Quinoa Phenotyping Methodologies: An International Consensus
Abstract
:1. Introduction
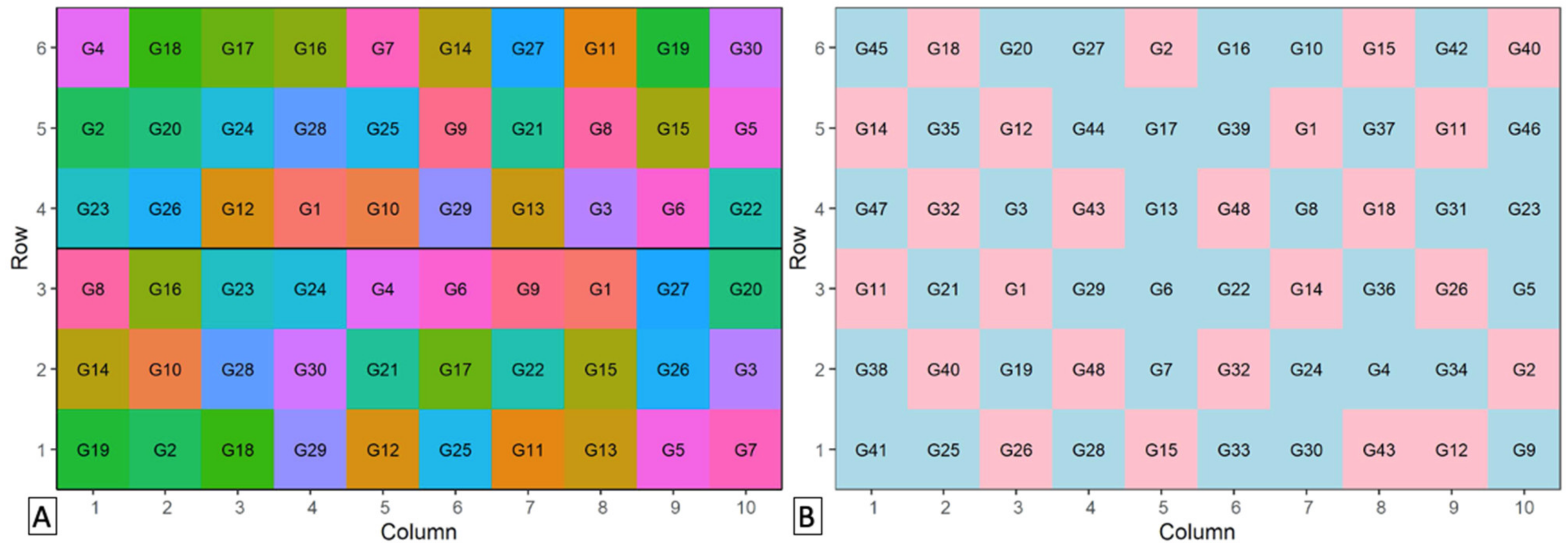
2. Quinoa Database
3. Germplasm Selection
4. Experimental Design and Crop Management
4.1. Planting
4.2. Irrigation
4.3. Fertilization
4.4. Weeding, Pest and Disease Controls
5. Environmental Variables
6. Observations during Growth
6.1. Phenology over Time
6.2. Radiation Capture and Efficiency of Use
6.3. Unmanned Aerial Vehicle-Based Phenotyping
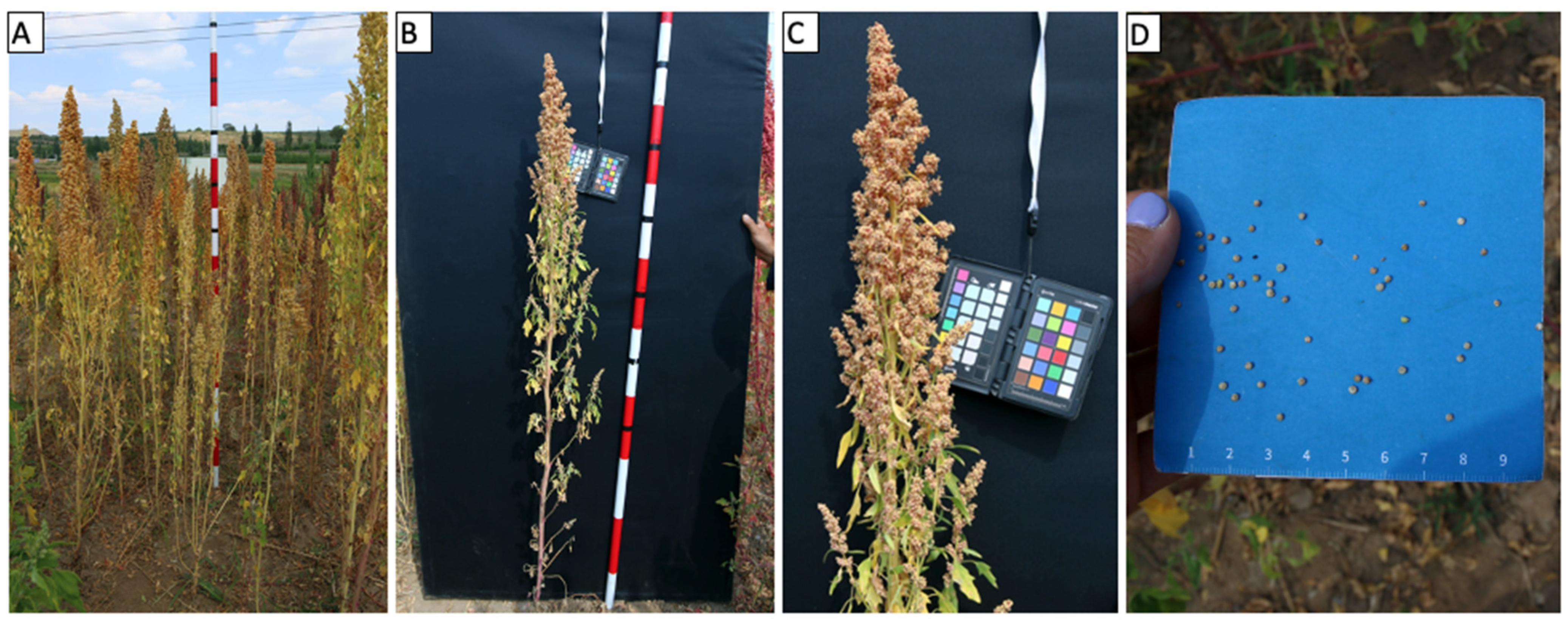
7. Phenotyping of Mature Plants
7.1. Assessing the Quality of Phenotypic Data
7.2. Plot-Level Phenotypes
7.2.1. Plot Population Homogeneity
- 1: Most plants are the same (up to 10% different).
- 3: Over half of plants are the same (10–30% different).
- 5: Less than half of plants are the same (30–50% different).
- 7: Over 50% of the plants are different, completely mixed plot; will need to be excluded from analysis.
7.2.2. Plot Coverage
- 1: Up to 20% of the plot is covered, plant establishment is very poor.
- 3: Less than half of the plot is covered, ~30% (20–40%).
- 5: Around half of the plot is covered, ~50% (40–60%).
- 7: Over half of the plot is covered, ~70% (60–80%).
- 9: Over 80% of the plot is covered, plant establishment is very good.
7.2.3. Stem Breakage Incidence
- 1: Up to 20% of the plot is affected.
- 3: Up to half of the plot is affected, ~30% (20–40%).
- 5: Around half of the plot is affected, ~50% (40–60%).
- 7: Over half of the plot is affected, ~70% (60–80%).
- 9: Over 80% of the plot is affected.
7.2.4. Stem Lodging and Stem Angle
- 1 < 22.5° inclination or deviation of the stem from the vertical (i.e., most plants are upright).
- 3 < 45°.
- 5 < 67.5°.
- 7 < 90° (i.e., most plants are on or very close to the ground).
7.2.5. Panicle Axis Angle
- 1 < 45° inclination or deviation of the panicle from the vertical (i.e., most panicles are upright).
- 3 < 90°.
- 5 < 135°.
- 7 < 180° (i.e., most panicles are pointing towards the ground).
7.2.6. Stem Lying Incidence
7.2.7. Growth Habit
- 1: Not branched at base, usually with a clearly defined terminal panicle.
- 3: Some branching from the base; no significant panicles on branches in the basal area (thus, this is not worth harvesting).
- 5: Branching from the base with more significant panicles.
- 7: Main panicle is difficult to identify.
7.2.8. Branchiness
- 1: Low number or no secondary branches.
- 3: Some branches (30–50% of the primary branch length has secondary branching).
- 5: Branched (50–70% of the primary branch length has secondary branching).
- 7: Highly branched (above 70% of the primary branch length has secondary branching).
7.3. Plant-Level Phenotypes
7.3.1. Quantitative Plant-Level Phenotypes
Plant Height
Panicle Length
Stem Diameter
Number of Significant Panicles
Categorical Plant-Level Phenotypes
Seed Shattering
- 1: No seeds falling.
- 3: Some seeds falling.
- 5: Many seeds falling.
- 7: Majority of seeds is falling, “raining” seeds, and a large number of seeds present on the ground at measurement.
Panicle Shape
- 1: Glomerulate—glomerules with globose shape, resembling “bulbous clusters”.
- 3: Intermediate—panicles have both amarantiform and glomerulate traits, resembling fingers with glomerules.
- 5: Amarantiform—glomerules with elongated shape, resembling “fingers”.
Panicle Density
- 1: Lax (loose)—glomerules sparsely spaced, panicle axes easily visible.
- 3: Intermediate—glomerules tighter but with panicle axes still visible.
- 5: Primary axis rarely visible.
- 7: Compact—glomerules tightly packed, no panicle axes visible.
Panicle Leafiness
- 1: Leaves are present in less than one-third of the panicles.
- 3: Leaves are present in more than one-third but less than three-fourths of the primary, sporadic, and not dense panicles.
- 5: Leaves present in three-fourths to of the entire primary axis, frequent but not dense leafiness.
- 7: Many leaves present throughout the primary axis.
Panicle Color
- Green (13);
- Green with Purple (16);
- Pink/Purple/Red (4);
- Orange/Yellow (5);
- Dark colored (7);
- Beige/White (i.e., no pigmentation, mostly for mature plants) (15).
Stem Color
- Green (13);
- Red (4);
- No pigmentation (beige, white, yellow) (15).
Stem Striae and Axil Pigmentation
- Presence (1);
- Absence (0).
Stem Leaf Shape Characteristics
- Rhomboidal (1);
- Triangular (2).
- Entire (1);
- Dentate (3);
- Serrate (5).
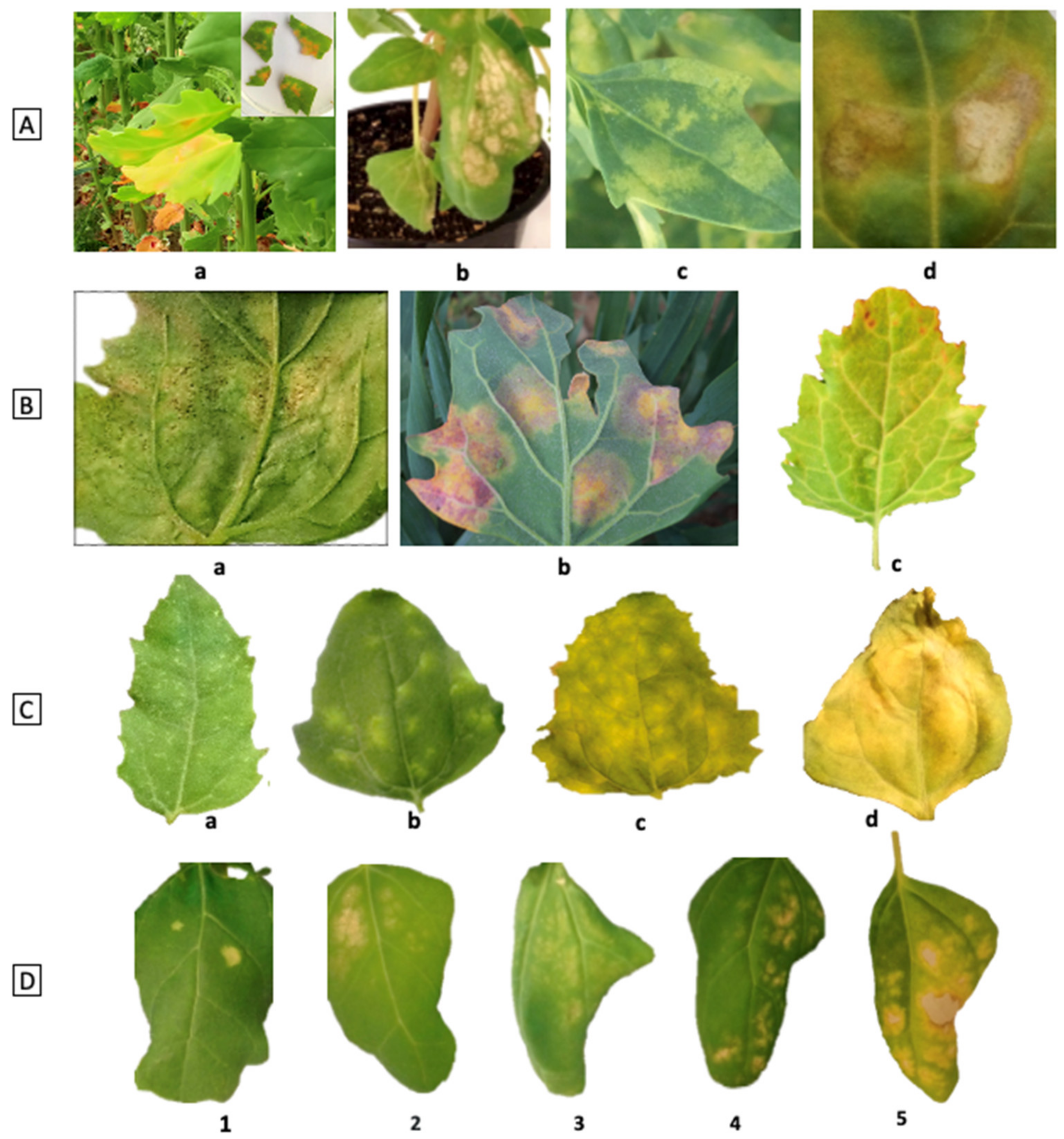
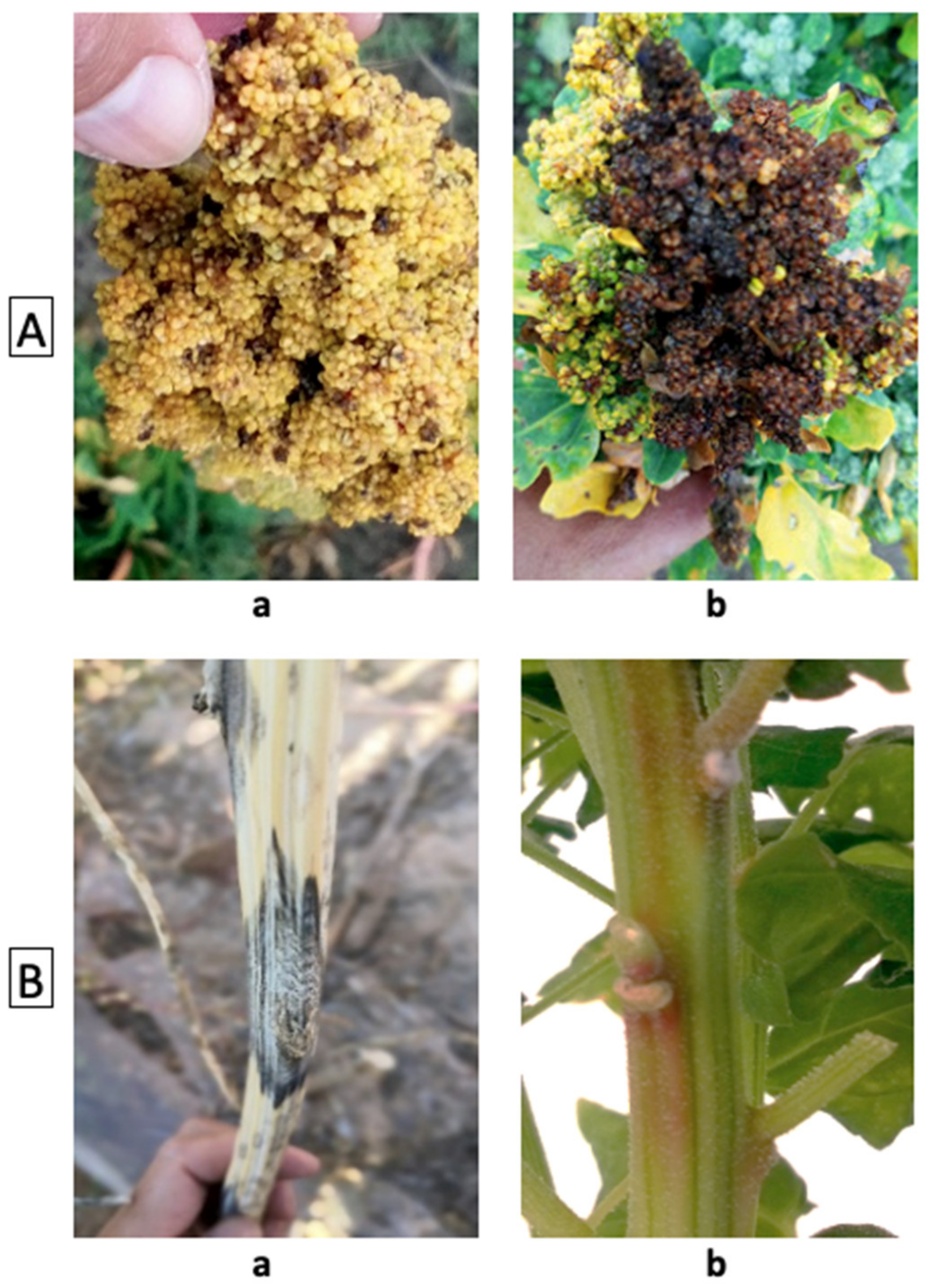
8. Phenotyping of Disease
9. Harvest and Post-Harvest
9.1. Harvest Protocols
9.2. Seed Phenotyping
9.2.1. In-Field Seed Morphology Descriptors
9.2.2. Seed Scanning
9.2.3. Seed Nutritional Phenotyping
Near-Infrared Spectroscopy
NIR—An Example of Calibration Development for Quinoa
The Nutritional Phenotyping Pipeline at Washington State University
9.2.4. Detection of Saponins in Quinoa
9.2.5. Quinoa Seed Longevity
10. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cho, S.J.; McCarl, B.A. Climate change influences on crop mix shifts in the United States. Sci. Rep. 2017, 7, 40845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, M.; Altdorff, D.; Li, P.; Galagedara, L.; Holden, J.; Unc, A. Northward shift of the agricultural climate zone under 21st-century global climate change. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Challinor, A.; Watson, J.; Lobell, D.; Howden, S.; Smith, D.R.; Chhetri, N. A meta-analysis of crop yield under climate change and adaptation. Nat. Clim. Chang. 2014, 4, 287–291. [Google Scholar] [CrossRef]
- Burchi, F.; Fanzo, J.; Frison, E. The Role of Food and Nutrition System Approaches in Tackling Hidden Hunger. Int. J. Environ. Res. Public Heal. 2011, 8, 358–373. [Google Scholar] [CrossRef]
- Boushey, C.J.; Coulston, A.M.; Delahanty, L.; Ferruzzi, M. Nutrition in the Prevention and Treatment of Disease, 4th ed.; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar]
- IPCC. 2019: Climate Change and Land: An IPCC Special Report on Climate Change, Desertification, Land Degradation, Sustainable Land Management, Food Security, and Greenhouse Gas Fluxes in Terrestrial Ecosystems, CH5; Shukla, P.R., Skea, J., Calvo Buendia, E., Masson-Delmotte, V., Pörtner, H.-O., Roberts, D.C., Zhai, P., Slade, R., Connors, S., van Diemen, R., et al., Eds.; IPCC: Geneva, Switzerland, 2019. [Google Scholar]
- Bazile, D.; Bertero, D.; Nieto, C. State of the Art Report of Quinoa in the World in 2013; FAO & CIRAD: Rome, Italy, 2015. [Google Scholar]
- Ruiz, K.B.; Biondi, S.; Oses, R.; Acuña-Rodríguez, I.S.; Antognoni, F.; Martinez-Mosqueira, E.A.; Coulibaly, A.; Canahua-Murillo, A.; Pinto, M.; Zurita-Silva, A.; et al. Quinoa biodiversity and sustainability for food security under climate change. A review. Agron. Sustain. Dev. 2014, 34, 349–359. [Google Scholar] [CrossRef] [Green Version]
- United Nations. SD Goal 2 Department of Economic and Social Affairs. Retrieved 22 December 2020. Available online: https://sdgs.un.org/goals/goal2 (accessed on 15 August 2021).
- FAO Secretariat, 2013 International Year of Quinoa. Distribution and Production. Retrieved 22 December 2020. Available online: http://www.fao.org/quinoa-2013/what-is-quinoa/distribution-and-production/en/ (accessed on 15 August 2021).
- Alandia, G.; Rodriguez, J.; Jacobsen, S.-E.; Bazile, D.; Condori, B. Global expansion of quinoa and challenges for the Andean region. Glob. Food Secur. 2020, 26, 100429. [Google Scholar] [CrossRef]
- Angeli, V.; Silva, P.M.; Massuela, D.C.; Khan, M.W.; Hamar, A.; Khajehei, F.; Graeff-Hönninger, S.; Piatti, C. Quinoa (Chenopodium quinoa Willd.): An Overview of the Potentials of the “Golden Grain” and Socio-Economic and Environmental Aspects of Its Cultivation and Marketization. Foods 2020, 9, 216. [Google Scholar] [CrossRef] [Green Version]
- Bazile, D.; Pulvento, C.; Verniau, A.; Al-Nusairi, M.S.; Ba, D.; Breidy, J.; Hassan, L.; Mohammed, M.; Mambetov, O.; Otambekova, M.; et al. Worldwide Evaluations of Quinoa: Preliminary Results from Post International Year of Quinoa FAO Projects in Nine Countries. Front. Plant. Sci. 2016, 7, 850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rey, E.; Jarvis, D.E. Structural and Functional Genomics of Chenopodium quinoa. In The Quinoa Genome; Schmöckel, S.M., Ed.; Springer: Cham, Switzerland, 2021. [Google Scholar] [CrossRef]
- Jarvis, D.E.; Ho, Y.S.; Lightfoot, D.; Schmöckel, S.M.; Li, B.; Borm, T.J.A.; Ohyanagi, H.; Mineta, K.; Michell, C.; Saber, N.; et al. The genome of Chenopodium quinoa. Nat. Cell Biol. 2017, 542, 307–312. [Google Scholar] [CrossRef] [Green Version]
- Cobb, J.N.; Declerck, G.; Greenberg, A.; Clark, R.; McCouch, S. Next-generation phenotyping: Requirements and strategies for enhancing our understanding of genotype–phenotype relationships and its relevance to crop improvement. Theor. Appl. Genet. 2013, 126, 867–887. [Google Scholar] [CrossRef] [Green Version]
- Reynolds, M.; Chapman, S.; Herrera, L.A.C.; Molero, G.; Mondal, S.; Pequeno, D.N.; Pinto, F.; Pinera-Chavez, F.J.; Poland, J.; Rivera-Amado, C.; et al. Breeder friendly phenotyping. Plant. Sci. 2020, 295, 110396. [Google Scholar] [CrossRef] [PubMed]
- Morton, M.J.L.; Awlia, M.; Al Tamimi, N.; Saade, S.; Pailles, Y.; Negrão, S.; Tester, M. Salt stress under the scalpel–Dissecting the genetics of salt tolerance. Plant. J. 2019, 97, 148–163. [Google Scholar] [CrossRef] [Green Version]
- Khush, G.S. Strategies for increasing the yield potential of cereals: Case of rice as an example. Plant. Breed. 2013, 132, 433–436. [Google Scholar] [CrossRef]
- Reynolds, M.; Foulkes, J.; Furbank, R.; Griffiths, S.; King, J.; Murchie, E.; Parry, M.; Slafer, G. Achieving yield gains in wheat. Plant. Cell Environ. 2012, 35, 1799–1823. [Google Scholar] [CrossRef]
- González, F.G.; Capella, M.; Ribichich, K.F.; Curin, F.; Giacomelli, J.I.; Ayala, F.; Watson, G.; Otegui, M.E.; Chan, R.L. Field-grown transgenic wheat expressing the sunflower gene HaHB4 significantly outyields the wild type. J. Exp. Bot. 2019, 70, 1669–1681. [Google Scholar] [CrossRef] [Green Version]
- González, F.G.; Rigalli, N.; Miranda, P.V.; Romagnoli, M.; Ribichich, K.F.; Trucco, F.; Portapila, M.; Otegui, M.E.; Chan, R.L. An Interdisciplinary Approach to Study the Performance of Second-generation Genetically Modified Crops in Field Trials: A Case Study With Soybean and Wheat Carrying the Sunflower HaHB4 Transcription Factor. Front. Plant. Sci. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Ertiro, B.T.; Olsen, M.; Das, B.; Gowda, M.; Labuschagne, M. Efficiency of indirect selection for grain yield in maize (Zea mays L.) under low nitrogen conditions through secondary traits under low nitrogen and grain yield under optimum conditions. Euphytica 2020, 216, 1–12. [Google Scholar] [CrossRef]
- Fernandes, S.B.; Dias, K.O.D.G.; Ferreira, D.F.; Brown, P.J. Efficiency of multi-trait, indirect, and trait-assisted genomic selection for improvement of biomass sorghum. Theor. Appl. Genet. 2018, 131, 747–755. [Google Scholar] [CrossRef] [Green Version]
- Lozada, D.N.; Godoy, J.V.; Ward, B.P.; Carter, A.H. Genomic Prediction and Indirect Selection for Grain Yield in US Pacific Northwest Winter Wheat Using Spectral Reflectance Indices from High-Throughput Phenotyping. Int. J. Mol. Sci. 2019, 21, 165. [Google Scholar] [CrossRef] [Green Version]
- Musvosvi, C.; Setimela, P.S.; Wali, M.C.; Gasura, E.; Channappagoudar, B.B.; Patil, S.S. Contribution of Secondary Traits for High Grain Yield and Stability of Tropical Maize Germplasm across Drought Stress and Non-Stress Conditions. Agron. J. 2018, 110, 819–832. [Google Scholar] [CrossRef]
- Sra, S.K.; Sharma, M.; Kaur, G.; Sharma, S.; Akhatar, J.; Sharma, A.; Banga, S.S. Evolutionary aspects of direct or indirect selection for seed size and seed metabolites in Brassica juncea and diploid progenitor species. Mol. Biol. Rep. 2019, 46, 1227–1238. [Google Scholar] [CrossRef]
- Zaman, S.U.; Malik, A.I.; Kaur, P.; Ribalta, F.M.; Erskine, W. Waterlogging Tolerance at Germination in Field Pea: Variability, Genetic Control, and Indirect Selection. Front. Plant. Sci. 2019, 10, 953. [Google Scholar] [CrossRef] [Green Version]
- Ziyomo, C.; Bernardo, R. Drought Tolerance in Maize: Indirect Selection through Secondary Traits versus Genomewide Selection. Crop. Sci. 2013, 53, 1269–1275. [Google Scholar] [CrossRef] [Green Version]
- Malosetti, M.; Ribaut, J.-M.; Van Eeuwijk, F.A. The statistical analysis of multi-environment data: Modeling genotype-by-environment interaction and its genetic basis. Front. Physiol. 2013, 4, 44. [Google Scholar] [CrossRef] [Green Version]
- White, J.W.; Andrade-Sanchez, P.; Gore, M.A.; Bronson, K.F.; Coffelt, T.A.; Conley, M.M.; Feldmann, K.A.; French, A.; Heun, J.T.; Hunsaker, D.J.; et al. Field-based phenomics for plant genetics research. Field Crop. Res. 2012, 133, 101–112. [Google Scholar] [CrossRef]
- Bertero, H.; de la Vega, A.; Correa, G.; Jacobsen, S.; Mujica, A. Genotype and genotype-by-environment interaction effects for grain yield and grain size of quinoa (Chenopodium quinoa Willd.) as revealed by pattern analysis of international multi-environment trials. Field Crop. Res. 2004, 89, 299–318. [Google Scholar] [CrossRef]
- Curti, R.; De La Vega, A.; Andrade, A.; Bramardi, S.; Bertero, H. Adaptive responses of quinoa to diverse agro-ecological environments along an altitudinal gradient in North West Argentina. Field Crop. Res. 2016, 189, 10–18. [Google Scholar] [CrossRef]
- Desclaux, D.; Nolot, J.M.; Chiffoleau, Y.; Gozé, E.; Leclerc, C. Changes in the concept of genotype × environment interactions to fit agriculture diversification and decentralized participatory plant breeding: Pluridisciplinary point of view. Euphytica 2008, 163, 533–546. [Google Scholar] [CrossRef]
- Leclerc, C.; D’Eeckenbrugge, G.C. Social Organization of Crop Genetic Diversity. The G × E × S Interaction Model. Divers 2011, 4, 1–32. [Google Scholar] [CrossRef] [Green Version]
- Yan, W.; Hunt, L.; Sheng, Q.; Szlavnics, Z. Cultivar Evaluation and Mega-Environment Investigation Based on the GGE Biplot. Crop. Sci. 2000, 40, 597–605. [Google Scholar] [CrossRef]
- Yan, W.; Kang, M.S. GGE Biplot Analysis. 2002. Available online: https://doi.org/10.1201/9781420040371 (accessed on 15 August 2021).
- Bioversity International; FAO. Descriptors for quinoa (Chenopodium quinoa Willd.) and wild relatives. In Bioversity International, FAO, PROINPA, INIAF and IFAD. 2013. Descriptors for Quinoa (Chenopodium quinoa Willd.) and Wild Relatives; Bioversity International: Rome, Italy; Fundación PROINPA: La Paz, Bolivia; Instituto Nacional de Innovación Agropecuaria y: La Paz, Bolivia, 2013; Volume 1. [Google Scholar]
- CPVO. Protocol for Tests on Distinctness, Uniformity and Stability Chenopodium Quinoa Willd. 2021. Available online: https://cpvo.europa.eu/sites/default/files/documents/chenopodium.pdf (accessed on 15 August 2021).
- Papoutsoglou, E.A.; Faria, D.; Arend, D.; Arnaud, E.; Athanasiadis, I.N.; Chaves, I.; Coppens, F.; Cornut, G.; Costa, B.V.; Ćwiek-Kupczyńska, H.; et al. Enabling reusability of plant phenomic datasets with MIAPPE 1.1. New Phytol. 2020, 227, 260–273. [Google Scholar] [CrossRef] [Green Version]
- Sosa-Zuniga, V.; Brito, V.; Fuentes, F.; Steinfort, U. Phenological growth stages of quinoa (Chenopodium quinoa) based on the BBCH scale. Ann. Appl. Biol. 2017, 171, 117–124. [Google Scholar] [CrossRef]
- Raubach, S.; Kilian, B.; Dreher, K.; Amri, A.; Bassi, F.M.; Boukar, O.; Cook, D.; Cruickshank, A.; Fatokun, C.; El Haddad, N.; et al. From bits to bites: Advancement of the Germinate platform to support prebreeding informatics for crop wild relatives. Crop. Sci. 2020, 61, 1538–1566. [Google Scholar] [CrossRef]
- Shaw, P.D.; Raubach, S.; Hearne, S.; Dreher, K.; Bryan, G.; McKenzie, G.; Milne, I.; Stephen, G.; Marshall, D. Germinate 3: Development of a Common Platform to Support the Distribution of Experimental Data on Crop Wild Relatives. Crop. Sci. 2017, 57, 1259–1273. [Google Scholar] [CrossRef]
- Shrestha, R.; Matteis, L.; Skofic, M.; Portugal, A.; McLaren, G.; Hyman, G.; Arnaud, E. Bridging the phenotypic and genetic data useful for integrated breeding through a data annotation using the Crop Ontology developed by the crop communities of practice. Front. Physiol. 2012, 3, 326. [Google Scholar] [CrossRef] [Green Version]
- UN. Resolution Adopted by the General Assembly on 22 December 2011. 2012. Available online: https://www.un.org/ga/search/view_doc.asp?symbol=A/RES/66/221&referer=/english/&Lang=E (accessed on 15 August 2021).
- Assessment of the International Year of Quinoa 2013 Executive Summary. 2014. Available online: http://www.fao.org/quinoa-2013/iyq/en/ (accessed on 15 August 2021).
- Rojas, W.; Pinto, M.; Alanoca, C.; Gómez-Pando, L.; León-Lobos, P.; Alercia, A.; Diulgheroff, S.; Padulosi, S.; Bazile, D. Quinoa genetic resources and ex situ conservation. In State of the Art Report on Quinoa around the World in 2013; Didier, B., Daniel, B.H., Carlos, N., Eds.; FAO: Rome, Italy, 2015; pp. 56–82. [Google Scholar]
- Bazile, D. Fair and sustainable expansion of traditional crops-lessons from quinoa. Farming Matters 2016, 32.2, 36–39. [Google Scholar]
- Bazile, D.; Jacobsen, S.-E.; Verniau, A. The Global Expansion of Quinoa: Trends and Limits. Front. Plant. Sci. 2016, 7, 622. [Google Scholar] [CrossRef] [Green Version]
- Christensen, S.A.; Pratt, D.B.; Pratt, C.; Nelson, P.T.; Stevens, M.R.; Jellen, E.; Coleman, C.E.; Fairbanks, D.J.; Bonifacio, A.; Maughan, P.J. Assessment of genetic diversity in the USDA and CIP-FAO international nursery collections of quinoa (Chenopodium quinoa Willd.) using microsatellite markers. Plant. Genet. Resour. 2007, 5, 82–95. [Google Scholar] [CrossRef] [Green Version]
- Colque-Little, C.; Abondano, M.C.; Lund, O.S.; Amby, D.B.; Piepho, H.-P.; Andreasen, C.; Schmöckel, S.; Schmid, K. Genetic variation for tolerance to the downy mildew pathogen Peronospora variabilis in genetic resources of quinoa (Chenopodium quinoa). BMC Plant. Biol. 2021, 21, 1–19. [Google Scholar] [CrossRef]
- Tártara, S.M.C.; Manifesto, M.M.; Bramardi, S.J.; Bertero, H.D. Genetic structure in cultivated quinoa (Chenopodium quinoa Willd.), a reflection of landscape structure in Northwest Argentina. Conserv. Genet. 2012, 13, 1027–1038. [Google Scholar] [CrossRef]
- Fuentes, F.; Martinez, E.A.; Hinrichsen, P.; Jellen, E.; Maughan, P.J. Assessment of genetic diversity patterns in Chilean quinoa (Chenopodium quinoa Willd.) germplasm using multiplex fluorescent microsatellite markers. Conserv. Genet. 2009, 10, 369–377. [Google Scholar] [CrossRef]
- Mason, S.L.; Stevens, M.R.; Jellen, E.N.; Bonifacio, A.; Fairbanks, D.J.; Coleman, C.E.; Mccarty, R.R.; Rasmussen, A.G.; Maughan, P.J. Development and Use of Microsatellite Markers for Germplasm Characterization in Quinoa (Chenopodium quinoa Willd.). Crop. Sci. 2005, 45, 1618–1630. [Google Scholar] [CrossRef]
- Mizuno, N.; Toyoshima, M.; Fujita, M.; Fukuda, S.; Kobayashi, Y.; Ueno, M.; Tanaka, K.; Tanaka, T.; Nishihara, E.; Mizukoshi, H.; et al. The genotype-dependent phenotypic landscape of quinoa in salt tolerance and key growth traits. DNA Res. 2020, 27. [Google Scholar] [CrossRef]
- Patiranage, D.S.R.; Rey, E.; Emrani, N.; Wellman, G.; Schmid, K.; Schmöckel, S.M.; Tester, M.; Jung, C. Genome-wide association study in the pseudocereal quinoa reveals selection pattern typical for crops with a short breeding history. bioRxiv 2020. [Google Scholar] [CrossRef]
- Rana, T.S.; Narzary, D.; Ohri, D. Genetic diversity and relationships among some wild and cultivated species of Chenopodium L. (Amaranthaceae) using RAPD and DAMD methods. Curr. Sci. 2010, 98, 840–846. [Google Scholar]
- Salazar, J.; Torres, M.D.L.; Gutierrez, B.; Torres, A.F. Molecular characterization of Ecuadorian quinoa (Chenopodium quinoa Willd.) diversity: Implications for conservation and breeding. Euphytica 2019, 215, 60. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Gu, M.; Liu, Y.; Lv, Y.; Zhou, L.; Lu, H.; Liang, S.; Bao, H.; Zhao, H. Development of novel InDel markers and genetic diversity in Chenopodium quinoa through whole-genome re-sequencing. BMC Genom. 2017, 18, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Tapia, M.E.; Mujica, A.; Canahua, A. Origin, Geographic Distribution and Production System of Quinoa (Chenopodium Quinoa); Publicacion-Universidad Nacional Tecnica del Altiplano: Puno, Peru, 1980. [Google Scholar]
- Chable, V.; Thommen, A.; Goldringer, I.; Infante, V.; Levillain, T.; Lammerts Van Bueren, E. Report on the Definitions of Varieties in Europe, of Local Adaptation, and of Varieties Threatened by Genetic Erosion. 2008. Available online: https://hal.inrae.fr/hal-02820022 (accessed on 15 August 2021).
- Murphy, K.M.; Bazile, D.; Kellogg, J.; Rahmanian, M. Development of a Worldwide Consortium on Evolutionary Participatory Breeding in Quinoa. Front. Plant. Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Bonifacio, A.; Proinpa, F. Improvement of Quinoa (Chenopodium quinoa Willd.) and Qañawa (Chenopodium pallidicaule Aellen) in the context of climate change in the high Andes. Cien. Inv. Agr. 2019, 46, 113–124. [Google Scholar] [CrossRef]
- Jacobsen, S.-E. Developmental stability of quinoa under European conditions. Ind. Crop. Prod. 1998, 7, 169–174. [Google Scholar] [CrossRef]
- Mackay, I.; Piepho, H.; Garcia, A.A.F. Statistical Methods for Plant Breeding. In Handbook of Statistical Genomics; Wily: Hoboken, NJ, USA, 2019; pp. 501–520. [Google Scholar] [CrossRef]
- Molenaar, H.; Boehm, R.; Piepho, H.-P. Phenotypic Selection in Ornamental Breeding: It’s Better to Have the BLUPs Than to Have the BLUEs. Front. Plant. Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Piepho, H.P.; Möhring, J.; Melchinger, A.E.; Büchse, A. BLUP for phenotypic selection in plant breeding and variety testing. Euphytica 2008, 161, 209–228. [Google Scholar] [CrossRef]
- Welham, S.J.; Gezan, S.A.; Clark, S.J.; Mead, A. Statistical Methods in Biology: Design and Analysis of Experiments and Regression; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- Cullis, B.R.; Smith, A.B.; Cocks, N.A.; Butler, D.G. The Design of Early-Stage Plant Breeding Trials Using Genetic Relatedness. J. Agric. Biol. Environ. Stat. 2020, 25, 553–578. [Google Scholar] [CrossRef]
- Podlich, D.W.; Cooper, M. QU-GENE: A simulation platform for quantitative analysis of genetic models. Bioinformatics 1998, 14, 632–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faux, A.; Gorjanc, G.; Gaynor, R.; Battagin, M.; Høj-Edwards, S.; Wilson, D.L.; Hearne, S.; Gonen, S.; Hickey, J.M. AlphaSim: Software for Breeding Program Simulation. Plant. Genome 2016, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jahufer, M.Z.Z.; Luo, D. DeltaGen: A Comprehensive Decision Support Tool for Plant Breeders. Crop. Sci. 2018, 58, 1118–1131. [Google Scholar] [CrossRef] [Green Version]
- Cullis, B.R.; Smith, A.B.; Coombes, N.E. On the design of early generation variety trials with correlated data. J. Agric. Biol. Environ. Stat. 2006, 11, 381–393. [Google Scholar] [CrossRef]
- Falconer, D.S.; Mackay, T.F.C. Introduction to Quantitative Genetics (Fourth Edition). In Trends in Genetics; Elsevier: Amsterdam, The Netherlands, 1996. [Google Scholar]
- Oakey, H.; Verbyla, A.; Pitchford, W.; Cullis, B.; Kuchel, H. Joint modeling of additive and non-additive genetic line effects in single field trials. Theor. Appl. Genet. 2006, 113, 809–819. [Google Scholar] [CrossRef]
- Hong, E.P.; Park, J.W. Sample Size and Statistical Power Calculation in Genetic Association Studies. Genom. Inform. 2012, 10, 117–122. [Google Scholar] [CrossRef]
- Coombes, N.E. DiGGer, a spatial design program. In Biometric Bulletin; NSW Department of Primary Industries: Orange, Australia, 2009. [Google Scholar]
- van Ittersum, M.; Rabbinge, R. Concepts in production ecology for analysis and quantification of agricultural input-output combinations. Field Crop. Res. 1997, 52, 197–208. [Google Scholar] [CrossRef]
- Quinoa: Improvement and Sustainable Production. 2015; Available online: https://doi.org/10.1002/9781118628041 (accessed on 15 August 2021).
- Sellami, M.H.; Pulvento, C.; Lavini, A. Agronomic Practices and Performances of Quinoa under Field Conditions: A Systematic Review. Plants 2020, 10, 72. [Google Scholar] [CrossRef]
- Eisa, S.S.; Abd El-Samad, E.H.; Hussin, S.A.; Ali, E.A.; Ebrahim, M.; González, J.A.; Ordano, M.; Erazzú, L.E.; El-Bordeny, N.E.; Abdel-Ati, A.A. Quinoa in Egypt-Plant Density Effects on Seed Yield and Nutritional Quality in Marginal Regions. Middle East J. Appl. Sci. 2018, 8, 515–522. [Google Scholar]
- Ahmadi, S.H.; Solgi, S.; Sepaskhah, A.R. Quinoa: A super or pseudo-super crop? Evidences from evapotranspiration, root growth, crop coefficients, and water productivity in a hot and semi-arid area under three planting densities. Agric. Water Manag. 2019, 225, 105784. [Google Scholar] [CrossRef]
- Aguilar, P.C.; Jacobsen, S.-E. Cultivation of Quinoa on the Peruvian Altiplano. Food Rev. Int. 2003, 19, 31–41. [Google Scholar] [CrossRef]
- Abdelaziz, H.; Redouane, C.-A. Phenotyping the Combined Effect of Heat and Water Stress on Quinoa; Springer: Cham, Switzerland, 2020; pp. 163–183. [Google Scholar] [CrossRef]
- Aufhammer, W.; Kaul, H.-P.; Kruse, M.; Lee, J.; Schwesig, D. Effects of sowing depth and soil conditions on seedling emergence of amaranth and quinoa. Eur. J. Agron. 1994, 3, 205–210. [Google Scholar] [CrossRef]
- Hinojosa, L.; González, J.A.; Barrios-Masias, F.H.; Fuentes, F.; Murphy, K.M. Quinoa Abiotic Stress Responses: A Review. Plants 2018, 7, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oelke, E.A.; Putnam, D.H.; Teynor, T.M.; Oplinger, E.S. Alternative Field Crops Manual: Quinoa; University of Wisconsin-Extension 1992, Cooperative Extension University of Minnesota: Center for Alternative Plant & Animal Products and the Minnesota Extension Service. 1992. Available online: https://hort.purdue.edu/newcrop/afcm/quinoa.html (accessed on 15 August 2021).
- Hirich, A.; Jacobsen, S.-E.; Choukr-Allah, R. Quinoa in Morocco-Effect of Sowing Dates on Development and Yield. J. Agron. Crop. Sci. 2014, 200, 371–377. [Google Scholar] [CrossRef]
- Yang, A.; Akhtar, S.S.; Amjad, M.; Iqbal, S.; Jacobsen, S.-E. Growth and Physiological Responses of Quinoa to Drought and Temperature Stress. J. Agron. Crop. Sci. 2016, 202, 445–453. [Google Scholar] [CrossRef]
- Präger, A.; Munz, S.; Nkebiwe, P.M.; Mast, B.; Graeff-Hönninger, S. Yield and Quality Characteristics of Different Quinoa (Chenopodium quinoa Willd.) Cultivars Grown under Field Conditions in Southwestern Germany. Agronomy 2018, 8, 197. [Google Scholar] [CrossRef] [Green Version]
- Garcia, M.; Raes, D.; Jacobsen, S.-E. Evapotranspiration analysis and irrigation requirements of quinoa (Chenopodium quinoa) in the Bolivian highlands. Agric. Water Manag. 2003, 60, 119–134. [Google Scholar] [CrossRef]
- Jayme-Oliveira, A.; Júnior, W.Q.R.; Ramos, M.L.G.; Ziviani, A.C.; Jakelaitis, A. Amaranth, quinoa, and millet growth and development under different water regimes in the Brazilian Cerrado. Pesquisa Agropecuária Brasileira 2017, 52, 561–571. [Google Scholar] [CrossRef]
- Allen, R.G.; Pereira, L.S.; Raes, D.; Smith, M. Crop evapotranspiration: Guidelines for computing crop requirements. In Irrigation and Drainage Paper No. 56; FAO: Rome, Italy, 1998. [Google Scholar] [CrossRef]
- FAO. ETo Calculator. In Land and Water Digital Media Series; FAO: Rome, Italy, 2012. [Google Scholar]
- Geerts, S.; Raes, D.; Garcia, M.; Vacher, J.; Mamani, R.; Mendoza, J.; Huanca, R.; Morales, B.; Miranda, R.; Cusicanqui, J.; et al. Introducing deficit irrigation to stabilize yields of quinoa (Chenopodium quinoa Willd.). Eur. J. Agron. 2008, 28, 427–436. [Google Scholar] [CrossRef]
- Razzaghi, F.; Plauborg, F.; Jacobsen, S.-E.; Jensen, C.R.; Andersen, M.N. Effect of nitrogen and water availability of three soil types on yield, radiation use efficiency and evapotranspiration in field-grown quinoa. Agric. Water Manag. 2012, 109, 20–29. [Google Scholar] [CrossRef]
- Fghire, R.; Wahbi, S.; Anaya, F.; Ali, O.I.; Benlhabib, O.; Ragab, R. Response of Quinoa to Different Water Management Strategies: Field Experiments and Saltmed Model Application Results. Irrig. Drain. 2015, 64, 29–40. [Google Scholar] [CrossRef]
- Pulvento, C.; Riccardi, M.; Lavini, A.; D’Andria, R.; Ragab, R. Saltmed Model to Simulate Yield And Dry Matter for Quinoa Crop And Soil Moisture Content Under Different Irrigation Strategies In South Italy. Irrig. Drain. 2013, 62, 229–238. [Google Scholar] [CrossRef] [Green Version]
- Bertero, H.D. Quinoa. In Crop Physiology Case Histories for Major Crops; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar] [CrossRef]
- Geren, H.; Kavut, Y.; Altınbaș, M. Effect of different row spacings on the grain yield and some yield characteristics of quinoa (Chenopodium quinoa Wild.) under Bornova ecological conditions. Ege Üniversitesi Ziraat Fakültesi Dergisi 2015, 52, 69–78. [Google Scholar]
- Erley, G.S.A.; Kaul, H.-P.; Kruse, M.; Aufhammer, W. Yield and nitrogen utilization efficiency of the pseudocereals amaranth, quinoa, and buckwheat under differing nitrogen fertilization. Eur. J. Agron. 2005, 22, 95–100. [Google Scholar] [CrossRef]
- Alandia, G.; Jacobsen, S.-E.; Kyvsgaard, N.C.; Condori, B.; Liu, F. Nitrogen Sustains Seed Yield of Quinoa Under Intermediate Drought. J. Agron. Crop. Sci. 2016, 202, 281–291. [Google Scholar] [CrossRef]
- Bascuñán-Godoy, L.; Sanhueza, C.; Pinto, K.; Cifuentes, L.; Reguera, M.; Briones-Labarca, V.; Zurita-Silva, A.; Álvarez, R.; Morales, A.; Silva, H. Nitrogen physiology of contrasting genotypes of Chenopodium quinoa Willd. (Amaranthaceae). Sci. Rep. 2018, 8, 17524. [Google Scholar] [CrossRef]
- Cruces, L.; Delgado, P.; Santivañez, T.; Jara, B.; Vernal, P. Guía de Identificación y Control de las Principales Plagas que Afectan a la Quinua en la Zona Andina. 2016. Available online: https://bivica.org/files/quinua-plagas.pdf (accessed on 15 August 2021).
- Rupavatharam, S.; Kennepohl, A.; Kummer, B.; Parimi, V. Automated plant disease diagnosis using innovative android App (Plantix) for farmers in Indian state of Andhra Pradesh. Phytopathology TSI 2018, 108, 10. [Google Scholar]
- Brachi, B.; Aime, C.; Glorieux, C.; Cuguen, J.; Roux, F. Adaptive Value of Phenological Traits in Stressful Environments: Predictions Based on Seed Production and Laboratory Natural Selection. PLoS ONE 2012, 7, e32069. [Google Scholar] [CrossRef] [Green Version]
- Rausher, M.D. The Measurement of Selection on Quantitative Traits: Biases Due to Environmental Covariances between Traits and Fitness. Evolution 1992, 46, 616–626. [Google Scholar] [CrossRef] [PubMed]
- De Leon, N.; Jannink, J.-L.; Edwards, J.W.; Kaeppler, S.M. Introduction to a Special Issue on Genotype by Environment Interaction. Crop. Sci. 2016, 56, 2081–2089. [Google Scholar] [CrossRef] [Green Version]
- Condon, J. Effective Soil Sampling–High and Low Cost Options to Gain Soil Fertility Information for Management. GRDC. 2019. Available online: https://grdc.com.au/resources-and-publications/grdc-update-papers/tab-content/grdc-update-papers/2019/02/effective-soil-sampling-high-and-low-cost-options-to-gain-soil-fertility-information-for-management. (accessed on 15 August 2021).
- Xu, Y. Envirotyping for deciphering environmental impacts on crop plants. Theor. Appl. Genet. 2016, 129, 653–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fricke, W. Water transport and energy. Plant. Cell Environ. 2017, 40, 977–994. [Google Scholar] [CrossRef]
- Zhang, D.; Du, Q.; Zhang, Z.; Jiao, X.; Song, X.; Li, J. Vapour pressure deficit control in relation to water transport and water productivity in greenhouse tomato production during summer. Sci. Rep. 2017, 7, 43461. [Google Scholar] [CrossRef]
- Kargas, G.; Londra, P.; Anastasatou, M.; Moustakas, N. The Effect of Soil Iron on the Estimation of Soil Water Content Using Dielectric Sensors. Water 2020, 12, 598. [Google Scholar] [CrossRef] [Green Version]
- Präger, A.; Boote, K.J.; Munz, S.; Graeff-Hönninger, S. Simulating Growth and Development Processes of Quinoa (Chenopodium quinoa Willd.): Adaptation and Evaluation of the CSM-CROPGRO Model. Agronomy 2019, 9, 832. [Google Scholar] [CrossRef] [Green Version]
- Alvar-Beltrán, J.; Gobin, A.; Orlandini, S.; Marta, A.D. AquaCrop parametrisation for quinoa in arid environments. Ital. J. Agron. 2020, 16. [Google Scholar] [CrossRef]
- Geerts, S.; Raes, D.; Garcia, M. Using AquaCrop to derive deficit irrigation schedules. Agric. Water Manag. 2010, 98, 213–216. [Google Scholar] [CrossRef]
- Geerts, S.; Raes, D.; Garcia, M.; Miranda, R.; Cusicanqui, J.A.; Taboada, C.; Mendoza, J.; Huanca, R.; Mamani, A.; Condori, O.; et al. Simulating Yield Response of Quinoa to Water Availability with AquaCrop. Agron. J. 2009, 101, 499–508. [Google Scholar] [CrossRef]
- Geerts, S.; Raes, D.; Garcia, M.; Taboada, C.; Miranda, R.; Cusicanqui, J.; Mhizha, T.; Vacher, J. Modeling the potential for closing quinoa yield gaps under varying water availability in the Bolivian Altiplano. Agric. Water Manag. 2009, 96, 1652–1658. [Google Scholar] [CrossRef]
- Kaoutar, F.; Abdelaziz, H.; Ouafae, B.; Redouane, C.-A.; Ragab, R. Yield and Dry Matter Simulation Using the Saltmed Model for Five Quinoa (Chenopodium Quinoa) Accessions Under Deficit Irrigation in South Morocco. Irrig. Drain. 2017, 66, 340–350. [Google Scholar] [CrossRef]
- FAO. Required Input for Simulations with AquaCrop. 2016. Available online: http://www.fao.org/3/i6050e/i6050e.pdf (accessed on 15 August 2021).
- Vanuytrecht, E.; Raes, D.; Steduto, P.; Hsiao, T.C.; Fereres, E.; Heng, L.K.; Garcia-Vila, M.; Moreno, P.M. AquaCrop: FAO’s crop water productivity and yield response model. Environ. Model. Softw. 2014, 62, 351–360. [Google Scholar] [CrossRef]
- Ragab, R. A holistic generic integrated approach for irrigation, crop and field management: The SALTMED model. Environ. Model. Softw. 2002, 17, 345–361. [Google Scholar] [CrossRef]
- Bertero, D.; Medan, D.; Hall, A.J. Changes in Apical Morphology during Floral Initiation and Reproductive Development in Quinoa (Chenopodium quinoaWilld.). Ann. Bot. 1996, 78, 317–324. [Google Scholar] [CrossRef] [Green Version]
- Curti, R.; de la Vega, A.; Andrade, A.; Bramardi, S.; Bertero, H. Multi-environmental evaluation for grain yield and its physiological determinants of quinoa genotypes across Northwest Argentina. Field Crop. Res. 2014, 166, 46–57. [Google Scholar] [CrossRef]
- Jacobsen, S.-E.; Stølen, O. Quinoa-Morphology, phenology and prospects for its production as a new crop in Europe. Eur. J. Agron. 1993, 2, 19–29. [Google Scholar] [CrossRef]
- Mujica, A.; Canahua, A. Fases fenológicas del cultivo de la quínua (Chenopodium quinoa Willd.). In Proceedings of the Curso Taller 1989, Fenología de Cultivos Andinos y Uso de La Información Agrometeorológica, Salcedo, Puno, Peru 7–10 August 1989; pp. 23–27. [Google Scholar]
- Ruiz, R.; Bertero, H. Light interception and radiation use efficiency in temperate quinoa (Chenopodium quinoa Willd.) cultivars. Eur. J. Agron. 2008, 29, 144–152. [Google Scholar] [CrossRef]
- Tardieu, F. Plant response to environmental conditions: Assessing potential production, water demand, and negative effects of water deficit. Front. Physiol. 2013, 4, 17. [Google Scholar] [CrossRef] [Green Version]
- Tardieu, F.; Bosquet, L.C.; Welcker, C. Model assisted dissection of the Genotype x Environment interaction. In Proceedings of the ASA 2012, CSSA and SSSA International Annual Meetings, Cincinnati, OH, USA, 21–24 October 2012. [Google Scholar]
- Tardieu, F.; Tuberosa, R. Dissection and modelling of abiotic stress tolerance in plants. Curr. Opin. Plant. Biol. 2010, 13, 206–212. [Google Scholar] [CrossRef]
- Passioura, J.; Angus, J. Improving Productivity of Crops in Water-Limited Environments. Adv. Agron. 2010, 106, 37–75. [Google Scholar] [CrossRef]
- Trapani, N.; Hall, A.; Sadras, V.; Vilella, F. Ontogenetic changes in radiation use efficiency of sunflower (Helianthus annuus L.) crops. Field Crop. Res. 1992, 29, 301–316. [Google Scholar] [CrossRef]
- Grimes, D.J. Koch’s Postulates—Then and Now. Microbe Mag. 2006, 1, 223–228. [Google Scholar] [CrossRef] [Green Version]
- Gómez, M.B.; Castro, P.A.; Mignone, C.; Bertero, H.D. Can yield potential be increased by manipulation of reproductive partitioning in quinoa (Chenopodium quinoa)? Evidence from gibberellic acid synthesis inhibition using Paclobutrazol. Funct. Plant. Biol. 2011, 38, 420–430. [Google Scholar] [CrossRef] [PubMed]
- Garbulsky, M.; Peñuelas, J.; Gamon, J.; Inoue, Y.; Filella, I. The photochemical reflectance index (PRI) and the Remote Sensing of leaf, canopy and ecosystem radiation use efficiencies: A review and meta-analysis. Remote Sens. Environ. 2011, 115, 281–297. [Google Scholar] [CrossRef]
- Hinojosa, L.; Kumar, N.; Gill, K.S.; Murphy, K.M. Spectral Reflectance Indices and Physiological Parameters in Quinoa under Contrasting Irrigation Regimes. Crop. Sci. 2019, 59, 1927–1944. [Google Scholar] [CrossRef] [Green Version]
- Sankaran, S.; Espinoza, C.Z.; Hinojosa, L.; Ma, X.; Murphy, K. High-Throughput Field Phenotyping to Assess Irrigation Treatment Effects in Quinoa. Age 2019, 2, 1–7. [Google Scholar] [CrossRef]
- Danielsen, S.; Munk, L. Evaluation of disease assessment methods in quinoa for their ability to predict yield loss caused by downy mildew. Crop. Prot. 2004, 23, 219–228. [Google Scholar] [CrossRef]
- Xie, C.; Yang, C. A review on plant high-throughput phenotyping traits using UAV-based sensors. Comput. Electron. Agric. 2020, 178, 105731. [Google Scholar] [CrossRef]
- Tmušić, G.; Manfreda, S.; Aasen, H.; James, M.R.; Gonçalves, G.; Ben-Dor, E.; Brook, A.; Polinova, M.; Arranz, J.J.; Mészáros, J.; et al. Current Practices in UAS-based Environmental Monitoring. Remote Sens. 2020, 12, 1001. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Liu, J.; Zhao, C.; Li, Z.; Huang, Y.; Yu, H.; Xu, B.; Yang, X.; Zhu, D.; Zhang, X.; et al. Unmanned Aerial Vehicle Remote Sens.ing for Field-Based Crop Phenotyping: Current Status and Perspectives. Front. Plant. Sci. 2017, 8, 1111. [Google Scholar] [CrossRef] [PubMed]
- Ziliani, M.G.; Parkes, S.D.; Hoteit, I.; McCabe, M.F. Intra-Season Crop Height Variability at Commercial Farm Scales Using a Fixed-Wing UAV. Remote Sens. 2018, 10, 2007. [Google Scholar] [CrossRef] [Green Version]
- Johansen, K.; Morton, M.J.L.; Malbeteau, Y.; Aragon, B.; Al-Mashharawi, S.; Ziliani, M.G.; Angel, Y.; Fiene, G.; Negrão, S.; Mousa, M.A.A.; et al. Predicting Biomass and Yield in a Tomato Phenotyping Experiment Using UAV Imagery and Random Forest. Front. Artif. Intell. 2020, 3. [Google Scholar] [CrossRef]
- Di Gennaro, S.F.; Rizza, F.; Badeck, F.-W.; Berton, A.; Delbono, S.; Gioli, B.; Toscano, P.; Zaldei, A.; Matese, A. UAV-based high-throughput phenotyping to discriminate barley vigour with visible and near-infrared vegetation indices. Int. J. Remote Sens. 2018, 39, 5330–5344. [Google Scholar] [CrossRef]
- Wang, T.; Thomasson, J.A.; Yang, C.; Isakeit, T.; Nichols, R.L. Automatic Classification of Cotton Root Rot Disease Based on UAV Remote Sensing. Remote Sens. 2020, 12, 1310. [Google Scholar] [CrossRef] [Green Version]
- Su, J.; Liu, C.; Coombes, M.; Hu, X.; Wang, C.; Xu, X.; Li, Q.; Guo, L.; Chen, W.-H. Wheat yellow rust monitoring by learning from multispectral UAV aerial imagery. Comput. Electron. Agric. 2018, 155, 157–166. [Google Scholar] [CrossRef]
- Chivasa, W.; Mutanga, O.; Biradar, C. UAV-Based Multispectral Phenotyping for Disease Resistance to Accelerate Crop Improvement under Changing Climate Conditions. Remote Sens. 2020, 12, 2445. [Google Scholar] [CrossRef]
- Holman, F.H.; Riche, A.B.; Castle, M.; Wooster, M.J.; Hawkesford, M.J. Radiometric Calibration of ‘Commercial off the Shelf’ Cameras for UAV-Based High-Resolution Temporal Crop Phenotyping of Reflectance and NDVI. Remote Sens. 2019, 11, 1657. [Google Scholar] [CrossRef] [Green Version]
- Jackson, R.D.; Idso, S.B.; Reginato, R.J.; Pinter, P.J. Canopy temperature as a crop water stress indicator. Water Resour. Res. 1981, 17, 1133–1138. [Google Scholar] [CrossRef]
- Turner, D.; Lucieer, A.; Watson, C. An Automated Technique for Generating Georectified Mosaics from Ultra-High Resolution Unmanned Aerial Vehicle (UAV) Imagery, Based on Structure from Motion (SfM) Point Clouds. Remote Sens. 2012, 4, 1392–1410. [Google Scholar] [CrossRef] [Green Version]
- Shendryk, Y.; Sofonia, J.; Garrard, R.; Rist, Y.; Skocaj, D.; Thorburn, P. Fine-scale prediction of biomass and leaf nitrogen content in sugarcane using UAV LiDAR and multispectral imaging. Int. J. Appl. Earth Obs. Geoinf. 2020, 92, 102177. [Google Scholar] [CrossRef]
- Messina, G.; Modica, G. Applications of UAV Thermal Imagery in Precision Agriculture: State of the Art and Future Research Outlook. Remote Sens. 2020, 12, 1491. [Google Scholar] [CrossRef]
- Aragon, B.; Johansen, K.; Parkes, S.; Malbeteau, Y.; Al-Mashharawi, S.; Al-Amoudi, T.; Andrade, C.F.; Turner, D.; Lucieer, A.; McCabe, M.F. A Calibration Procedure for Field and UAV-Based Uncooled Thermal Infrared Instruments. Sensors 2020, 20, 3316. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.; Kljun, N.; Olsson, P.-O.; Mihai, L.; Liljeblad, B.; Weslien, P.; Klemedtsson, L.; Eklundh, L. Challenges and Best Practices for Deriving Temperature Data from an Uncalibrated UAV Thermal Infrared Camera. Remote Sens. 2019, 11, 567. [Google Scholar] [CrossRef] [Green Version]
- Malbeteau, Y.; Johansen, K.; Aragon, B.; Al-Mashhawari, S.K.; McCabe, M.F. Overcoming the challenges of thermal infrared orthomosaics using a swath-based approach to correct for dynamic temperature and wind effects. Remote Sens. 2021, 13, 3255. [Google Scholar] [CrossRef]
- Aasen, H.; Bolten, A. Multi-temporal high-resolution imaging spectroscopy with hyperspectral 2D imagers-From theory to application. Remote Sens. Environ. 2018, 205, 374–389. [Google Scholar] [CrossRef]
- Adão, T.; Hruška, J.; Pádua, L.; Bessa, J.; Peres, E.; Morais, R.; Sousa, J.J. Hyperspectral Imaging: A Review on UAV-Based Sensors, Data Processing and Applications for Agriculture and Forestry. Remote Sens. 2017, 9, 1110. [Google Scholar] [CrossRef] [Green Version]
- Angel, Y.; Turner, D.; Parkes, S.; Malbeteau, Y.; Lucieer, A.; McCabe, M.F. Automated Georectification and Mosaicking of UAV-Based Hyperspectral Imagery from Push-Broom Sensors. Remote Sens. 2020, 12, 34. [Google Scholar] [CrossRef] [Green Version]
- Barreto, M.A.P.; Johansen, K.; Angel, Y.; McCabe, M.F. Radiometric Assessment of a UAV-Based Push-Broom Hyperspectral Camera. Sensors 2019, 19, 4699. [Google Scholar] [CrossRef] [Green Version]
- Hassler, S.C.; Baysal-Gurel, F. Unmanned Aircraft System (UAS) Technology and Applications in Agriculture. Agronomy 2019, 9, 618. [Google Scholar] [CrossRef] [Green Version]
- Ivushkin, K.; Bartholomeus, H.; Bregt, A.K.; Pulatov, A.; Franceschini, M.H.; Kramer, H.; van Loo, E.N.; Roman, V.J.; Finkers, R. UAV based soil salinity assessment of cropland. Geoderma 2019, 338, 502–512. [Google Scholar] [CrossRef]
- Holman, F.H.; Riche, A.B.; Michalski, A.; Castle, M.; Wooster, M.J.; Hawkesford, M.J. High Throughput Field Phenotyping of Wheat Plant Height and Growth Rate in Field Plot Trials Using UAV Based Remote Sensing. Remote Sens. 2016, 8, 1031. [Google Scholar] [CrossRef]
- Galli, G.; Sabadin, F.; Costa-Neto, G.M.F.; Fritsche-Neto, R. A novel way to validate UAS-based high-throughput phenotyping protocols using in silico experiments for plant breeding purposes. Theor. Appl. Genet. 2020, 134, 715–730. [Google Scholar] [CrossRef]
- Keller, B.; Matsubara, S.; Rascher, U.; Pieruschka, R.; Steier, A.; Kraska, T.; Muller, O. Genotype Specific Photosynthesis x Environment Interactions Captured by Automated Fluorescence Canopy Scans Over Two Fluctuating Growing Seasons. Front. Plant. Sci. 2019, 10, 1482. [Google Scholar] [CrossRef] [Green Version]
- Raesch, A.R.; Muller, O.; Pieruschka, R.; Rascher, U. Field Observations with Laser-Induced Fluorescence Transient (LIFT) Method in Barley and Sugar Beet. Agriculture 2014, 4, 159–169. [Google Scholar] [CrossRef] [Green Version]
- Pieruschka, R.; Schurr, U. Plant Phenotyping: Past, Present, and Future. Plant. Phenomics 2019, 2019, 1–6. [Google Scholar] [CrossRef]
- Mochida, K.; Koda, S.; Inoue, K.; Hirayama, T.; Tanaka, S.; Nishii, R.; Melgani, F. Computer vision-based phenotyping for improvement of plant productivity: A machine learning perspective. GigaScience 2018, 8. [Google Scholar] [CrossRef] [Green Version]
- Ubbens, J.; Cieslak, M.; Prusinkiewicz, P.; Parkin, I.; Ebersbach, J.; Stavness, I. Latent Space Phenotyping: Automatic Image-Based Phenotyping for Treatment Studies. Plant. Phenomics 2020, 2020, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Brunner, G.; Liu, Y.; Pascual, D.; Richter, O.; Ciaramita, M.; Wattenhofer, R. On Identifiability in Transformers. arXiv 2019, arXiv:1908.04211. [Google Scholar]
- Chefer, H.; Gur, S.; Wolf, L. Transformer Interpretability Beyond Attention Visualization. 2021. Available online: https://openaccess.thecvf.com/content/CVPR2021/html/Chefer_Transformer_Interpretability_Beyond_Attention_Visualization_CVPR_2021_paper.html (accessed on 15 August 2021).
- Mastebroek, H.; van Loo, E.; Dolstra, O. Combining ability for seed yield traits of Chenopodium quinoa breeding lines. Euphytica 2002, 125, 427–432. [Google Scholar] [CrossRef]
- Ploschuk, E.L.; Hall, A. Capitulum position in sunflower affects grain temperature and duration of grain filling. Field Crop. Res. 1995, 44, 111–117. [Google Scholar] [CrossRef]
- Dong, Y.; Ewang, Y.-Z. Seed shattering: From models to crops. Front. Plant. Sci. 2015, 6, 476. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.; Jacobsen, S.-E.; Bonifacio, A.; Murphy, K. A Crossing Method for Quinoa. Sustainability 2015, 7, 3230–3243. [Google Scholar] [CrossRef] [Green Version]
- Colque-Little, C.; Amby, D.; Andreasen, C. A Review of Chenopodium quinoa (Willd.) Diseases—An Updated Perspective. Plants 2021, 10, 1228. [Google Scholar] [CrossRef] [PubMed]
- Agrios, G. Plant Pathology; Elsevier Academic Press: Cambridge, MA, USA, 2005. [Google Scholar]
- Lamichhane, J.R.; Venturi, V. Synergisms between microbial pathogens in plant disease complexes: A growing trend. Front. Plant. Sci. 2015, 6, 385. [Google Scholar] [CrossRef] [Green Version]
- Danielsen, S.; Jacobsen, S.-E.; Hockenhull, J. First Report of Downy Mildew of Quinoa Caused by Peronospora farinosa f. sp. chenopodii in Denmark. Plant. Dis. 2002, 86, 1175. [Google Scholar] [CrossRef] [PubMed]
- Testen, A.; McKemy, J.M.; Backman, P.A. First Report of Passalora Leaf Spot of Quinoa Caused by Passalora dubia in the United States. Plant. Dis. 2013, 97, 139. [Google Scholar] [CrossRef]
- Testen, A.; McKemy, J.M.; Backman, P.A. First Report of Ascochyta Leaf Spot of Quinoa Caused by Ascochyta sp. in the United States. Plant. Dis. 2013, 97, 844. [Google Scholar] [CrossRef]
- Testen, A.; Jiménez-Gasco, M.D.M.; Ochoa, J.B.; Backman, P.A. Molecular Detection of Peronospora variabilis in Quinoa Seed and Phylogeny of the Quinoa Downy Mildew Pathogen in South America and the United States. Phytopathology 2014, 104, 379–386. [Google Scholar] [CrossRef] [Green Version]
- Yin, H.; Zhou, J.; Lv, H.; Qin, N.; Chang, F.J.; Zhao, X.J. Identification, Pathogenicity, and Fungicide Sensitivity of Ascochyta caulina (Teleomorph: Neocamarosporium calvescens) Associated with Black Stem on Quinoa in China. Plant. Dis. 2020, 104. [Google Scholar] [CrossRef] [PubMed]
- Dřímalková, M.; Veverka, K. Seedlings damping-off of Chenopodium quinoa Willd. Plant. Prot. Sci. 2004, 40, 5–10. [Google Scholar] [CrossRef] [Green Version]
- Fonseca-Guerra, I.; Chiquillo, C.; Padilla, M.J.; Benavides-Rozo, M. First report of bacterial leaf spot on Chenopodium quinoa caused by Pseudomonas syringae in Colombia. J. Plant. Dis. Prot. 2021, 128, 871–874. [Google Scholar] [CrossRef]
- Isobe, K.; Sugiyama, T.; Katagiri, M.; Ishizuka, C.; Tamura, Y.; Higo, M.; Fujita, Y. Study on the Cause Damping-off in Quinoa (Chenopodium quinoa Willd.) and a Method for Suppressing its Occurrence. Jpn. J. Crop. Sci. 2019, 88, 117–124. [Google Scholar] [CrossRef]
- Pal, N.; Testen, A.L. First Report of Quinoa Anthracnose Caused by Colletotrichum nigrum and C. truncatum in the United States. Plant. Dis. 2021, 105, 705. [Google Scholar] [CrossRef]
- Danielsen, S.; Ames, T. Mildew (Peronospora farinosa) of quinua (Chenopodium quinoa) in the Andean Region: Practical Manual for the Study of the Disease and Pathogen; International Potato Center: Lima, Peru, 2004. [Google Scholar]
- Staub, J.; Bacher, J.; Poetter, K. Sources of Potential Errors in the Application of Random Amplified Polymorphic DNAs in Cucumber. HortScience 1996, 31, 262–266. [Google Scholar] [CrossRef] [Green Version]
- Conrath, U. Molecular aspects of defence priming. Trends Plant. Sci. 2011, 16, 524–531. [Google Scholar] [CrossRef]
- Hammerschmidt, R.; M’Etraux, J.-P.; Van Loon, L. Inducing Resistance: A Summary of Papers Presented at the First International Symposium on Induced Resistance to Plant Diseases, Corfu, May 2000. Eur. J. Plant. Pathol. 2001, 107, 1–6. [Google Scholar] [CrossRef]
- Grogan, R.G. The Science and Art of Plant-Disease Diagnosis. Annu. Rev. Phytopathol. 1981, 19, 333–351. [Google Scholar] [CrossRef]
- Pereira, E.; Encina-Zelada, C.; Barros, L.; Gonzales-Barron, U.; Cadavez, V.; Ferreira, I.C. Chemical and nutritional characterization of Chenopodium quinoa Willd (quinoa) grains: A good alternative to nutritious food. Food Chem. 2019, 280, 110–114. [Google Scholar] [CrossRef] [Green Version]
- Merchant, N.; Lyons, E.; Goff, S.; Vaughn, M.; Ware, D.; Micklos, D.; Antin, P. The iPlant Collaborative: Cyberinfrastructure for Enabling Data to Discovery for the Life Sciences. PLoS Biol. 2016, 14, e1002342. [Google Scholar] [CrossRef] [Green Version]
- Nowak, V.; Du, J.; Charrondière, U.R. Assessment of the nutritional composition of quinoa (Chenopodium quinoa Willd.). Food Chem. 2016, 193, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Valencia-Chamorro, S.A. QUINOA. In Encyclopedia of Food Sciences and Nutrition; Elsevier: Amsterdam, The Netherlands, 2003; pp. 4895–4902. [Google Scholar]
- Aluwi, N.A.; Gu, B.-J.; Dhumal, G.S.; Medina-Meza, I.G.; Murphy, K.M.; Ganjyal, G.M. Impacts of Scarification and Degermination on the Expansion Characteristics of Select Quinoa Varieties during Extrusion Processing. J. Food Sci. 2016, 81, E2939–E2949. [Google Scholar] [CrossRef] [PubMed]
- Foley, W.J.; McIlwee, A.; Lawler, I.; Aragones, L.; Woolnough, A.P.; Berding, N. Ecological applications of near infrared reflectance spectroscopy-a tool for rapid, cost-effective prediction of the composition of plant and animal tissues and aspects of animal performance. Oecologia 1998, 116, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Lane, H.M.; Murray, S.C.; Montesinos-López, O.A.; Montesinos-López, A.; Crossa, J.; Rooney, D.K.; Barrero-Farfan, I.D.; De La Fuente, G.N.; Morgan, C.L.S. Phenomic selection and prediction of maize grain yield from near-infrared reflectance spectroscopy of kernels. TPPJ 2020, 3, e20002. [Google Scholar] [CrossRef] [Green Version]
- Escuredo, O.; Martín, M.I.G.; Moncada, G.W.; Fischer, S.; Hierro, J.M.H. Amino acid profile of the quinoa (Chenopodium quinoa Willd.) using near infrared spectroscopy and chemometric techniques. J. Cereal Sci. 2014, 60, 67–74. [Google Scholar] [CrossRef]
- Rodríguez, S.D.; Rolandelli, G.; Buera, M.P. Detection of quinoa flour adulteration by means of FT-MIR spectroscopy combined with chemometric methods. Food Chem. 2019, 274, 392–401. [Google Scholar] [CrossRef]
- Roa-Acosta, D.F.; Bravo-Gómez, J.E.; García-Parra, M.A.; Rodríguez-Herrera, R.; Solanilla-Duque, J.F. Hyper-protein quinoa flour (Chenopodium Quinoa Wild): Monitoring and study of structural and rheological properties. LWT 2020, 121, 108952. [Google Scholar] [CrossRef]
- Morais, C.L.M.; Santos, M.C.D.; Lima, K.M.G.; Martin, F.L. Improving data splitting for classification applications in spectrochemical analyses employing a random-mutation Kennard-Stone algorithm approach. Bioinformatics 2019, 35, 5257–5263. [Google Scholar] [CrossRef]
- Horwitz, W. Official Methods of Analysis of AOAC International; Association of Official Analytical Chemists International: Gaithersburg, MD, USA, 2019. [Google Scholar]
- Agelet, L.E.; Hurburgh, C.R. A Tutorial on Near Infrared Spectroscopy and Its Calibration. Crit. Rev. Anal. Chem. 2010, 40, 246–260. [Google Scholar] [CrossRef]
- Rinnan, Å.; Berg, F.V.D.; Engelsen, S.B. Review of the most common pre-processing techniques for near-infrared spectra. TrAC Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Craine, E.B.; Murphy, K.M. Seed Composition and Amino Acid Profiles for Quinoa Grown in Washington State. Front. Nutr. 2020, 7, 126. [Google Scholar] [CrossRef]
- Kuljanabhagavad, T.; Thongphasuk, P.; Chamulitrat, W.; Wink, M. Triterpene saponins from Chenopodium quinoa Willd. Phytochemistry 2008, 69, 1919–1926. [Google Scholar] [CrossRef]
- Madl, T.; Sterk, H.; Mittelbach, M.; Rechberger, G.N. Tandem mass spectrometric analysis of a complex triterpene saponin mixture of Chenopodium quinoa. J. Am. Soc. Mass Spectrom. 2006, 17, 795–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woldemichael, G.M.; Wink, M. Identification and biological activities of triterpenoid saponins from Chenopodium quinoa. J. Agric. Food Chem. 2001, 49, 2327–2332. [Google Scholar] [CrossRef]
- Otterbach, S.; Wellman, G.; Schmöckel, S.M. Saponins of Quinoa: Structure, Function and Opportunities. In The Quinoa Genome; Schmöckel, S.M., Ed.; Springer: Cham, Swizterland, 2021; pp. 119–138. [Google Scholar] [CrossRef]
- Koziol, M.J. Afrosimetric estimation of threshold saponin concentration for bitterness in quinoa (Chenopodium quinoa Willd). J. Sci. Food Agric. 1991, 54, 211–219. [Google Scholar] [CrossRef]
- Hirich, A.; Rafik, S.; Rahmani, M.; Fetouab, A.; Azaykou, F.; Filali, K.; Ahmadzai, H.; Jnaoui, Y.; Soulaimani, A.; Moussafir, M.; et al. Development of Quinoa Value Chain to Improve Food and Nutritional Security in Rural Communities in Rehamna, Morocco: Lessons Learned and Perspectives. Plants 2021, 10, 301. [Google Scholar] [CrossRef] [PubMed]
- Afzal, I.; Kamran, M.; Basra, S.M.A.; Khan, S.H.U.; Mahmood, A.; Farooq, M.; Tan, D.K. Harvesting and post-harvest management approaches for preserving cottonseed quality. Ind. Crop. Prod. 2020, 155, 112842. [Google Scholar] [CrossRef]
- Hong, T.D.; Linington, S.; Ellis, R.H. Seed Storage Behaviour: A Compendium Handbooks for Genebanks No. 4. In Ecology and Classification of North American Freshwater Invertebrates; International Plant Genetic Resources Institute: Rome, Italy, 1996; Available online: https://cgspace.cgiar.org/handle/10568/105158 (accessed on 15 August 2021).
- Hong, T.D.; Ellis, R.H. A Protocol to Determine Seed Storage Behaviour; IPGRI Technical Bulletin No. 1; International Plant Genetic Resources Institute (IPGRI): Rome, Italy, 1996. [Google Scholar]
- Roberts, E.H.; Ellis, R. Water and Seed Survival. Ann. Bot. 1989, 63, 39. [Google Scholar] [CrossRef]
- Afzal, I.; Bakhtavar, M.A.; Ishfaq, M.; Sagheer, M.; Baributsa, D. Maintaining dryness during storage contributes to higher maize seed quality. J. Stored Prod. Res. 2017, 72, 49–53. [Google Scholar] [CrossRef]
- Bradford, K.J.; Dahal, P.; Van Asbrouck, J.; Kunusoth, K.; Bello, P.; Thompson, J.; Wu, F. The dry chain: Reducing postharvest losses and improving food safety in humid climates. Trends Food Sci. Technol. 2018, 71, 84–93. [Google Scholar] [CrossRef]
- De Vitis, M.; Hay, F.; Dickie, J.B.; Trivedi, C.; Choi, J.; Fiegener, R. Seed storage: Maintaining seed viability and vigor for restoration use. Restor. Ecol. 2020, 28. [Google Scholar] [CrossRef]
- Ceccato, D.; Delatorre-Herrera, J.; Burrieza, H.; Bertero, D.; Martínez, E.; Delfino, I.; Moncada, S.; Bazile, D.; Castellión, M. Seed physiology and response to germination conditions. In State of the Art Report on Quinoa around the World in 2013; FAO: Santiago, Chile, 2015; pp. 131–142. [Google Scholar]
- Ceccato, D.V.; Bertero, H.D.; Batlla, D. Environmental control of dormancy in quinoa (Chenopodium quinoa) seeds: Two potential genetic resources for pre-harvest sprouting tolerance. Seed Sci. Res. 2011, 21, 133–141. [Google Scholar] [CrossRef]
- McGinty, E.; Murphy, K.; Hauvermale, A. Seed Dormancy and Preharvest Sprouting in Quinoa (Chenopodium quinoa Willd). Plants 2021, 10, 458. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.H.; Hong, T.D.; Jackson, M.T. Seed Production Environment, Time of Harvest, and the Potential Longevity of Seeds of Three Cultivars of Rice (Oryza sativa L.). Ann. Bot. 1993, 72, 583–590. [Google Scholar] [CrossRef]
- Romero, G.; Heredia, A.; Chaparro-Zambrano, H.N. Germinative potential in quinoa (Chenopodium quinoa Willd.) seeds stored under cool conditions. Rev. UDCA Actual. Divulg. Científ. 2018, 21, 341–350. [Google Scholar] [CrossRef] [Green Version]
- Spehar, C.R.; Santos, R.L.D.B. Quinoa BRS Piabiru: Alternative for diversification of cropping systems. Pesqui. Agropecuária Brasileira. 2002, 37, 809–893. [Google Scholar] [CrossRef] [Green Version]
- Castellión, M.; Matiacevich, S.; Buera, P.; Maldonado, S. Protein deterioration and longevity of quinoa seeds during long-term storage. Food Chem. 2010, 121, 952–958. [Google Scholar] [CrossRef]
- Prego, I.; Maldonado, S.; Otegui, M. Seed Structure and Localization of Reserves inChenopodium quinoa. Ann. Bot. 1998, 82, 481–488. [Google Scholar] [CrossRef] [Green Version]
- Ng, S.-C.; Anderson, A.; Coker, J.; Ondrus, M. Characterization of lipid oxidation products in quinoa (Chenopodium quinoa). Food Chem. 2007, 101, 185–192. [Google Scholar] [CrossRef]
- Baributsa, D.; Njoroge, A. The use and profitability of hermetic technologies for grain storage among smallholder farmers in eastern Kenya. J. Stored Prod. Res. 2020, 87, 101618. [Google Scholar] [CrossRef]
- Kiobia, D.; Silayo, V.; Mutabazi, K.; Graef, F.; Mourice, S. Performance of hermetic storage bags for maize grains under farmer-managed conditions: Good practice versus local reality. J. Stored Prod. Res. 2020, 87, 101586. [Google Scholar] [CrossRef]
- Bakhtavar, M.A.; Afzal, I. Climate smart Dry Chain Technology for safe storage of quinoa seeds. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- de Jesus Souza, F.I.; Devilla, I.A.; de Souza, R.T.; Teixeira, I.R.; Spehar, C.R. Physiological quality of quinoa seeds submitted to different storage conditions. Afr. J. Agric. Res. 2016, 11, 1299–1308. [Google Scholar] [CrossRef] [Green Version]
- Mammadi, A.; Afshari, R.T. Modeling of quinoa (Chenopodium quinoa) seed viability with probit analysis. Iran. J. Field Crop. Sci. 2018, 49, 49–57. [Google Scholar] [CrossRef]
- Ellis, R.; Hong, T.D.; Roberts, E.H. A Low-Moisture-Content Limit to Logarithmic Relations Between Seed Moisture Content and Longevity. Ann. Bot. 1988, 61, 405–408. [Google Scholar] [CrossRef]






| Soil, To Be Measured Before and After the Field Season |
|---|
| Watering regime |
| Water holding capacity |
| Composition in terms of % sand, silt, organic matter, etc. |
| Nutrient and mineral composition—total nitrogen, organic carbon, phosphorus, potassium, sulfur, etc. Note: when measuring nitrates, the soil sample must be kept cold because nitrates are unstable |
| Soil physical properties affecting plant growth |
| pH |
| Apparent density |
| Electrical conductivity (EC), especially for salinity trials |
| Weather |
| Precipitation, and irrigation schedule |
| Temperature, at least daily Tmax and Tmin, but preferably recorded continuously throughout the day to enable calculation of degree-days to flowering and to maturity |
| Humidity—relative humidity/dewpoint temperature |
| Daily irradiance (mol m−2 d−1), recorded continuously throughout the day |
| Wind speed (average daily speed) |
| Day length (including twilight time) |
| Plot-Level Phenotype | Scoring Metric | Description |
|---|---|---|
| Plot coverage | 1,3,5,7,9 * | Percentage of the plot covered, 1 = poor to 9 = good establishment |
| Plot population homogeneity | 1,3,5,7 | Judgment of homogeneity of the accession, 1 = homogeneous to 7 = mixed |
| Branchiness | 1,3,5,7 | Score for the overall amount of side branches along the entire length of the stem, ignoring very small and spindly branches, ranging from 1 = no branches to 7 = bushy plant with many (i.e., greater than 7) major lateral branches |
| Growth habit | 1,3,5,7 | Four categories of growth habit described in images on the phenotyping card. Here the focus lies on whether branching is present in the bottom third of the stem from the base of the plant and if a main inflorescence can be identified |
| Stem breakage incidence | 1,3,5,7,9 * | Stems are broken or detached, assessing the percentage of the plot affected |
| Stem lodging incidence | 1,3,5,7,9 * | Plants are prostrate, on or near the ground, with intact stems; assessing the percentage of the plot affected |
| Stem lying incidence | 1,3,5,7,9 * | Stem of the plant is not emerging straight up from the soil but has a kink at the base, growing along the ground before rising; assessing the percentage of the plot affected |
| Stem angle | 1,3,5,7 | The angle at which the majority of plants are leaning, measured between the vertical axis and the horizontal axis |
| Panicle axis angle | 1,3,5,7 | The angle at which the majority of panicle axes are leaning, measured between an upright panicle on the vertical and a panicle pointing towards the ground |
| Plant-Level Phenotype | Unit | Description |
|---|---|---|
| Plant height | cm | Height of the most representative plants of the plot, usually from the middle of the plot, measured with a long measuring stick from soil to the tip of the panicle. If more than one distinct phenotype is present, more than one plant may be recorded in a new row of the spreadsheet, with all phenotypes that are differing recorded separately |
| Panicle length | cm | Length of the primary panicle measured with the same stick. Measured from the base of the panicle to the tip |
| Stem diameter near plant base | mm | Thickness of the stem measured with calipers at the middle of the bottom third of the plant stem |
| Stem diameter under panicle | mm | Thickness of the stem measured just underneath the panicle |
| Number of significant panicles | count | Count of the number of significant panicles, i.e., larger panicles, near the top of the plant, harvestable, that provide a major contribution to the seed harvested from the plant |
| Plant-Level Phenotype | Scoring Metric | Description |
|---|---|---|
| Growth stage | BBCH scale | Phenological growth stage; very important to record at mature phenotyping |
| Seed shattering | 1,3,5,7 | Grain persistence in the plant at physiological maturity. Assessing how easy seeds fall off the panicle upon light touch: 1 = no seeds falling to 7 = majority of seeds falling |
| Panicle shape | 1,3,5 | Classified into one of the three categories: glomerulate, intermediate, or amarantiform |
| Panicle density | 1,3,5,7 | Scored from 1 = lax (loose) with panicle axes easily visible to 7 = tight and compact panicles |
| Panicle leafiness | 1,3,5,7 | Scored from 1 = no leaves to 7 = many leaves |
| Panicle color | 13,4,15,16,5,7 | Categorized according to the color phenotyping card |
| Stem color | 13,4,15 | Categorized into green (13), red (4), or no pigmentation (15) |
| Stem striae | 0,1 | Presence (1) absence (0) scoring of stem streaks or stripes |
| Axil pigmentation | 0,1 | Presence (1) absence (0) scoring of pigmented axils |
| Stem leaf shape | 1,2 | Leaves of the stem are categorized into two groups: rhomboidal (1) and triangular (2) |
| Harvest and Post-Harvest | Unit | Description |
|---|---|---|
| Number of plants harvested | count | |
| Above-ground dry biomass | grams | Cutting plants at the very base with secateurs and drying the entire plant in an oven until mass is constant. Recording total dry weight |
| Below-ground biomass | grams | If possible, root biomass could also be measured (especially when plants are growing in sandy soil) |
| Seed yield for representative plants | grams | Seed mass of approximately four representative plants that were harvested from the center of the plot (seed should be dried to constant weight) |
| Total seed yield per plot | grams × m−2 | Harvesting the panicles remaining per plot while excluding borders, and adding the weight to that from the four representative plants, seed dried in oven to constant weight |
| Seed yield per plant | grams | Total harvested seed mass per plant may be calculated from the seed weight of all plants in the plot divided by the number of plants harvested |
| Harvest index | Yield/ above-ground biomass | |
| Seed weight (TGW) | grams/1000 seeds | Thousand Grain Weight (TGW), the weight of 1000 seeds |
| Seed hectoliter weight | grams/100mL | Estimation of density, determined by weighing all seeds fitting into a 100 mL volume |
| Seed size (average area; average perimeter) | millimeter | Seed size outputs from image analysis separated by semicolon (method options described in Section 9.2) |
| Seed color (average red; average green; average blue) | Numeric RGB equivalent | Seed color output values for red, green, and blue components, semi colon separated (obtained from image analysis methods, see Section 9.2) |
| Stats from WSU Calibration V3 Data (g 100g−1 Protein) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Range | Min | Max | RMSECV | SECV | Robust SECV | RPDCV | R2CV | |
| Alanine | 1.99 | 2.89 | 4.88 | 0.022 | 0.022 | 0.018 | 3.036 | 0.892 |
| Arginine | 4.68 | 4.58 | 9.25 | 0.053 | 0.053 | 0.044 | 4.308 | 0.946 |
| Aspartic acid | 3.22 | 5.51 | 8.73 | 0.039 | 0.040 | 0.036 | 3.768 | 0.930 |
| Cysteine | 0.76 | 1.31 | 2.07 | 0.010 | 0.010 | 0.010 | 3.188 | 0.902 |
| Glutamic acid | 7.04 | 8.22 | 15.26 | 0.093 | 0.093 | 0.086 | 3.802 | 0.931 |
| Glycine | 1.33 | 4.78 | 6.11 | 0.041 | 0.041 | 0.036 | 2.447 | 0.834 |
| Histidine | 1.05 | 1.96 | 3.01 | 0.015 | 0.015 | 0.014 | 4.564 | 0.952 |
| Isoleucine | 1.51 | 2.89 | 4.41 | 0.021 | 0.022 | 0.019 | 3.392 | 0.913 |
| Leucine | 2.55 | 4.3 | 6.85 | 0.031 | 0.032 | 0.029 | 3.473 | 0.917 |
| Lysine | 3.14 | 3.45 | 6.59 | 0.029 | 0.029 | 0.033 | 3.290 | 0.908 |
| Methionine | 1.15 | 1.31 | 2.46 | 0.012 | 0.012 | 0.009 | 2.955 | 0.886 |
| Phenylalanine | 1.57 | 2.71 | 4.28 | 0.019 | 0.019 | 0.018 | 3.889 | 0.934 |
| Proline | 1.68 | 2.80 | 4.48 | 0.023 | 0.023 | 0.018 | 2.556 | 0.847 |
| Serine | 1.39 | 2.89 | 4.28 | 0.019 | 0.019 | 0.016 | 3.176 | 0.901 |
| Taurine | 1.96 | 0.82 | 2.79 | 0.012 | 0.012 | 0.009 | 1.669 | 0.645 |
| Threonine | 1.60 | 2.43 | 4.02 | 0.017 | 0.017 | 0.016 | 3.015 | 0.890 |
| Tryptophan | 0.93 | 0.55 | 1.48 | 0.012 | 0.012 | 0.009 | 1.681 | 0.647 |
| Tyrosine | 0.93 | 2.12 | 3.05 | 0.014 | 0.014 | 0.013 | 3.393 | 0.913 |
| Valine | 1.84 | 3.36 | 5.20 | 0.024 | 0.024 | 0.023 | 3.260 | 0.906 |
| Hydroxylysine | 0.18 | 0.05 | 0.23 | 0.004 | 0.004 | 0.003 | 1.591 | 0.605 |
| Hydroxyproline | 0.93 | 0.29 | 1.21 | 0.010 | 0.010 | 0.011 | 1.821 | 0.699 |
| Stats from WSU calibration V3 data (g 100g−1 sample) | ||||||||
| Crude protein | 11.95 | 6.82 | 18.77 | 0.394 | 0.395 | 0.406 | 5.521 | 0.967 |
| Ash | 3.32 | 2.21 | 5.53 | 0.154 | 0.154 | 0.129 | 3.084 | 0.895 |
| Crude fat | 6.95 | 0.00 | 6.95 | 0.310 | 0.311 | 0.316 | 3.883 | 0.934 |
| Crude fiber | 13.67 | 1.44 | 15.11 | 0.442 | 0.443 | 0.377 | 4.904 | 0.958 |
| Moisture | 3.76 | 6.41 | 10.17 | 0.183 | 0.183 | 0.159 | 6.579 | 0.977 |
| TotalAA | 10.06 | 5.84 | 15.90 | 0.413 | 0.413 | 0.328 | 4.018 | 0.938 |
| Range | Min | Max | RMSECV | SECV | Robust SECV | RPDCV | R2CV | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stanschewski, C.S.; Rey, E.; Fiene, G.; Craine, E.B.; Wellman, G.; Melino, V.J.; S. R. Patiranage, D.; Johansen, K.; Schmöckel, S.M.; Bertero, D.; et al. Quinoa Phenotyping Methodologies: An International Consensus. Plants 2021, 10, 1759. https://doi.org/10.3390/plants10091759
Stanschewski CS, Rey E, Fiene G, Craine EB, Wellman G, Melino VJ, S. R. Patiranage D, Johansen K, Schmöckel SM, Bertero D, et al. Quinoa Phenotyping Methodologies: An International Consensus. Plants. 2021; 10(9):1759. https://doi.org/10.3390/plants10091759
Chicago/Turabian StyleStanschewski, Clara S., Elodie Rey, Gabriele Fiene, Evan B. Craine, Gordon Wellman, Vanessa J. Melino, Dilan S. R. Patiranage, Kasper Johansen, Sandra M. Schmöckel, Daniel Bertero, and et al. 2021. "Quinoa Phenotyping Methodologies: An International Consensus" Plants 10, no. 9: 1759. https://doi.org/10.3390/plants10091759
APA StyleStanschewski, C. S., Rey, E., Fiene, G., Craine, E. B., Wellman, G., Melino, V. J., S. R. Patiranage, D., Johansen, K., Schmöckel, S. M., Bertero, D., Oakey, H., Colque-Little, C., Afzal, I., Raubach, S., Miller, N., Streich, J., Amby, D. B., Emrani, N., Warmington, M., ... on behalf of the Quinoa Phenotyping Consortium. (2021). Quinoa Phenotyping Methodologies: An International Consensus. Plants, 10(9), 1759. https://doi.org/10.3390/plants10091759









