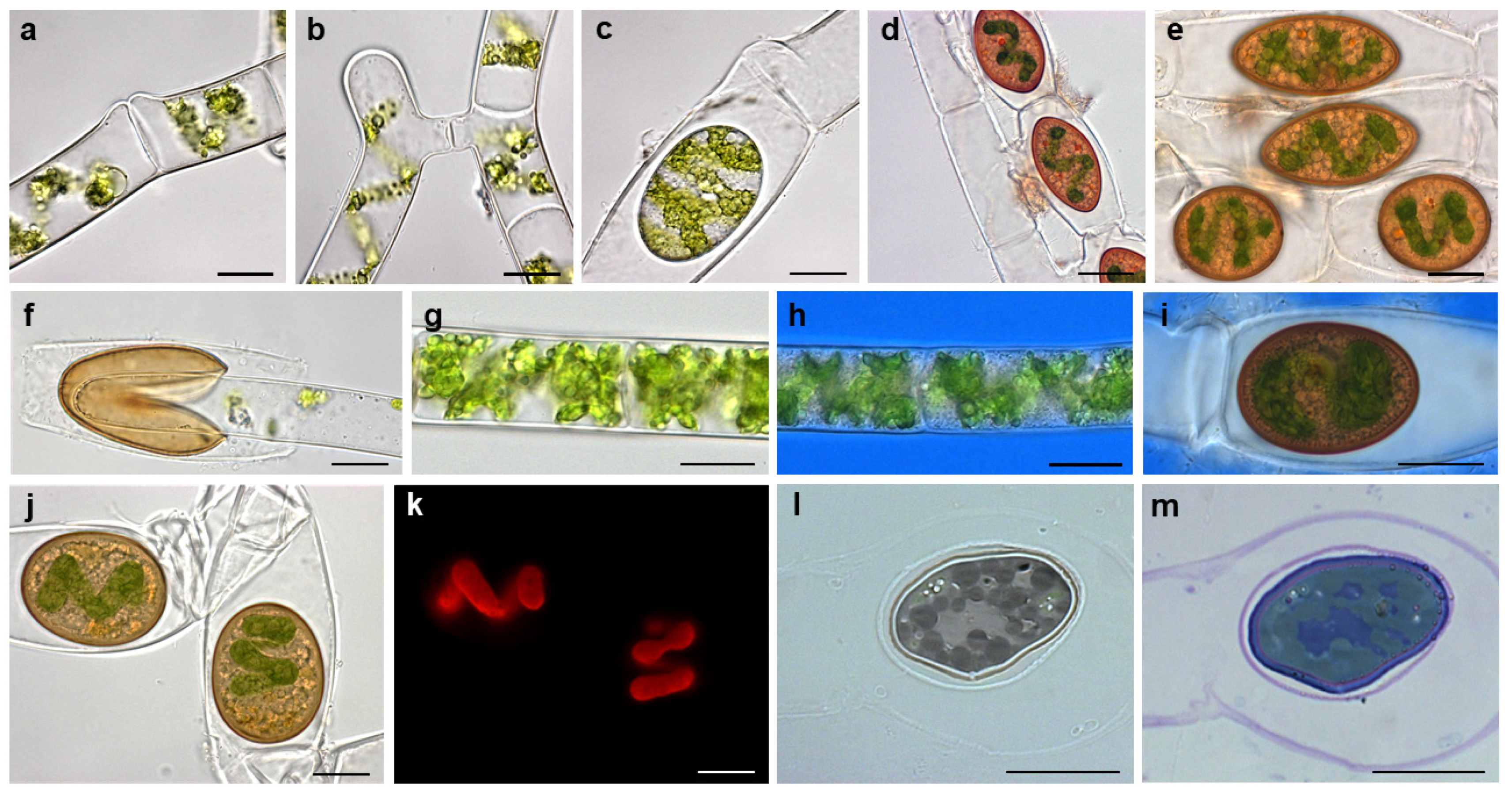
2.1. Morphological Observations on Field Sampled Spirogyra sp. and Establishment of Cultures
2.2. Conjugation in Spirogyra mirabilis Can Be Induced under Laboratory Conditions, but May Be Dependent on Internal Factors
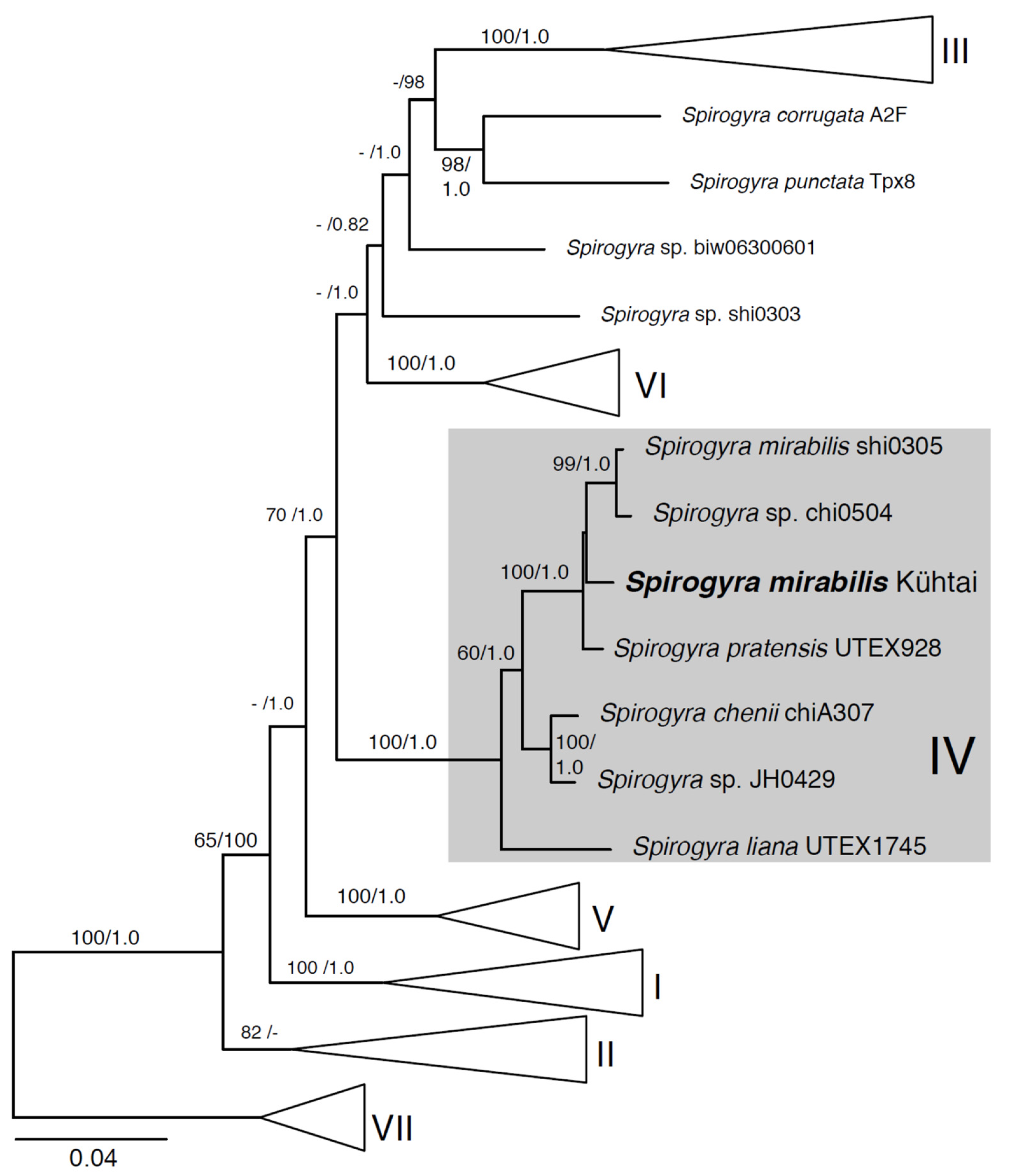
2.3. Phylogenetic Position of the Investigated Spirogyra
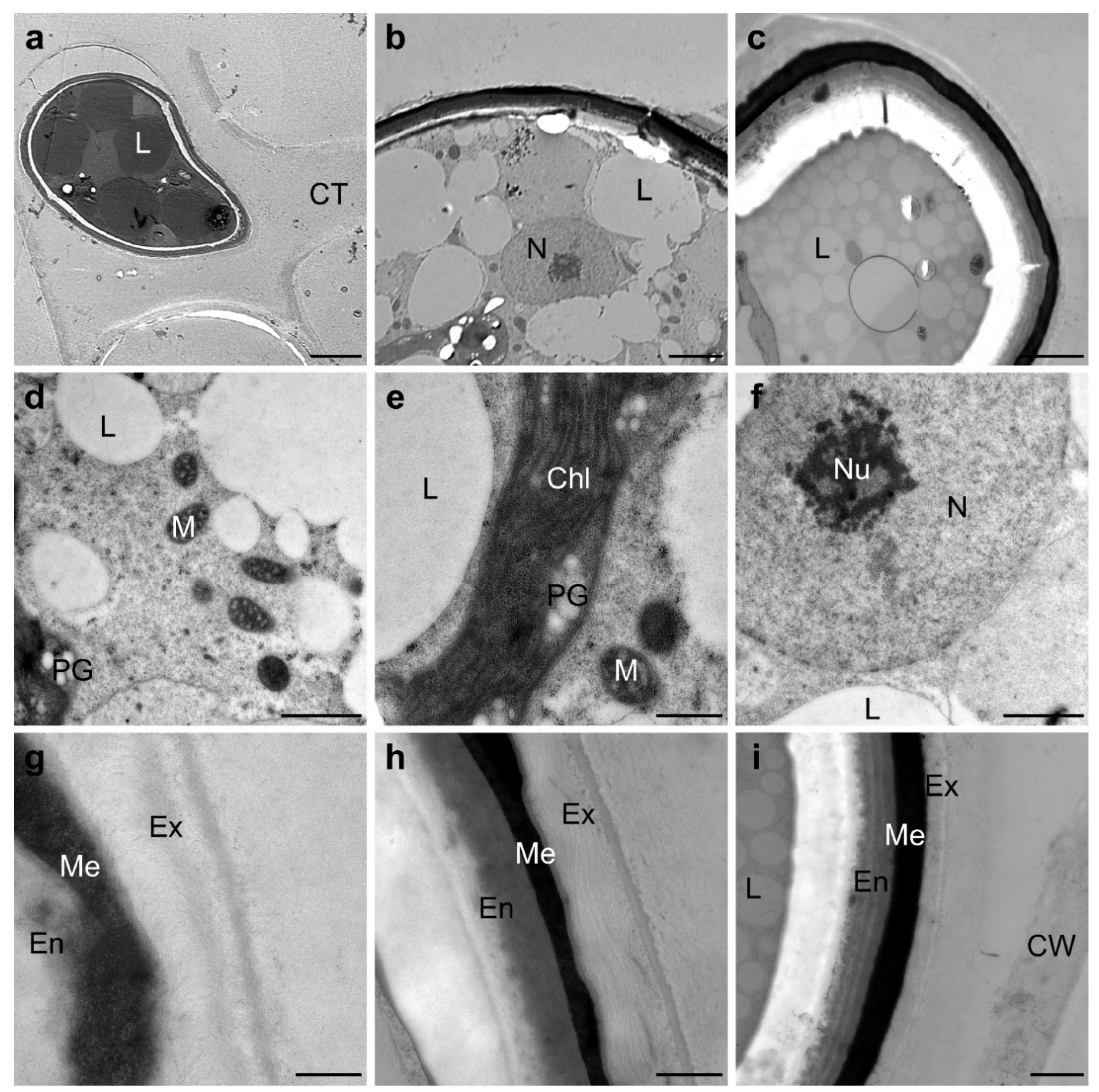
2.4. Spirogyra Zygospore Ultrastructure
2.5. Spirogyra Cell Walls Are Rich in Pectins, Hemicelluloses, and Glycoproteins
2.6. Immunostainings Reveal Arabinogalactan Proteins as Part of the Resistant Zygospore Wall
2.7. Confocal Raman Microscopy Detects Aromatic Compounds Similar to Lycopodium Spore Cell Walls
3. Discussion
3.1. Successful Induction of Conjugation in Spirogyra Isolates
3.2. Phylogenetic Analysis Contradicts Initial Morphological Species Assignment
3.3. The Cell Wall Contains Various Polysaccharides, Glycoproteins, and Aromatic Compounds
3.4. Prominent Role of AGPs and Extensins in Spirogyra
3.5. Complex Arrangement of the Zygospore Cell Wall Suggests Maximal Protection
4. Conclusions
5. Material and Methods
5.1. Sampling Site/Plant Material and Isolation
5.2. Induction of Conjugation
5.3. Light-, Fluorescence, and Confocal Laser Scanning Microscopy
5.4. Phylogenetic Analysis
5.5. High Pressure Freeze Fixation
5.6. Transmission Electron Microscopy
5.7. Cell Wall Extraction and Glycan Micro Array Analysis
5.8. Immunostaining of Selected Cell Wall Components
5.9. Confocal Raman Spectroscopy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AGP | arabinogalactan protein |
| APW | artificial pond water |
| BBM | Bold’s basal medium |
| BI | Bayesian inference |
| HG | Homogalacturonan |
| ML | maximum likelihood |
| PAR | photosynthetically active radiation |
| PBS | phosphate-buffered saline |
| RGI | rhamnogalacturonan I |
| TEM | transmission electron microscopy |
References
- Wodniok, S.; Brinkmann, H.; Glöckner, G.; Heidel, A.J.; Philippe, H.; Melkonian, M.; Becker, B. Origin of land plants: Do conjugating green algae hold the key? BMC Evol Biol. 2011, 11, 104. [Google Scholar] [CrossRef] [Green Version]
- de Vries, J.; Archibald, J.M. Plant evolution: Landmarks on the path to terrestrial life. New Phytol. 2018, 217, 1428–1434. [Google Scholar] [CrossRef]
- Leebens-Mack, J.H.; Barker, M.S.; Carpenter, E.J.; Deyholos, M.K.; Gitzendanner, M.A.; Graham, S.W.; Grosse, I.; Li, Z.; Melkonian, M.; Mirarab, S.; et al. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 2019, 574, 679–685. [Google Scholar] [CrossRef] [Green Version]
- Cheng, S.; Xian, W.; Fu, Y.; Marin, B.; Keller, J.; Wu, T.; Sun, W.; Li, X.; Xu, Y.; Zhang, Y.; et al. Genomes of subaerial Zygnematophyceae provide insights into land plant evolution. Cell 2019, 179, 1057–1067.e14. [Google Scholar] [CrossRef] [PubMed]
- Jiao, C.; Sørensen, I.; Sun, X.; Sun, H.; Behar, H.; Alseekh, S.; Philippe, G.; Lopez, K.P.; Sun, L.; Reed, R.; et al. The Penium margaritaceum genome: Hallmarks of the origins of land plants. Cell 2020, 181, 1097–1111. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.H.; Klochkova, T.A.; Kang, S.H. Notes on freshwater and terrestrial algae from Ny-Alesund, Svalbard (high Arctic sea area). J. Environ. Biol. 2008, 29, 485–491. [Google Scholar] [PubMed]
- Kim, G.H.; Klochkova, T.A.; Han, J.W.; Kang, S.-H.; Choi, H.-G.; Chung, K.W.; kim, S.J. Freshwater and terrestrial algae from Ny-Alesund and Blomstrandhalvoya island (Svalbard). Arctic 2011, 64, 25–31. [Google Scholar] [CrossRef] [Green Version]
- Holzinger, A.; Roleda, M.Y.; Lütz, C. The vegetative arctic freshwater green alga Zygnema is insensitive to experimental UV exposure. Micron 2009, 40, 831–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pichrtová, M.; Hájek, T.; Elster, J. Annual development of mat-forming conjugating green algae Zygnema spp. in hydro-terrestrial habitats in the Arctic. Polar Biol. 2016, 39, 1653–1662. [Google Scholar] [CrossRef]
- Skácelová, K.; Barták, M.; Coufalík, P.; Nývlt, D.; Trnková, K. Biodiversity of freshwater algae and cyanobacteria on deglaciated northern part of James Ross Island, Antarctica. A preliminary study. Czech Polar Rep. 2013, 3, 93–106. [Google Scholar] [CrossRef]
- Kaplan, F.; Lewis, L.A.; Herburger, K.; Holzinger, A. Osmotic stress in Arctic and Antarctic strains of the green alga Zygnema (Zygnematales, Streptophyta): Effects on photosynthesis and ultrastructure. Micron 2013, 44, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Rippin, M.; Becker, B.; Holzinger, A. Enhanced desiccation tolerance in mature cultures of the streptophytic green alga Zygnema circumcarinatum revealed by transcriptomics. Plant Cell Physiol. 2017, 58, 2067–2084. [Google Scholar] [CrossRef] [PubMed]
- Rippin, M.; Pichrtová, M.; Arc, E.; Kranner, I.; Becker, B.; Holzinger, A. Metatranscriptomic and metabolite profiling reveals vertical heterogeneity within a Zygnema green algal mat from Svalbard (High Arctic). Environ. Microbiol. 2019, 21, 4283–4299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Vries, J.; Curtis, B.A.; Gould, S.B.; Archibald, J.M. Embryophyte stress signaling evolved in the algal progenitors of land plants. Proc. Natl. Acad. Sci. USA 2018, 115, E3471–E3480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pichrtová, M.; Remias, D.; Lewis, L.; Holzinger, A. Changes in phenolic compounds and cellular ultrastructure of Arctic and Antarctic strains of Zygnema (Zygnematophyceae, Streptophyta) after exposure to experimentally enhanced UV to PAR ratio. Microb. Ecol. 2013, 65, 68–83. [Google Scholar] [CrossRef] [Green Version]
- Pichrtová, M.; Kulichová, J.; Holzinger, A. Nitrogen limitation and slow drying induce desiccation tolerance in conjugating green algae (Zygnematophyceae, Streptophyta) from polar habitats. PLoS ONE 2014, 9, e113137. [Google Scholar] [CrossRef] [PubMed]
- Holzinger, A.; Albert, A.; Aigner, S.; Uhl, J.; Schmitt-Kopplin, P.; Pichrtová, M. Arctic, Antarctic, and temperate green algae Zygnema spp. under UV-B stress: Vegetative cells perform better than pre-akinetes. Protoplasma 2018, 255, 1239–1252. [Google Scholar] [CrossRef] [Green Version]
- Trumhová, K.; Holzinger, A.; Obwegeser, S.; Neuner, G.; Pichrtová, M. The conjugating green alga Zygnema sp. (Zygnematophyceae) from the Arctic shows high frost tolerance in mature cells (pre-akinetes). Protoplasma 2019, 256, 1681–1694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arc, E.; Pichrtová, M.; Kranner, I.; Holzinger, A. Pre-akinete formation in Zygnema sp. from polar habitats is associated with metabolite re-arrangement. J. Exp. Bot. 2020, 71, 3314–3322. [Google Scholar] [CrossRef] [Green Version]
- Pichrtová, M.; Arc, E.; Stöggl, W.; Kranner, I.; Hájek, T.; Hackl, H.; Holzinger, A. Formation of lipid bodies and changes in fatty acid composition upon pre-akinete formation in Arctic and Antarctic Zygnema (Zygnematophyceae, Streptophyta) strains. FEMS Microbiol. Ecol. 2016, 92, fiw096. [Google Scholar] [CrossRef] [Green Version]
- Pichrtová, M.; Holzinger, A.; Kulichová, J.; Ryšánek, D.; Šoljaková, T.; Trumhová, K.; Nemcova, Y. Molecular and morphological diversity of Zygnema and Zygnemopsis (Zygenmatophyceae, Streptophyta) on Svalbard (High Arctic). Eur. J. Phycol. 2018, 53, 492–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadlubowska, J.Z. Süßwasserflora von Mitteleuropa, Bd. 16: Chlorophyta VIII, 1st ed.; Springer Spektrum: Wiesbaden, Germany, 1984. [Google Scholar]
- Permann, C.; Herburger, K.; Niedermeier, M.; Felhofer, M.; Gierlinger, N.; Holzinger, A. Cell wall characteristics during sexual reproduction of Mougeotia sp. (Zygnematophyceae) revealed by electron microscopy, glycan microarrays and RAMAN spectroscopy. Protoplasma 2021, 1–15. [Google Scholar] [CrossRef]
- Poulícková, A.; Zizka, Z.; Hasler, P.; Benada, O. Zygnematalean zygospores: Morphological features and use in species identification. Folia Microbiol. 2007, 52, 135–145. [Google Scholar] [CrossRef]
- Czurda, V. Experimentelle Untersuchungen über die Sexualitätsverhältnisse der Zygnemalen. Beih. Bot. Zentralbl. 1930, 47, 15–68. [Google Scholar]
- Stancheva, R.; Sheath, R.; Hall, J.D. Systematics of the genus Zygnema (Zygnematophyceae, Charophyta) from Californian watersheds. J. Phycol. 2012, 48, 409–422. [Google Scholar] [CrossRef]
- Stancheva, R.; Hall, J.D.; Mccourt, R.M.; Sheath, R.G. Identity and phylogenetic placement of Spirogyra species (Zygnematophyceae, Charophyta) from California streams and elsewhere. J. Phycol. 2013, 49, 588–607. [Google Scholar] [CrossRef] [PubMed]
- Schagerl, M.; Zwirn, M. A brief introduction into the morphological species concept of Spirogyra and emanating problems. Algol. Stud. 2015, 148, 67–86. [Google Scholar] [CrossRef]
- Gauch, H.G. Studies on the Life Cycle and Genetics of Zygnema. Master’s Thesis, Cornell University, Ithaca, NY, USA, 1966. [Google Scholar]
- Grote, M. On sexual reproduction of Spirogyra majuscula (green algae) under define culture conditions. Z. Pflanzenphysiol. 1977, 83, 95–107. [Google Scholar] [CrossRef]
- Simons, J.; Van Beem, A.P.; de Vries, P.J.R. Induction of conjugation and spore formation in species of Spirogyra (Chlorophyceae, Zygnematales). Acta Bot. Neerl. 1984, 33, 323–334. [Google Scholar] [CrossRef]
- Stabenau, H.; Säftel, W. Induction of conjugation in Mougeotia. Can. J. Bot. 1989, 67, 198–199. [Google Scholar] [CrossRef]
- Yamashita, T.; Sasaki, K. Conditions for the induction of the mating process and changes in contents of carbohydrates and nitrogen compounds during the mating process of Spirogyra. J. Fac. Sci. Hokkaido Univ. Ser. V 1979, 11, 279–287. [Google Scholar]
- Kim, Y.H.; Kim, G.H. A Biology of the Green Algae, Spirogyra; Gaesin: Chungju, Korean, 2002. [Google Scholar]
- Allen, M.A. The Biology of a Species Complex in Spirogyra. Ph.D. Thesis, Indiana University, Bloomington, IN, USA, 1958. [Google Scholar]
- Ikegaya, H.; Nakase, T.; Iwata, K.; Tsuchida, H.; Sonobe, S.; Shimmen, T. Studies on conjugation of Spirogyra using monoclonal culture. J. Plant Res. 2012, 125, 457–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Sheek, M.; Gharieb, M.M.; Schagerl, M. Induction of sexual reproduction and zygospore patterns in the filamentous green alga Spirogyra (Conjugatophyceae: Zygnrmatales). J. BioSci. Biotechnol. 2018, 6, 147–154. [Google Scholar]
- Takano, T.; Higuchi, S.; Ikegaya, H.; Matsuzaki, R.; Kawachi, M.; Takahashi, F.; Nozaki, H. Identification of 13 Spirogyra species (Zygnemataceae) by traits of sexual reproduction induced under laboratory culture conditions. Sci. Rep. 2019, 9, 7458. [Google Scholar] [CrossRef] [PubMed]
- Zwirn, M.; Chen, C.; Uher, B.; Schagerl, M. Induction of sexual reproduction in Spirogyra clones-does an universal trigger exist? Fottea Olomouc. 2013, 13, 77–85. [Google Scholar] [CrossRef]
- Zhou, H.; von Schwartzenberg, K. Zygnematophyceae—From living Algae collections to the establishment of future models. J. Exp. Bot. 2020, 71, 296–3304. [Google Scholar] [CrossRef]
- de Vries, J.; de Vries, S.; Curtis, B.A.; Zhou, H.; Penny, S.; Feussner, K.; Pinto, D.M.; Steinert, M.; Cohen, A.M.; von Schwartzenberg, K.; et al. Heat stress response in the closest algal relatives of land plants reveals conserved stress signaling circuits. Plant J. 2020, 103, 1025–1048. [Google Scholar] [CrossRef]
- Ju, C.; van de Poel, B.; Cooper, E.D.; Thierer, J.H.; Gibbons, T.R.; Delwiche, C.F.; Chang, C. Conservation of ethylene as a plant hormone over 450 million years of evolution. Nat. Plants 2015, 1, 14004. [Google Scholar] [CrossRef]
- Van de Poel, B.; Cooper, E.D.; van der Straeten, D.; Chang, C.; Delwiche, C.F. Transcriptome profiling of the green alga Spirogyra pratensis (Charophyta) suggests an ancestral role for ethylene in cell wall metabolism, photosynthesis, and abiotic stress responses. Plant Physiol. 2016, 172, 533–545. [Google Scholar] [CrossRef] [Green Version]
- Domozych, D.S.; Ciancia, M.; Fangel, J.U.; Mikkelsen, M.D.; Ulvskov, P.; Willats, W.G.T. The cell walls of green algae: A journey through evolution and diversity. Front. Plant. Sci. 2012, 3, 82. [Google Scholar] [CrossRef] [Green Version]
- Popper, Z.A.; Michel, G.; Hervé, C.; Domozych, D.S.; Willats, W.G.T.; Tuohy, M.G.; Kloareg, B.; Stengel, D.B. Evolution and diversity of plant cell walls: From algae to flowering plants. Annu. Rev. Plant Biol. 2011, 62, 567–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikegaya, H.; Hayashi, T.; Kaku, T.; Iwata, K.; Sonobe, S.; Shimmen, T. Presence of xyloglucan-like polysaccharide in Spirogyra and possible involvement in cell-cell attachment. Phycol. Res. 2008, 56, 216–222. [Google Scholar] [CrossRef]
- Palacio-López, K.; Tinaz, B.; Holzinger, A.; Domozych, D.S. Arabinogalactan proteins and the extracellular matrix of charophytes: A sticky business. Front. Plant Sci. 2019, 10, 447. [Google Scholar] [CrossRef]
- Yoon, M.; Kim, M.K.; Kim, G.H. Conjugation process in Spirogyra varians monitored with FITC-lectins (Zygnemataceae, Chlorophyta). ALGAE 2009, 24, 39–45. [Google Scholar] [CrossRef]
- Blokker, P. Structural analysis of resistant polymers in extant algae and ancient sediments. Geol. Ultraiectina 2000, 193, 1–145. [Google Scholar]
- Fry, S.C.; York, W.S.; Albersheim, P.; Darvill, A.; Hayashi, T.; Joseleau, J.P.; Kato, Y.; Lorences, E.P.; Maclachlan, G.A.; McNeil, M.; et al. An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol. Plant. 1993, 89, 1–3. [Google Scholar] [CrossRef]
- Agarwal, P.; Landolt, D. Protection of steel by aromatic carboxylic acid corrosion inhibitors. MSF 1998, 289–292, 1229–1236. [Google Scholar] [CrossRef]
- Varsányi, G. Vibrational Spectra of Benzene Derivatives; Academic Press: Cambridge, MA, USA, 1969. [Google Scholar]
- Pati, Y.C.; Rezaiifar, R.; Krishnaprasad, P.S. Orthogo-nal matching pursuit: Recursive function approximationwith applications to wavelet decomposition. In Proceedings of the Conference Record of The Twenty-Seventh Asilomar Conference on Signals, Systems and Computers, Pacific Grove, CA, USA, 1–3 November 1993. [Google Scholar]
- Barcytė, D.; Pilátová, J.; Mojzes, P.; Nedbalová, L. The Arctic Cylindrocystis (Zygnematophyceae, Streptophyta) green algae are genetically and morphologically diverse and exhibit effective accumulation of polyphosphate. J. Phycol. 2020, 56, 217–232. [Google Scholar] [CrossRef]
- Chen, C.; Barfuss, M.H.J.; Pröschold, T.; Schagerl, M. Hidden genetic diversity in the green alga Spirogyra (Zygnematophyceae, Streptophyta). BMC Evol. Biol. 2012, 12, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gontcharov, A.A.; Marin, B.; Melkonian, M. Are combined analyses better than single gene phylogenies? A case study using SSU rDNA and rbcL sequence comparisons in the Zygnematophyceae (Streptophyta). Mol. Biol. Evol. 2004, 21, 612–624. [Google Scholar] [CrossRef] [PubMed]
- Pascher, A. Die Süsswasser-Flora Deutschlands, Österreichs und der Schweiz; Gustav Fischer: Jena, Germany, 1913. [Google Scholar]
- Prescott, G.W. Algae of the Western Great Lakes Area; Wm, C., Ed.; Brown Co.: Dubuque, IA, USA, 1962. [Google Scholar]
- Geitler, L. Zur Morphologie von Spirogyra mirabilis. Österreich. Bot. Z. 1970, 118, 297–300. [Google Scholar] [CrossRef]
- Popper, Z.A.; Fry, S.C. Primary cell wall composition of bryophytes and charophytes. Ann. Bot. 2003, 91, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Sørensen, I.; Pettolino, F.A.; Bacic, A.; Ralph, J.; Lu, F.; O’Neill, M.A.; Fei, Z.; Rose, J.K.C.; Domozych, D.S.; Willats, W.G.T. The charophycean green algae provide insights into the early origins of plant cell walls. Plant J. 2011, 68, 201–211. [Google Scholar] [CrossRef]
- Domozych, D.S.; Sørensen, I.; Popper, Z.A.; Ochs, J.; Andreas, A.; Fangel, J.U.; Pielach, A.; Sacks, C.; Brechka, H.; Ruisi-Besares, P.; et al. Pectin metabolism and assembly in the cell gall of the Charophyte Green alga Penium margaritaceum. Plant Physiol. 2014, 165, 105–118. [Google Scholar] [CrossRef] [Green Version]
- Herburger, K.; Ryan, L.M.; Popper, Z.; Holzinger, A. Localisation and substrate specificities of transglycanases in charophyte algae relate to development and morphology. J. Cell Sci. 2018, 131, jcs203208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harolt, J.; Moestrup, O.; Ulvskov, P. Why plants were terrestrial from the beginning. Trends Plant. Sci. 2016, 21, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, M.D.; Harholt, J.; Westereng, B.; Domozych, D.; Fry, S.C.; Johansen, I.E.; Fangel, J.U.; Łężyk, M.; Feng, T.; Nancke, L.; et al. Ancient origin of fucosylated xyloglucan in charophycean green algae. Commun. Biol. 2021, 4, 754. [Google Scholar] [CrossRef]
- Popper, Z.A. Evolution and diversity of green plant cell walls. Curr. Opin. Plant Biol. 2008, 11, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Marcus, S.E.; Blake, A.W.; Benians, T.A.; Lee, K.J.; Poyser, C.; Donaldson, L.; Leroux, O.; Rogowski, A.; Petersen, H.L.; Boraston, A.; et al. Restricted access of proteins to mannan polysaccharides in intact plant cell walls. Plant J. 2010, 64, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Lamport, D.T.A.; Kieliszewski, M.J.; Chen, Y.; Cannon, M.C. Role of the extensin superfamily in primary cell wall architecture. Plant Physiol. 2011, 156, 11–19. [Google Scholar] [CrossRef] [Green Version]
- Herburger, K.; Xin, A.; Holzinger, A. Homogalacturonan accumulation in cell walls of the green alga Zygnema sp. (Charophyta) increases desiccation resistance. Front. Plant Sci. 2019, 10, 540. [Google Scholar] [CrossRef] [Green Version]
- Pereira, A.M.; Lopes, A.L.; Coimbra, S. Arabinogalactan Proteins as Interactors along the Crosstalk between the Pollen Tube and the Female Tissues. Front. Plant Sci. 2016, 7, 1895. [Google Scholar] [CrossRef]
- Blokker, P.; Schouten, S.; van den Enden, H.; Leeuw, J.W.; Hatcher, P.G.; Sinninghe Damste, J.S. Chemical structure of algaenans from the fresh water algae Tetraedron minimum, Scenedesmus communis and Pediastrum boryanum. Org. Geochem. 1998, 29, 1453–1468. [Google Scholar] [CrossRef]
- Bischoff, H.W.; Bold, H.C. Some Soil Algae from Enchanted Rock and Related Algal Species, Phycological Sudies IV; Publ. No. 6318; University of Texas: Austin, TX, USA, 1963; pp. 1–95. [Google Scholar]
- Sakayama, H.; Nozaki, H.; Kasaki, H.; Hara, Y. Taxonomic re-examination of Nitella (Charales, Charophyceae) from Japan, based on microscopical studies of oospore wall ornamentation and rbcL gene sequences. Phycologia 2002, 41, 397–408. [Google Scholar] [CrossRef] [Green Version]
- Shimada, A.; Kanai, S.; Maruyama, T. Partial sequence of ribulose-1,5-biphosphate carboxylase/oxygenase and phylogenie of Prochloran and Prochlorococcus (Prochlorales). J. Mol. Evol. 1995, 40, 671–677. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aichinger, N.; Lütz-Meindl, U. Organelle interactions and possible degradation pathways visualized in high-pressure frozen algal cells. J. Microsc. 2005, 219, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Moller, I.; Susan, E.M.; Haeger, A.; Verhertbruggen, Y.; Verhoef, R.; Schols, H.; Ulvskov, P.; Mikkelsen, J.D.; Knox, J.P.; Willats, W. High-throughput screening of monoclonal antibodies against plant cell wall glycans by hierarchical clustering of their carbohydrate microarray binding profiles. Glycoconj. J. 2007, 25, 37–48. [Google Scholar] [CrossRef] [Green Version]
- Kračun, S.K.; Fangel, J.U.; Rydahl, M.G.; Pedersen, H.L.; Vidal-Melgosa, S.; Willats, W.G.T. Carbohydrate Microarray Technology Applied to High-Throughput Mapping of Plant Cell Wall Glycans Using Comprehensive Microarray Polymer Profiling (CoMPP). In High-Throughput Glycomics and Glycoproteomics. Methods in Molecular Biology; Laucm, G., Wuhrer, M., Eds.; Humana Press: New York, NY, USA, 2017; Volume 1503. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Permann, C.; Herburger, K.; Felhofer, M.; Gierlinger, N.; Lewis, L.A.; Holzinger, A. Induction of Conjugation and Zygospore Cell Wall Characteristics in the Alpine Spirogyra mirabilis (Zygnematophyceae, Charophyta): Advantage under Climate Change Scenarios? Plants 2021, 10, 1740. https://doi.org/10.3390/plants10081740
Permann C, Herburger K, Felhofer M, Gierlinger N, Lewis LA, Holzinger A. Induction of Conjugation and Zygospore Cell Wall Characteristics in the Alpine Spirogyra mirabilis (Zygnematophyceae, Charophyta): Advantage under Climate Change Scenarios? Plants. 2021; 10(8):1740. https://doi.org/10.3390/plants10081740
Chicago/Turabian StylePermann, Charlotte, Klaus Herburger, Martin Felhofer, Notburga Gierlinger, Louise A. Lewis, and Andreas Holzinger. 2021. "Induction of Conjugation and Zygospore Cell Wall Characteristics in the Alpine Spirogyra mirabilis (Zygnematophyceae, Charophyta): Advantage under Climate Change Scenarios?" Plants 10, no. 8: 1740. https://doi.org/10.3390/plants10081740
APA StylePermann, C., Herburger, K., Felhofer, M., Gierlinger, N., Lewis, L. A., & Holzinger, A. (2021). Induction of Conjugation and Zygospore Cell Wall Characteristics in the Alpine Spirogyra mirabilis (Zygnematophyceae, Charophyta): Advantage under Climate Change Scenarios? Plants, 10(8), 1740. https://doi.org/10.3390/plants10081740





