Eragrostis curvula, a Model Species for Diplosporous Apomixis
Abstract
:1. Introduction
2. Diplosporous Apomixis and Sexuality in E. curvula
3. Advantages of the E. curvula Apomictic Model
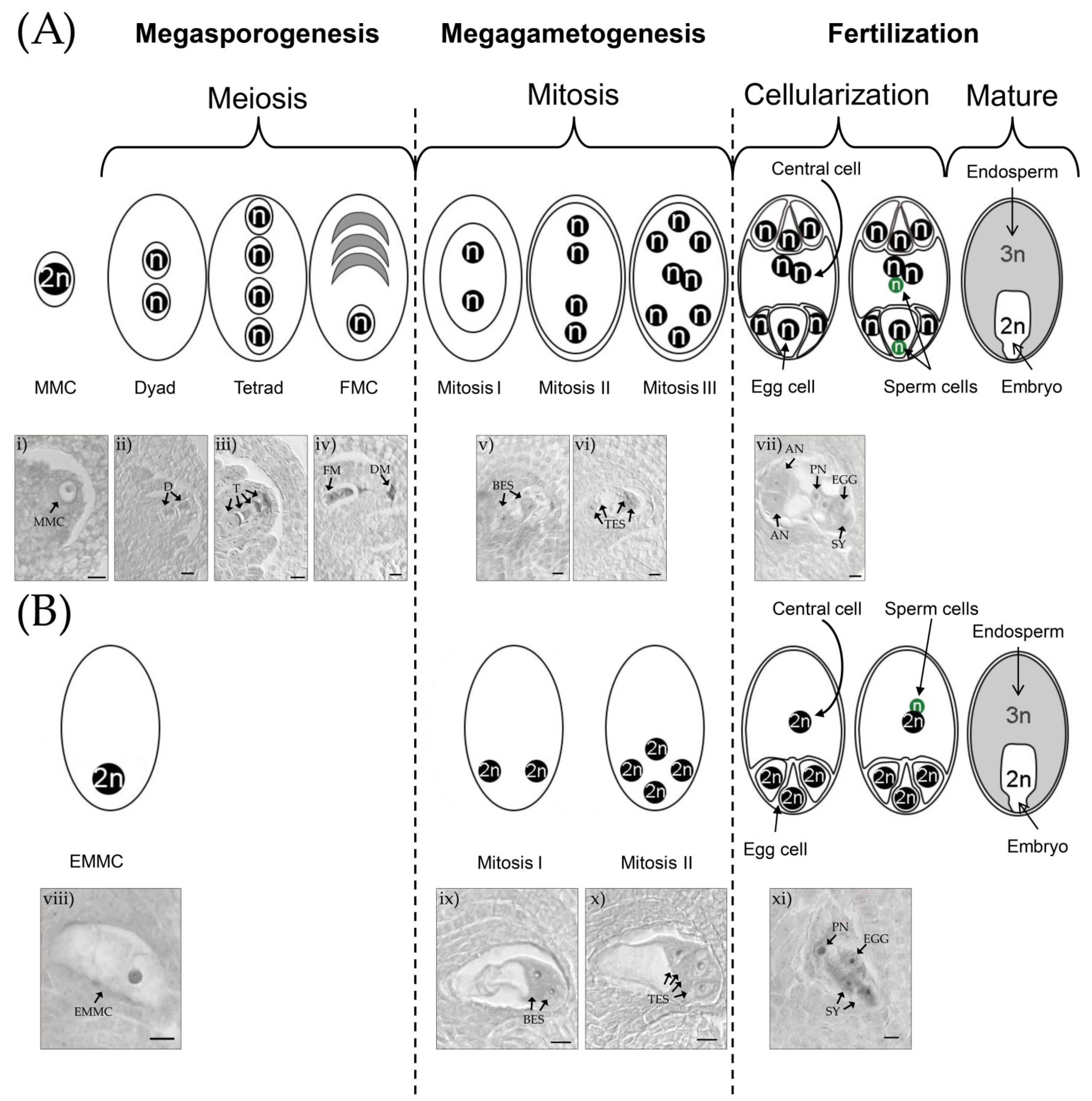
4. Methods for Assessing the Reproductive Mode. in E. curvula
5. Ploidy
6. Apomixis Inheritance in E. curvula
7. The Diplospory Genomic Region
8. Plasticity in the Apomictic: Sexual Frequency
9. Gene Expression in Apomictic and Sexual Genotypes
10. Epigenetics Regulation of Apomixis
11. State of the Art of Diplosporous Apomixis in E. curvula
12. Conclusions
- -
- Forty-two transcriptomes from leaves and inflorescences;
- -
- Four small-RNAs libraries;
- -
- The first linkage map for the species, highly saturated, with GBS-SNPs;
- -
- Three genome assemblies, one diploid and two tetraploids;
- -
- More than 4500 SSRs;
- -
- The whole methylation profiles of full apomictic, facultative, and sexual genotypes;
- -
- One genomic region and candidate genes for apomixis.
Author Contributions
Funding
Conflicts of Interest
References
- Peterson, P.M.; Romaschenko, K.; Johnson, G. A classification of the Chloridoideae (Poaceae) based on multi-gene phylogenetic trees. Mol. Phylogenet. Evol. 2010, 55, 580–598. [Google Scholar] [CrossRef]
- Ingram, A.L.; Doyle, J.J. Eragrostis (Poaceae): Monophyly and infrageneric classification. Aliso J. Syst. Evol. Bot. 2007, 23, 595–604. [Google Scholar] [CrossRef] [Green Version]
- Peterson, P.M.; Giraldo-Canas, D. The Genus Eragrostis (Poaceae: Chloridoideae) in Northwestern South America (Colombia, Ecuador, and Peru): Morphological and Taxonomic Studies; Biblioteca José Jerónimo Triana: Bogota, Colombia, 2012; Volume 24, pp. 1–195. [Google Scholar]
- Cochrane, L.; Bekele, Y.W. Average crop yield (2001–2017) in Ethiopia: Trends at national, regional and zonal levels. Data Brief 2018, 16, 1025. [Google Scholar] [CrossRef]
- Roberts, J.; Singarayer, F.; van Etten, E.; Turville, C. Germination biology of four climatically varied populations of the invasive species African lovegrass (Eragrostis curvula). Weed Sci. 2021, 69, 210–218. [Google Scholar] [CrossRef]
- Hartley, W.; Slater, C. Studies on the origin, evolution, and distribution of the Gramineae. III. The tribes of the subfamily Eragrostoideae. Aust. J. Bot. 1960, 8, 256–276. [Google Scholar] [CrossRef]
- Pickering, J.; Pickering, J. Discover Life. University of Georgia Athens. Available online: http://discoverlife.org (accessed on 15 March 2021).
- McKiernan, S.; Gill, N.; Atchison, J. Watching the grass grow: How landholders learn to live with an invasive plant in conditions of uncertainty. In Routledge Handbook of Biosecurity and Invasive Species, 1st ed.; Barker, K., Francis, R.A., Eds.; Routledge: London, UK, 2021; pp. 77–90. [Google Scholar] [CrossRef]
- Yoshioka, A.; Kadoya, T.; Suda, S.I.; Washitani, I. Impacts of weeping lovegrass (Eragrostis curvula) invasion on native grasshoppers: Responses of habitat generalist and specialist species. Biol. Invasions 2010, 12, 531–539. [Google Scholar] [CrossRef]
- Voigt, P.W.; Rethman, N.; Poverene, M. Lovegrasses. Warm-Season (C4) Grasses, 1st ed.; Moser, L.E., Burson, B.L., Sollenberger, L.E., Eds.; American Society of Agronomy: Fitchburg, MA, USA, 2004; Volume 45, pp. 1027–1055. [Google Scholar] [CrossRef]
- Ingram, A.L.; Doyle, J.J. The origin and evolution of Eragrostis tef (Poaceae) and related polyploids: Evidence from nuclear waxy and plastid rps16. Am. J. Bot. 2003, 90, 116–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colom, M.R.; Vazzana, C. Drought stress effects on three cultivars of Eragrostis curvula: Photosynthesis and water relations. Plant Growth Regul. 2001, 34, 195–202. [Google Scholar] [CrossRef]
- Colom, M.R.; Vazzana, C. Water stress effects on three cultivars of Eragrostis curvula. Ital. J. Agron. 2002, 6, 127–132. [Google Scholar]
- Msiza, N.H.; Ravhuhali, K.E.; Mokoboki, H.K.; Mavengahama, S.; Motsei, L.E. Ranking species for veld restoration in semi-arid regions using agronomic, morphological and chemical parameters of selected grass species at different developmental stages under controlled environment. Agronomy 2021, 11, 52. [Google Scholar] [CrossRef]
- Sáenz-Ceja, J.E.; Sáenz-Reyes, J.; Castillo-Quiroz, D.; Castillo-Reyes, F.; Muñoz-Flores, H.J.; Rueda-Sánchez, A. Potential areas for silvopastoral systems based on the ecological niche of two forage crops and three species of conifers. Rev. Chapingo Ser. Hortic. 2021, 27, 289–308. [Google Scholar] [CrossRef]
- Svinurai, W.; Hassen, A.; Tesfamariam, E.; Ramoelo, A.; Cullen, B. Calibration and evaluation of the Sustainable Grazing Systems pasture model for predicting native grass aboveground biomass production in southern Africa. Afr. J. Range Forage Sci. 2021, 8, 1–13. [Google Scholar] [CrossRef]
- Leigh, J.H. The relative palatability of various varieties of weeping lovegrass (Eragrostis curvula (Schrad) Nees). Grass Forage Sci. 1961, 16, 135–140. [Google Scholar] [CrossRef]
- Voigt, P.W.; Kneebone, W.R.; McIlvain, E.H.; Shoop, M.C.; Webster, J.E. Palatability, chemical composition, and animal gains from selections of weeping lovegrass, Eragrostis curvula (Schrad.) Nees. Agron. J. 1970, 62, 673–676. [Google Scholar] [CrossRef]
- Gargano, A.O.; Adúriz, M.A.; Arelovich, H.M.; Amela, M.I. Forage yield and nutritive value of Eragrostis curvula and Digitaria eriantha in central-south semi-arid Argentina. Trop. Grassl. 2001, 35, 161–167. [Google Scholar]
- Adejoro, F.A.; Hassen, A. Effect of supplementing or treating Eragrostis curvula hay with urea or nitrate on its digestibility and in vitro fermentation. S. Afr. J. Anim. Sci. 2017, 47, 168–177. [Google Scholar] [CrossRef] [Green Version]
- Follett, R.F.; Stewart, C.E.; Bradford, J.; Pruessner, E.G.; Sims, P.L.; Vigil, M.F. Long-term pasture management impacts on eolian sand soils in the southern mixed-grass prairie. Quat. Int. 2020, 565, 84–93. [Google Scholar] [CrossRef]
- Johnston, W.H.; Aveyard, J.M. Testing and selection of African lovegrass (Eragrostis curvula) for soil conservation in south-western New South Wales. Aust. Plant Introd. Rev. 1977, 12, 27–40. [Google Scholar]
- Gomes, P.I.; Asaeda, T. Spatial and temporal heterogeneity of Eragrostis curvula in the downstream flood meadow of a regulated river. Ann. Limnol.-Int. J. Limnol. 2009, 45, 181–193. [Google Scholar] [CrossRef] [Green Version]
- Brown, W.V.; Emery, W.H.P. Apomixis in the Gramineae: Panicoideae. Am. J. Bot. 1958, 253–263. [Google Scholar] [CrossRef]
- Streetman, L. Reproduction of the lovegrass, the genus Eragrostis-I E. chloromelas Steud, E. curvula (Schrad.) Nees, E. Lehmanniana Nees and E. superba Peyr. Wrightia 1963, 3, 41–51. [Google Scholar]
- Voigt, P.W.; Bashaw, E.C. Apomixis and sexuality in Eragrostis curvula. Crop. Sci. 1972, 12, 843–847. [Google Scholar] [CrossRef]
- Voigt, P.W. Registration of OTA-S weeping lovegrass germplasm (Reg. No. GP 8). Crop. Sci. 1976, 16, 886. [Google Scholar] [CrossRef]
- Voigt, P.W.; Bashaw, E.C. Facultative apomixis in Eragrostis curvula. Crop. Sci. 1976, 16, 803–806. [Google Scholar] [CrossRef]
- Echenique, V.; Díaz, M.; Polci, P.A.; Mroginski, L.A. Embryogenic cell suspensions from different explants and cultivars of Eragrostis curvula (Schrad.) Nees. Biocell 2001, 25, 131–138. [Google Scholar]
- Zappacosta, D.; Meier, M.; Carrera, A.; Pacheco, G.; Cardone, S.; Selva, J.P.; Echenique, V. Molecular markers to study the variability within the Eragrostis curvula complex. Phyton 2011, 80, 211–220. [Google Scholar]
- Cardone, S.; Polci, P.; Selva, J.P.; Mecchia, M.; Pessino, S.; Hermann, P.; Cambi, V.; Voigt, P.; Spangenberg, G.; Echenique, V. Novel genotypes of the subtropical grass Eragrostis curvula for the study of apomixis (diplospory). Euphytica 2006, 151, 263–272. [Google Scholar] [CrossRef]
- Mecchia, M.A.; Ochogavía, A.; Selva, J.P.; Laspina, N.; Felitti, S.; Martelotto, L.G.; Spangenberg, G.; Echenique, V.; Pessino, S.C. Genome polymorphisms and gene differential expression in a ‘back-and-forth’ ploidy-altered series of weeping lovegrass (Eragrostis curvula). J. Plant Physiol. 2007, 164, 1051–1061. [Google Scholar] [CrossRef]
- Cervigni, G.D.; Paniego, N.; Díaz, M.; Selva, J.P.; Zappacosta, D.; Zanazzi, D.; Landerreche, I.; Martelotto, L.; Felitti, S.; Pessino, S.; et al. Expressed sequence tag analysis and development of gene associated markers in a near-isogenic plant system of Eragrostis curvula. Plant Mol. Biol. 2008, 67, 1–10. [Google Scholar] [CrossRef]
- Cervigni, G.D.; Paniego, N.; Pessino, S.; Selva, J.P.; Díaz, M.; Spangenberg, G.; Echenique, V. Gene expression in diplosporous and sexual Eragrostis curvula genotypes with differing ploidy levels. Plant Mol. Biol. 2008, 67, 11–23. [Google Scholar] [CrossRef]
- Garbus, I.; Romero, J.R.; Selva, J.P.; Pasten, M.C.; Chinestra, C.; Carballo, J.; Zappacosta, D.; Echenique, V. De novo transcriptome sequencing and assembly from apomictic and sexual Eragrostis curvula genotypes. PLoS ONE 2017, 12, e0185595. [Google Scholar] [CrossRef]
- Meier, M.; Zappacosta, D.; Selva, J.P.; Pessino, S.; Echenique, V. Evaluation of different methods for assessing the reproductive mode of weeping lovegrass plants, Eragrostis curvula (Schrad.) Nees. Aust. J. Bot. 2011, 59, 253–261. [Google Scholar] [CrossRef]
- Zappacosta, D.; Gallardo, J.; Carballo, J.; Meier, M.; Rodrigo, J.M.; Gallo, C.A.; Selva, J.P.; Stein, J.; Ortiz, J.P.; Albertini, E.; et al. A high-density linkage map of the forage grass Eragrostis curvula and localization of the diplospory locus. Front. Plant Sci. 2019, 10, 918. [Google Scholar] [CrossRef] [Green Version]
- Romero, J.; Selva, J.P.; Pessino, S.; Echenique, V.; Garbus, I. Repetitive sequences in Eragrostis curvula cDNA EST libraries obtained from genotypes with different ploidy. Biol. Plant. 2016, 60, 55–67. [Google Scholar] [CrossRef] [Green Version]
- Carballo, J.; Santos, B.A.C.M.; Zappacosta, D.; Garbus, I.; Selva, J.P.; Gallo, C.A.; Díaz, A.; Albertini, E.; Caccamo, M.; Echenique, V. A high-quality genome of Eragrostis curvula grass provides insights into Poaceae evolution and supports new strategies to enhance forage quality. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Ochogavía, A.C.; Cervigni, G.; Selva, J.P.; Echenique, V.; Pessino, S.C. Variation in cytosine methylation patterns during ploidy level conversions in Eragrostis curvula. Plant Mol. Biol. 2009, 70, 17–29. [Google Scholar] [CrossRef]
- Zappacosta, D.; Ochogavía, A.; Rodrigo, J.M.; Romero, J.R.; Meier, M.; Garbus, I.; Pessino, S.C.; Echenique, V. Increased apomixis expression concurrent with genetic and epigenetic variation in a newly synthesized Eragrostis curvula polyploid. Sci. Rep. 2014, 4, 1–7. [Google Scholar] [CrossRef]
- Rodrigo, J.M.; Zappacosta, D.C.; Selva, J.P.; Garbus, I.; Albertini, E.; Echenique, V. Apomixis frequency under stress conditions in weeping lovegrass (Eragrostis curvula). PLoS ONE 2017, 12, 0175852. [Google Scholar] [CrossRef]
- Selva, J.P.; Siena, L.; Rodrigo, J.M.; Garbus, I.; Zappacosta, D.; Romero, J.R.; Ortiz, J.P.A.; Pessino, S.C.; Leblanc, O.; Echenique, V. Temporal and spatial expression of genes involved in DNA methylation during reproductive development of sexual and apomictic Eragrostis curvula. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Garbus, I.; Selva, J.P.; Pasten, M.C.; Bellido, A.M.; Carballo, J.; Albertini, E.; Echenique, V. Characterization and discovery of miRNA and miRNA targets from apomictic and sexual genotypes of Eragrostis curvula. BMC Genom. 2019, 20, 839. [Google Scholar] [CrossRef] [Green Version]
- Selva, J.P.; Zappacosta, D.; Carballo, J.; Rodrigo, J.M.; Bellido, A.; Gallo, C.A.; Gallardo, J.; Echenique, V. Genes modulating the increase in sexuality in the facultative diplosporous grass Eragrostis curvula under water stress conditions. Genes 2020, 11, 969. [Google Scholar] [CrossRef] [PubMed]
- Carballo, J.; Zappacosta, D.; Marconi, G.; Gallardo, J.; Di Marsico, M.; Gallo, C.A.; Caccamo, M.; Albertini, E.; Echenique, V. Differential methylation patterns in apomictic vs. sexual genotypes of the diplosporous grass Eragrostis curvula. Plants 2021, 10, 946. [Google Scholar] [CrossRef]
- Savidan, Y. Apomixis: Genetics and breeding. In Plant Breeding Reviews, 1st ed.; Janick, J., Ed.; Wiley: New York, NY, USA, 2000; Volume 18, pp. 13–86. [Google Scholar]
- Fei, X.; Shi, J.; Liu, Y.; Niu, J.; Wei, A. The steps from sexual reproduction to apomixis. Planta 2019, 249, 1715–1730. [Google Scholar] [CrossRef] [Green Version]
- Hojsgaard, D.; Klatt, S.; Baier, R.; Carman, J.G.; Hörandl, E. Taxonomy and biogeography of apomixis in angiosperms and associated biodiversity characteristics. Crit. Rev. Plant Sci. 2014, 33, 414–427. [Google Scholar] [CrossRef] [Green Version]
- Mengesha, M.H.; Guard, A.T. Development of the embryo sac and embryo of teff, Eragrostis tef. Can. J. Bot. 1966, 44, 1071–1075. [Google Scholar] [CrossRef]
- Crane, C.F. Classification of apomictic mechanisms. In The Flowering of Apomixis: From Mechanisms to Genetic Engineering; Savidan, Y., Carman, J.G., Dresselhaus, T., Eds.; ClMMYT, IRD, European Commission OC VI (FAIR): Mexico, Mexico, 2001; pp. 24–43. [Google Scholar]
- Khanday, I.; Skinner, D.; Yang, B.; Mercier, R.; Sundaresan, V. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 2019, 565, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Wang, K. Fixation of hybrid vigor in rice: Synthetic apomixis generated by genome editing. aBIOTECH 2020, 1, 15–20. [Google Scholar] [CrossRef] [Green Version]
- Calzada, J.P.; Crane, C.F.; Stelly, D.M. Apomixis—The asexual revolution. Science 1996, 274, 1322–1323. [Google Scholar] [CrossRef]
- Barcaccia, G.; Albertini, E. Apomixis in plant reproduction: A novel perspective on an old dilemma. Plant Reprod. 2013, 26, 159–179. [Google Scholar] [CrossRef] [Green Version]
- Noyes, R.D. Sexual devolution in plants: Apomixis uncloaked? Bioessays 2008, 30, 798–801. [Google Scholar] [CrossRef]
- Albertini, E.; Barcaccia, G.; Carman, J.G.; Pupilli, F. Did apomixis evolve from sex or was it the other way around? J. Exp. Bot. 2019, 70, 2951–2964. [Google Scholar] [CrossRef] [PubMed]
- Carman, J.G. Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol. J. Linn. Soc. 1997, 61, 51–94. [Google Scholar] [CrossRef]
- Hand, M.L.; Koltunow, A.M. The genetic control of apomixis: Asexual seed formation. Genetics 2014, 197, 441–450. [Google Scholar] [CrossRef] [Green Version]
- Vorster, T.B. Cytogenetic studies in the Eragrostis curvula complex. Bothalia 1977, 12, 215–221. [Google Scholar] [CrossRef] [Green Version]
- Voigt, P.W. Discovery of sexuality in Eragrostis curvula (Schrad.) Nees. Crop. Sci. 1971, 11, 424–425. [Google Scholar] [CrossRef]
- Brix, K. Sexual reproduction in Eragrostis curvula (Schrad.) Nees. Z. Pflanzenzuchtg 1974, 71, 25–32. [Google Scholar]
- Stalker, H.T.; Wright, L.N. Reproduction of Eragrostis curvula (Schrad.) Nees. J. Ariz. Acad. Sci. 1975, 10, 106–110. [Google Scholar] [CrossRef]
- Poverene, M. Contribución Citogenética y Quimiosistemática a la Taxonomía del Pasto Llorón, Eragrostis curvula (Schrad.) Nees s. lat. Ph.D. Thesis, Universidad Nacional del Sur, Bahía Blanca, Argentina, 1988. [Google Scholar]
- Winkler, H.U. Über parthenogenesis und apogamie im pflanzenreiche. Prog. Rei. Bot. 1908, 2, 293–454. [Google Scholar]
- Nogler, G.A. Gametophytic apomixis. In Embryology of Angiosperms; Johri, B.M., Ed.; Springer: Berlin, Germany, 1984; pp. 475–518. [Google Scholar]
- Vorster, T.B.; Liebenberg, H. Classification of embryo sacs in the Eragrostis curvula complex. Bothalia 1984, 15, 167–174. [Google Scholar] [CrossRef] [Green Version]
- Van Dijk, P.J.; Bakx-Schotman, J.T. Formation of unreduced megaspores (diplospory) in apomictic dandelions (Taraxacum officinale, sl) is controlled by a sex-specific dominant locus. Genetics 2004, 166, 483–492. [Google Scholar] [CrossRef] [Green Version]
- Nogler, G.A. Genetics of apospory in apomictic Ranunculus auricomus. V: Conclusion. Bot. Helv. 1984, 94, 411–422. [Google Scholar]
- Aliyu, O.M.; Schranz, M.E.; Sharbel, T.F. Quantitative variation for apomictic reproduction in the genus Boechera (Brassicaceae). Am. J. Bot. 2010, 97, 1719–1731. [Google Scholar] [CrossRef]
- Mateo de Arias, M. Effects of Plant Stress on Facultative Apomixis in Boechera (Brassicaceae). Ph.D. Thesis, Utah State University, Logan, UT, USA, 2015. [Google Scholar]
- Scott, R.J.; Spielman, M. Epigenetics: Imprinting in plants and mammals–the same but different? Curr. Biol. 2004, 14, R201–R203. [Google Scholar] [CrossRef]
- Kooter, J.M.; Matzke, M.A.; Meyer, P. Listening to the silent genes: Transgene silencing, gene regulation and pathogen control. Trends Plant Sci. 1999, 4, 340–347. [Google Scholar] [CrossRef]
- Alleman, M.; Doctor, J. Genomic imprinting in plants: Observations and evolutionary implications. Plant Mol. Biol. 2000, 43, 147–161. [Google Scholar] [CrossRef]
- Brukhin, V. Molecular and genetic regulation of apomixis. Russ. J. Genet. 2017, 53, 943–964. [Google Scholar] [CrossRef]
- FAO. World Food and Agriculture Statistical Yearbook; FAO-Food & Agriculture Organization of the United Nation: Rome, Italy, 2020. [Google Scholar] [CrossRef]
- Lin, B.Y. Ploidy barrier to endosperm development in maize. Genetics 1984, 107, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Y.; Luo, M.; Johnson, S.D.; Zhu, X.W.; Liu, L.; Huang, F.; Liu, Y.T.; Xu, P.Z.; Wu, X.J. Parental genome imbalance causes post-zygotic seed lethality and deregulates imprinting in rice. Rice 2016, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dilkes, B.P.; Comai, L. A differential dosage hypothesis for parental effects in seed development. Plant Cell 2004, 16, 3174–3180. [Google Scholar] [CrossRef] [Green Version]
- Naumova, T.N. Apomixis in Angiosperms, 1st ed.; Nucellar and Integumentary Embryony; CRC Press: Boca Raton, FL, USA; London, UK, 1993; pp. 1–152. [Google Scholar] [CrossRef]
- Asker, S.; Jerling, L. Apomixis in Plants, 1st ed.; CRC Press: Boca Raton, FL, USA, 1992; pp. 1–298. [Google Scholar]
- Koltunow, A.M.; Grossniklaus, U. Apomixis: A developmental perspective. Annu. Rev. Plant Biol. 2003, 54, 547–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batygina, T.B.; Vinogradova, G.Y. Phenomenon of polyembryony. Genetic heterogeneity of seeds. Russ. J. Dev. Biol. 2007, 38, 126–151. [Google Scholar] [CrossRef]
- Rodkiewicz, B. Callose in cell walls during megasporogenesis in angiosperms. Planta 1970, 93, 39–47. [Google Scholar] [CrossRef]
- Peel, M.D.; Carman, J.G.; Leblanc, O. Megasporocyte callose in apomictic buffelgrass, Kentucky bluegrass, Pennisetum squamulatum Fresen, Tripsacum L., and weeping lovegrass. Crop. Sci. 1997, 37, 724–732. [Google Scholar] [CrossRef]
- Pienaar, R.d.V. The chromosome number of some South Africa and introduced graminae. In The Grasses and Pastures of South Africa; Meredith, D., Ed.; Central News Agency: Johannesburg, South Africa, 1955; pp. 551–570. [Google Scholar]
- Vorster, T.B. Sitogenetiese Ondersoek van die Eragrostis Curvula (Schrad.) Nees-Kompleks. Ph.D. Thesis, University of Pretoria, Pretoria, South Africa, 1978. [Google Scholar]
- Spies, J.J. Stomatal area as an anatomical criterion for the determination of chromosome number in the Eragrostis curvula complex. Bothalia 1982, 14, 119–122. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; McKibben, M.T.; Finch, G.S.; Blischak, P.D.; Sutherland, B.L.; Barker, M.S. Patterns and processes of diploidization in land plants. Annu. Rev. Plant Biol. 2021, 72. [Google Scholar] [CrossRef]
- Le Comber, S.C.; Ainouche, M.L.; Kovarik, A.; Leitch, A.R. Making a functional diploid: From polysomic to disomic inheritance. New Phytol. 2010, 186, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.; Gaeta, R.T.; Pires, J.C. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Natl. Acad. Sci. USA 2011, 108, 7908–7913. [Google Scholar] [CrossRef] [Green Version]
- Richards, A.J. Plant Breeding Systems; Chapman and Hall: London, UK, 1997; pp. 1–529. [Google Scholar]
- Lovell, J.T.; Aliyu, O.M.; Mau, M.; Schranz, M.E.; Koch, M.; Kiefer, C.; Song, B.H.; Mitchell-Olds, T.; Sharbel, T.F. On the origin and evolution of apomixis in Boechera. Plant Reprod. 2013, 26, 309–315. [Google Scholar] [CrossRef] [Green Version]
- Hojsgaard, D.; Hörandl, E. The rise of apomixis in natural plant populations. Front. Plant Sci. 2019, 10, 358. [Google Scholar] [CrossRef]
- Desrochers, A.M.; Rieseberg, L.H. Mentor effects in wild species of Helianthus (Asteraceae). Am. J. Bot. 1998, 85, 770–775. [Google Scholar] [CrossRef]
- Ozias-Akins, P.; Roche, D.; Hanna, W.W. Tight clustering and hemizygosity of apomixis-linked molecular markers in Pennisetum squamulatum implies genetic control of apospory by a divergent locus that may have no allelic form in sexual genotypes. Proc. Natl. Acad. Sci. USA 1998, 95, 5127–5132. [Google Scholar] [CrossRef] [Green Version]
- Hojsgaard, D.; Martínez, E.J.; Acuña, C.A.; Quarin, C.L.; Pupilli, F. A molecular map of the apomixis-control locus in Paspalum procurrens and its comparative analysis with other species of Paspalum. Theor. Appl. Genet. 2011, 123, 959–971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akiyama, Y.; Conner, J.A.; Goel, S.; Morishige, D.T.; Mullet, J.E.; Hanna, W.W.; Ozias-Akins, P. High-resolution physical mapping in Pennisetum squamulatum reveals extensive chromosomal heteromorphism of the genomic region associated with apomixis. Plant Physiol. 2004, 134, 1733–1741. [Google Scholar] [CrossRef] [Green Version]
- Goel, S.; Chen, Z.; Akiyama, Y.; Conner, J.A.; Basu, M.; Gualtieri, G.; Hanna, W.W.; Ozias-Akins, P. Comparative physical mapping of the apospory-specific genomic region in two apomictic grasses: Pennisetum squamulatum and Cenchrus ciliaris. Genetics 2006, 173, 389–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conner, J.A.; Goel, S.; Gunawan, G.; Cordonnier-Pratt, M.M.; Johnson, V.E.; Liang, C.; Wang, H.; Pratt, L.H.; Mullet, J.E.; DeBarry, J.; et al. Sequence analysis of bacterial artificial chromosome clones from the apospory-specific genomic region of Pennisetum and Cenchrus. Plant Physiol. 2008, 147, 1396–1411. [Google Scholar] [CrossRef] [Green Version]
- Galla, G.; Siena, L.A.; Ortiz, J.P.; Baumlein, H.; Barcaccia, G.; Pessino, S.C.; Bellucci, M.; Pupilli, F. A portion of the apomixis locus of Paspalum simplex is microsyntenic with an unstable chromosome segment highly conserved among Poaceae. Sci. Rep. 2019, 9, 3271. [Google Scholar] [CrossRef] [Green Version]
- Knox, R.B. Apomixis: Seasonal and population differences in a grass. Science 1967, 157, 325–326. [Google Scholar] [CrossRef]
- Evans, L.T.; Knox, R.B. Environmental control of reproduction in Themeda australis. Aust. J. Bot. 1969, 17, 375–389. [Google Scholar] [CrossRef]
- Gounaris, E.K.; Sherwood, R.T.; Gounaris, I.; Hamilton, R.H.; Gustine, D.L. Inorganic salts modify embryo sac development in sexual and aposporous Cenchrus ciliaris. Sex. Plant Reprod. 1991, 4, 188–192. [Google Scholar] [CrossRef]
- Houliston, G.J.; Chapman, H.M.; Bicknell, R.A. The influence of genotype and environment on the fecundity and facultative expression of apomixis in Hieracium pilosella. Folia Geobot. 2006, 41, 165–181. [Google Scholar] [CrossRef]
- Klatt, S.; Hadacek, F.; Hodač, L.; Brinkmann, G.; Eilerts, M.; Hojsgaard, D.; Hörandl, E. Photoperiod extension enhances sexual megaspore formation and triggers metabolic reprogramming in facultative apomictic Ranunculus auricomus. Front. Plant Sci. 2016, 7, 278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, Z.; Morales-Ruiz, T.; Ariza, R.R.; Roldán-Arjona, T.; David, L.; Zhu, J.K. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 2002, 111, 803–814. [Google Scholar] [CrossRef] [Green Version]
- Carballo, J.; Gallo, C.A.; Selva, J.P.; Garbus, I.; Zappacosta, D.; Santos, B.A.; Albertini, E.; Were, V.; Talbot, N.; Moscou, M.J.; et al. Novel genomic resources for the polyploid forage grass Eragrostis curvula. In Proceedings of the Plant and Animal Genome XXVIII Conference, San Diego, CA, USA, 11–15 January 2020. [Google Scholar]
- Tsaftaris, A.S.; Polidoros, A.N.; Koumproglou, R.A.; Tani, E.L.; Kovacevic, N.I.; Abatzidou, E.L. Epigenetic mechanisms in plants and their implications in plant breeding. In Proceedings of the Wake of the Double Helix: From the Green Revolution to the Gene Revolution Congress, Bologna, Italy, 27 May 2003; Tuberosa, R., Phillips, R.L., Gale, M., Eds.; Avenue Media: Bologna, Italy, 2005; pp. 157–171. [Google Scholar]
- Munshi, A.; Ahuja, Y.R.; Bahadur, B. Epigenetic mechanisms in plants: An overview. Plant Biol. Biotechnol. 2015, 265–278. [Google Scholar] [CrossRef]
- Jung, C.H.; O’brien, M.; Singh, M.B.; Bhalla, P.L. Epigenetic landscape of germline specific genes in the sporophyte cells of Arabidopsis thaliana. Front. Plant Sci. 2015, 3, 328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komiya, R.; Ohyanagi, H.; Niihama, M.; Watanabe, T.; Nakano, M.; Kurata, N.; Nonomura, K.I. Rice germline-specific Argonaute MEL 1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J. 2014, 78, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Bicknell, R.A.; Koltunow, A.M. Understanding apomixis: Recent advances and remaining conundrums. Plant Cell 2004, 16, S228–S245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soliman, M.; Podio, M.; Marconi, G.; Di Marsico, M.; Ortiz, J.P.A.; Albertini, E.; Delgado, L. Differential epigenetic marks are associated with apospory expressivity in diploid hybrids of Paspalum rufum. Plants 2021, 10, 793. [Google Scholar] [CrossRef] [PubMed]
- Podio, M.; Cáceres, M.E.; Samoluk, S.S.; Seijo, J.G.; Pessino, S.C.; Ortiz, J.P.; Pupilli, F. A methylation status analysis of the apomixis-specific region in Paspalum spp. suggests an epigenetic control of parthenogenesis. J. Exp. Bot. 2014, 65, 6411–6424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amiteye, S.; Corral, J.M.; Vogel, H.; Kuhlmann, M.; Mette, M.F.; Sharbel, T.F. Novel microRNAs and microsatellite-like small RNAs in sexual and apomictic Boechera species. MicroRNA 2013, 2, 46–63. [Google Scholar] [CrossRef]
- Sahu, P.P.; Pandey, G.; Sharma, N.; Puranik, S.; Muthamilarasan, M.; Prasad, M. Epigenetic mechanisms of plant stress responses and adaptation. Plant Cell Rep. 2013, 32, 1151–1159. [Google Scholar] [CrossRef] [PubMed]
- Kaeppler, S.M.; Phillips, R.L. Tissue culture-induced DNA methylation variation in maize. Proc. Natl. Acad. Sci. USA 1993, 90, 8773–8776. [Google Scholar] [CrossRef] [Green Version]
- Fang, X.; Qi, Y. RNAi in plants: An argonaute-centered view. Plant Cell 2016, 28, 272–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Lang, Z.; Zhu, J.K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef]
- Singh, M.; Goel, S.; Meeley, R.B.; Dantec, C.; Parrinello, H.; Michaud, C.; Leblanc, O.; Grimanelli, D. Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell 2011, 23, 443–458. [Google Scholar] [CrossRef] [Green Version]
- Papa, C.M.; Springer, N.M.; Muszynski, M.G.; Meeley, R.; Kaeppler, S.M. Maize chromomethylase Zea methyltransferase2 is required for CpNpG methylation. Plant Cell 2001, 13, 1919–1928. [Google Scholar] [CrossRef] [Green Version]
- Zemach, A.; Kim, M.Y.; Hsieh, P.H.; Coleman-Derr, D.; Eshed-Williams, L.; Thao, K.; Harmer, S.L.; Zilberman, D. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 2013, 153, 193–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, J.; Liang, W.; An, G.; Zhang, D. OsMADS6 controls flower development by activating rice FACTOR OF DNA METHYLATION LIKE1. Plant Physiol. 2018, 177, 713–727. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Wang, C.; Liu, H.; Zhou, Q.; Liu, Q.; Guo, Y.; Peng, T.; Song, J.; Zhang, J.; Chen, L.; et al. Identification and analysis of adenine N 6-methylation sites in the rice genome. Nat. Plants 2018, 4, 554–563. [Google Scholar] [CrossRef]
- Zühl, L.; Volkert, C.; Ibberson, D.; Schmidt, A. Differential activity of F-box genes and E3 ligases distinguishes sexual versus apomictic germline specification in Boechera. J. Exp. Bot. 2019, 70, 5643–5657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lechner, E.; Leonhardt, N.; Eisler, H.; Parmentier, Y.; Alioua, M.; Jacquet, H.; Leung, J.; Genschik, P. MATH/BTB CRL3 receptors target the homeodomain-leucine zipper ATHB6 to modulate abscisic acid signaling. Dev. Cell 2011, 21, 1116–1128. [Google Scholar] [CrossRef] [Green Version]
- Figueiredo, D.D.; Köhler, C. Auxin: A molecular trigger of seed development. Genes Dev. 2018, 32, 479–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Accession | Ploidy | Reproductive Mode | Molecular Resources |
|---|---|---|---|
| Victoria | 2× | Sexual | Genome [39]/ESTs [33]/SSRs [35,39] |
| PI299920 | 2× | Sexual | SSRs [35,39] |
| PI208214 | 2× | Sexual | SSRs [35,39] |
| PI219919 | 2× | Sexual | SSRs [35,39] |
| PI219928 | 2× | Sexual | SSRs [35,39] |
| Tanganyika USDA | 4× | Full apomictic | RNA-seq [35]/SSRs [35,39]/MCSeEd [46] |
| Tanganyika INTA | 4× | Facultative apomictic | ESTs [33]/Genome */SSRs [35,39] |
| Ermelo | 4× | Facultative apomictic | SSRs [35,39] |
| Morpa | 4× | Facultative apomictic | SSRs [35,39] |
| OTA-S | 4× | Sexual | RNA-seq [35]/SSRs [35,39]/GBS [37]/MCSeEd [46] |
| Catalina | 4× | Facultative apomictic | - |
| Don Walter | 4× | Facultative apomictic | RNA-seq [45]/SSRs [35,39]/GBS [37]/Genome */MCSeEd [46] |
| Bahiense | 4× | Facultative apomictic | ESTs [33]/SSRs [33] |
| 62 Hybrids (F1) | 4× | Apomictic:sexual (1:1) | GBS [37]/DArT-seq */AFLPs [37]/SSRs [37] |
| Don Eduardo | 6× | Apomictic | SSRs [35,39] |
| Don Luis | 6× | Apomictic | SSRs [35,39] |
| Kromdraai | 6× | Facultative apomictic | SSRs [35,39] |
| Don Pablo | 7× | Apomictic | SSRs [35,39] |
| Don Juan | 8× | Apomictic | SSRs [35,39] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carballo, J.; Zappacosta, D.; Selva, J.P.; Caccamo, M.; Echenique, V. Eragrostis curvula, a Model Species for Diplosporous Apomixis. Plants 2021, 10, 1818. https://doi.org/10.3390/plants10091818
Carballo J, Zappacosta D, Selva JP, Caccamo M, Echenique V. Eragrostis curvula, a Model Species for Diplosporous Apomixis. Plants. 2021; 10(9):1818. https://doi.org/10.3390/plants10091818
Chicago/Turabian StyleCarballo, Jose, Diego Zappacosta, Juan Pablo Selva, Mario Caccamo, and Viviana Echenique. 2021. "Eragrostis curvula, a Model Species for Diplosporous Apomixis" Plants 10, no. 9: 1818. https://doi.org/10.3390/plants10091818







