Prohydrojasmon Promotes the Accumulation of Phenolic Compounds in Red Leaf Lettuce
Abstract
:1. Introduction
2. Results
2.1. PDJ Treatments Increase the Phenolics Contents in Aerial Parts of Lettuce Plant
2.2. Extracts of PDJ-Treated Lettuce Plants Increased in Antioxidant Activities
2.3. Quantification and Identification of the Pheolics in the Leaf Extracts from Lettuce Plants Treated with PDJ
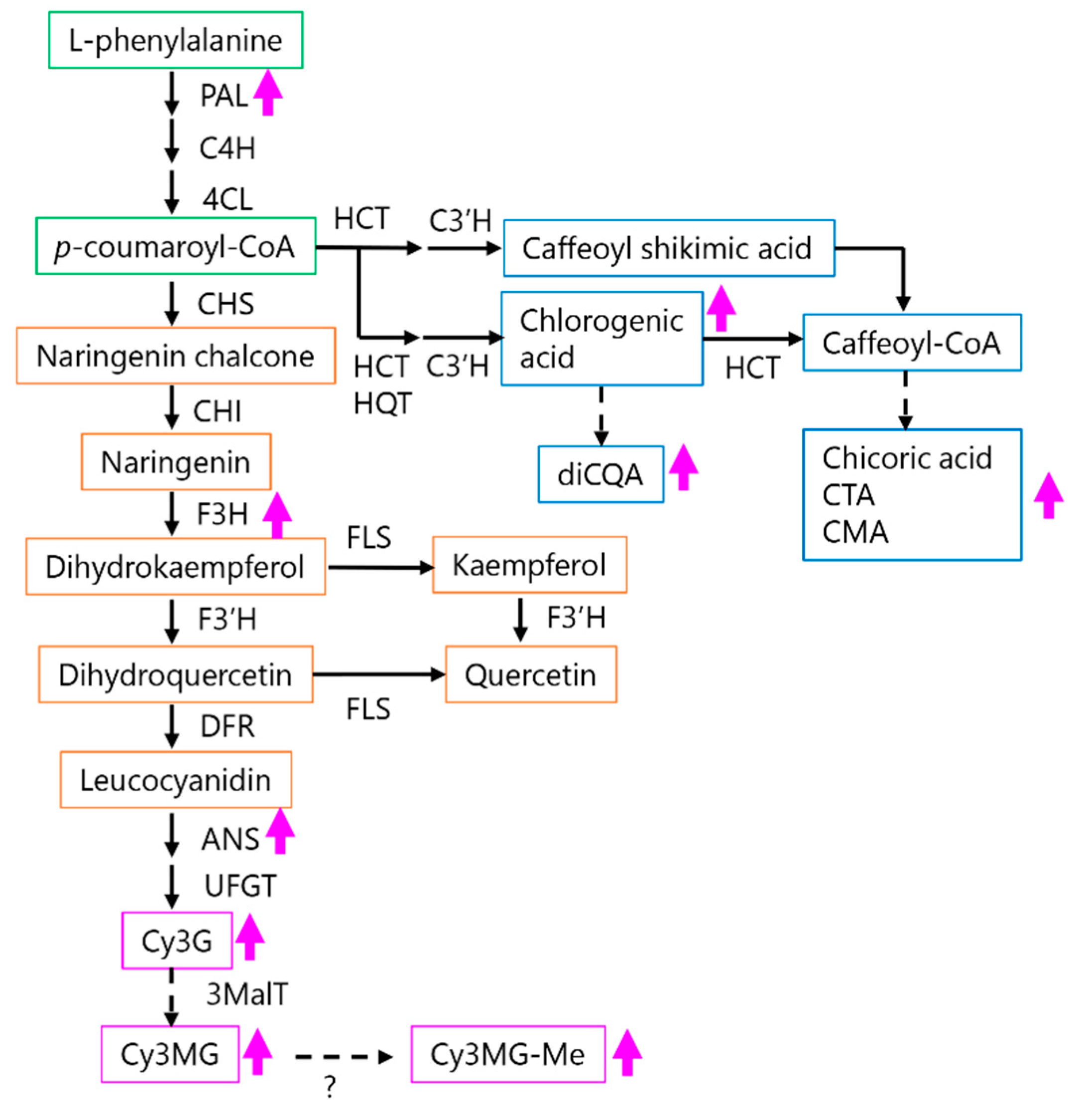
2.4. Expression Analysis of Lettuce Genes Involved in the Phenylpropanoid Derivative Synthesis Pathway in PDJ-Treated Lettuce
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. PDJ Treatments
4.3. Extraction of Phenolics from Lettuce Leaves
4.4. Measurement of the Total Phenolic Contents of Lettuce Extracts
4.5. Measurement of the Compounds Aabsorbing at 305 and 540 nm Contents of Lettuce Extracts
4.6. Measurement of the Antioxidant Activity of Lettuce Extracts
4.7. HPLC and LC-MS Analyses of the Extracts from PDJ-Treated Lettuce
4.8. Gene Expression Analysis of Phenylpropanoid Derivative Synthesis Genes in PDJ-Treated Lettuce
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cartea, M.E.; Francisco, M.; Soengas, P.; Velasco, P. Phenolic Compounds in Brassica Vegetables. Molecules 2011, 16, 251–280. [Google Scholar] [CrossRef] [PubMed]
- Arts, I.C.; Hollman, P.C.H. Polyphenols and disease risk in epidemiologic studies. Am. J. Clin. Nutr. 2005, 81, 317S–325S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jahangir, M.; Kim, H.K.; Choi, Y.H.; Verpoorte, R. Health-Affecting Compounds in Brassicaceae. Compr. Rev. Food Sci. Food Saf. 2009, 8, 31–43. [Google Scholar] [CrossRef]
- Liu, R.H. Health benefits of fruit and vegetables are from additive and synergistic combinations of phytochemicals. Am. J. Clin. Nutr. 2003, 78, 517S–520S. [Google Scholar] [CrossRef]
- Fraga, C.G.; Galleano, M.; Verstraeten, S.V.; Oteiza, P.I. Basic biochemical mechanisms behind the health benefits of polyphenols. Mol. Asp. Med. 2010, 31, 435–445. [Google Scholar] [CrossRef]
- Dupont, M.S.; Mondin, Z.; Williamson, G.; Price, K.R. Effect of Variety, Processing, and Storage on the Flavonoid Glycoside Content and Composition of Lettuce and Endive. J. Agric. Food Chem. 2000, 48, 3957–3964. [Google Scholar] [CrossRef] [PubMed]
- Nicolle, C.; Cardinault, N.; Gueux, E.; Jaffrelo, L.; Rock, E.; Mazur, A.; Amouroux, P.; Remesy, C. Health effect of vegetable-based diet: Lettuce consumption improves cholesterol metabolism and antioxidant status in the rat. Clin. Nutr. 2004, 23, 605–614. [Google Scholar] [CrossRef]
- Llorach, R.; Martinez-Sanchez, A.; Tomas-Barberan, F.A.; Gil, M.I.; Ferreres, F. Characterisation of polyphenols and antioxidant properties of five lettuce varieties and escarole. Food Chem. 2008, 108, 1028–1038. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, X.; Sandhu, A.K.; Gu, L.W. Effects of Exogenous Abscisic Acid on Yield, Antioxidant Capacities, and Phytochemical Contents of Greenhouse Grown Lettuces. J. Agric. Food Chem. 2010, 58, 6503–6509. [Google Scholar] [CrossRef]
- Becker, C.; Klaering, H.-P.; Kroh, L.W.; Krumbein, A. Cool-cultivated red leaf lettuce accumulates cyanidin-3-O-(6″-O-malonyl)-glucoside and caffeoylmalic acid. Food Chem. 2014, 146, 404–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitazaki, K.; Fukushima, A.; Nakabayashi, R.; Okazaki, Y.; Kobayashi, M.; Mori, T.; Nishizawa, T.; Reyes-Chin-Wo, S.; Michelmore, R.W.; Saito, K.; et al. Metabolic Reprogramming in Leaf Lettuce Grown Under Different Light Quality and Intensity Conditions Using Narrow-Band LEDs. Sci. Rep. 2018, 8, 7914. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-J.; Son, J.E.; Oh, M.-M. Growth and phenolic compounds of Lactuca sativa L. grown in a closed-type plant production system with UV-A, -B, or -C lamp. J. Sci. Food Agric. 2014, 94, 197–204. [Google Scholar] [CrossRef]
- Sparvoli, F.; Martin, C.; Scienza, A.; Gavazzi, G.; Tonelli, C. Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape (Vitis vinifera L.). Plant Mol. Biol. 1994, 24, 743–755. [Google Scholar] [CrossRef] [PubMed]
- Su, N.; Lu, Y.; Wu, Q.; Liu, Y.; Xia, Y.; Xia, K.; Cui, J. UV-B-induced anthocyanin accumulation in hypocotyls of radish sprouts continues in the dark after irradiation. J. Sci. Food Agric. 2015, 96, 886–892. [Google Scholar] [CrossRef]
- Zhou, W.W.; Liang, X.; Zhang, Y.X.; Dai, P.B.; Liang, B.; Li, J.L.; Sun, C.L.; Lin, X.Y. Role of sucrose in modulating the low-nitrogen-induced accumulation of phenolic compounds in lettuce (Lactuca sativa L.). J. Sci. Food Agric. 2020, 100, 5412–5421. [Google Scholar] [CrossRef]
- Li, T.; Jia, K.-P.; Lian, H.-L.; Yang, X.; Li, L.; Yang, H.-Q. Jasmonic acid enhancement of anthocyanin accumulation is dependent on phytochrome A signaling pathway under far-red light in Arabidopsis. Biochem. Biophys. Res. Commun. 2014, 454, 78–83. [Google Scholar] [CrossRef]
- Creelman, R.A.; Mullet, J.E. Jasmonic acid distribution and action in plants: Regulation during development and response to biotic and abiotic stress. Proc. Natl. Acad. Sci. USA 1995, 92, 4114–4119. [Google Scholar] [CrossRef] [Green Version]
- Rohwer, C.L.; Erwin, J.E. Horticultural applications of jasmonates: A review. J. Hortic. Sci. Biotech. 2008, 83, 283–304. [Google Scholar] [CrossRef]
- Mandour, N.S.; Kainoh, Y.; Ozawa, R.; Uefune, M.; Takabayashi, J. Effects of prohydrojasmon-treated corn plants on attractiveness to parasitoids and the performance of their hosts. J. Appl. Entomol. 2013, 137, 104–112. [Google Scholar] [CrossRef]
- Cetinbas, M.; Butar, S.; Sesli, Y.; Sekmen, S.; Yaman, B. The Effects of Pre-Harvest Prohydrojasmon Application on Yield and Fruit Quality of ‘Starking Delicious’ Apples in Different Locations. Fresen. Environ. Bull. 2018, 27, 7438–7445. [Google Scholar]
- Azis, H.R.; Takahashi, S.; Koshiyama, M.; Fujisawa, H.; Isoda, H. Effect of Prohydrojasmon on the Growth of Eggplant and Komatsuna. Plants 2020, 9, 1368. [Google Scholar] [CrossRef]
- Sato, K.; Ikoma, Y. Improvement in Handpicking Efficiency of Satsuma Mandarin Fruit with Combination Treatments of Gibberellin, Prohydrojasmon and Ethephon. Hortic. J. 2017, 86, 283–290. [Google Scholar] [CrossRef] [Green Version]
- Pu, C.-H.; Lin, S.-K.; Chuang, W.-C.; Shyu, T.-H. Modified QuEChERS method for 24 plant growth regulators in grapes using LC-MS/MS. J. Food Drug Anal. 2018, 26, 637–648. [Google Scholar] [CrossRef] [Green Version]
- Sudheeran, P.K.; Love, C.; Feygenberg, O.; Maurer, D.; Ovadia, R.; Oren-Shamir, M.; Alkan, N. Induction of red skin and improvement of fruit quality in ‘Kent’, ‘Shelly’ and ‘Maya’ mangoes by preharvest spraying of prohydrojasmon at the orchard. Postharvest Biol. Technol. 2019, 149, 18–26. [Google Scholar] [CrossRef]
- Kumar, S.P.; Maurer, D.; Feygenberg, O.; Love, C.; Alkan, N. Improving the Red Color and Fruit Quality of ‘Kent’ Mango Fruit by Pruning and Preharvest Spraying of Prohydrojasmon or Abscisic Acid. Agronomy 2020, 10, 944. [Google Scholar] [CrossRef]
- Wang, X.Q.; Cao, X.Y.; Shang, Y.; Bu, H.D.; Wang, T.Y.; Lyu, D.G.; Du, G.D. Preharvest application of prohydrojasmon affects color development, phenolic metabolism, and pigment-related gene expression in red pear (Pyrus ussuriensis). J. Sci. Food Agric. 2020, 100, 4766–4775. [Google Scholar] [CrossRef]
- Azis, H.R.; Etteieb, S.; Takahashi, S.; Koshiyama, M.; Fujisawa, H.; Isoda, H. Effect of prohydrojasmon on total phenolic content, anthocyanin accumulation and antioxidant activity in komatsuna and lettuce. Biosci. Biotechnol. Biochem. 2020, 84, 178–186. [Google Scholar] [CrossRef]
- Schütz, K.; Persike, M.; Carle, R.; Schieber, A. Characterization and quantification of anthocyanins in selected artichoke (Cynara scolymus L.) cultivars by HPLC–DAD–ESI–MSn. Anal. Bioanal. Chem. 2006, 384, 1511–1517. [Google Scholar] [CrossRef]
- Mulabagal, V.; Ngouajio, M.; Nair, A.; Zhang, Y.; Gottumukkala, A.L.; Nair, M.G. In vitro evaluation of red and green lettuce (Lactuca sativa) for functional food properties. Food Chem. 2010, 118, 300–306. [Google Scholar] [CrossRef]
- Carazzone, C.; Mascherpa, D.; Gazzani, G.; Papetti, A. Identification of phenolic constituents in red chicory salads (Cichorium intybus) by high-performance liquid chromatography with diode array detection and electrospray ionisation tandem mass spectrometry. Food Chem. 2013, 138, 1062–1071. [Google Scholar] [CrossRef]
- Brito, A.; Areche, C.; Sepulveda, B.; Kennelly, E.J.; Simirgiotis, M.J. Anthocyanin characterization, total phenolic quantification and antioxidant features of some Chilean edible berry extracts. Molecules 2014, 19, 10936–10955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Budryn, G.; Nebesny, E.; Podsędek, A.; Żyżelewicz, D.; Materska, M.; Jankowski, S.; Janda, B. Effect of different extraction methods on the recovery of chlorogenic acids, caffeine and Maillard reaction products in coffee beans. Eur. Food Res. Technol. 2009, 228, 913–922. [Google Scholar] [CrossRef]
- Liang, Y.-S.; Choi, Y.H.; Kim, H.K.; Linthorst, H.J.M.; Verpoorte, R. Metabolomic analysis of methyl jasmonate treated Brassica rapa leaves by 2-dimensional NMR spectroscopy. Phytochemistry 2006, 67, 2503–2511. [Google Scholar] [CrossRef]
- Materska, M.; Olszówka, K.; Chilczuk, B.; Stochmal, A.; Pecio, Ł.; Pacholczyk-Sienicka, B.; Piacente, S.; Pizza, C.; Masullo, M. Polyphenolic profiles in lettuce (Lactuca sativa L.) after CaCl2 treatment and cold storage. Eur. Food Res. Technol. 2018, 245, 733–744. [Google Scholar] [CrossRef] [Green Version]
- Sobolev, V.S.; Horn, B.W.; Potter, T.L.; Deyrup, S.; Gloer, J.B. Production of Stilbenoids and Phenolic Acids by the Peanut Plant at Early Stages of Growth. J. Agric. Food Chem. 2006, 54, 3505–3511. [Google Scholar] [CrossRef]
- Tong, J.; Ma, B.; Ge, L.; Mo, Q.; Zhou, G.; He, J.; Wang, Y. Dicaffeoylquinic Acid-Enriched Fraction of Cichorium glandulosum Seeds Attenuates Experimental Type 1 Diabetes via Multipathway Protection. J. Agric. Food Chem. 2015, 63, 10791–10802. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, W.; Lu, J.; Zhao, M. Phenolic profiles and antioxidant activities of commercial beers. Food Chem. 2010, 119, 1150–1158. [Google Scholar] [CrossRef]
- Ribas-Agusti, A.; Gratacos-Cubarsi, M.; Sarraga, C.; Garcia-Regueiro, J.A.; Castellari, M. Analysis of Eleven Phenolic Compounds Including Novel p-Coumaroyl Derivatives in Lettuce (Lactuca sativa L.) by Ultra-high-performance Liquid Chromatography with Photodiode Array and Mass Spectrometry Detection. Phytochem. Anal. 2011, 22, 555–563. [Google Scholar] [CrossRef]
- Damerum, A.; Selmes, S.L.; Biggi, G.F.; Clarkson, G.J.; Rothwell, S.D.; Truco, M.J.; Michelmore, R.W.; Hancock, R.; Shellcock, C.; Chapman, M.; et al. Elucidating the genetic basis of antioxidant status in lettuce (Lactuca sativa). Hortic. Res. 2015, 2, 15055. [Google Scholar] [CrossRef] [Green Version]
- Gurdon, C.; Poulev, A.; Armas, I.; Satorov, S.; Tsai, M.; Raskin, I. Genetic and Phytochemical Characterization of Lettuce Flavonoid Biosynthesis Mutants. Sci. Rep. 2019, 9, 3305. [Google Scholar] [CrossRef] [Green Version]
- Brugliera, F.; Tao, G.Q.; Tems, U.; Kalc, G.; Mouradova, E.; Price, K.; Stevenson, K.; Nakamura, N.; Stacey, I.; Katsumoto, Y.; et al. Violet/blue chrysanthemums--metabolic engineering of the anthocyanin biosynthetic pathway results in novel petal colors. Plant Cell Physiol. 2013, 54, 1696–1710. [Google Scholar] [CrossRef] [Green Version]
- Taguchi, G.; Ubukata, T.; Nozue, H.; Kobayashi, Y.; Takahi, M.; Yamamoto, H.; Hayashida, N. Malonylation is a key reaction in the metabolism of xenobiotic phenolic glucosides in Arabidopsis and tobacco. Plant J. 2010, 63, 1031–1041. [Google Scholar] [CrossRef]
- Neill, S.O.; Gould, K.S. Anthocyanins in leaves: Light attenuators or antioxidants? Funct. Plant Biol. 2003, 30, 865–873. [Google Scholar] [CrossRef] [Green Version]
- Becker, C.; Klaering, H.-P.; Schreiner, M.; Kroh, L.W.; Krumbein, A. Unlike Quercetin Glycosides, Cyanidin Glycoside in Red Leaf Lettuce Responds More Sensitively to Increasing Low Radiation Intensity before than after Head Formation Has Started. J. Agric. Food Chem. 2014, 62, 6911–6917. [Google Scholar] [CrossRef] [PubMed]
- Clifford, M.N.; Jaganath, I.B.; Ludwig, I.A.; Crozier, A. Chlorogenic acids and the acyl-quinic acids: Discovery, biosynthesis, bioavailability and bioactivity. Nat. Prod. Rep. 2017, 34, 1391–1421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.-J.; Chen, F.; Wang, X.; Choi, J.-H. Effect of Methyl Jasmonate on Phenolics, Isothiocyanate, and Metabolic Enzymes in Radish Sprout (Raphanus sativus L.). J. Agric. Food Chem. 2006, 54, 7263–7269. [Google Scholar] [CrossRef] [PubMed]
- Flores, G.; del Castillo, M.L.R. Influence of preharvest and postharvest methyl jasmonate treatments on flavonoid content and metabolomic enzymes in red raspberry. Postharvest Biol. Technol. 2014, 97, 77–82. [Google Scholar] [CrossRef] [Green Version]
- Shan, X.Y.; Zhang, Y.S.; Peng, W.; Wang, Z.L.; Xie, D.X. Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J. Exp. Bot. 2009, 60, 3849–3860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Milmanda, G.L.; Crocco, C.D.; Reichelt, M.; Mazza, C.A.; Kollner, T.G.; Zhang, T.; Cargnel, M.D.; Lichy, M.Z.; Fiorucci, A.S.; Fankhauser, C.; et al. A light-dependent molecular link between competition cues and defence responses in plants. Nat. Plants 2020, 6, 223–230. [Google Scholar] [CrossRef] [Green Version]
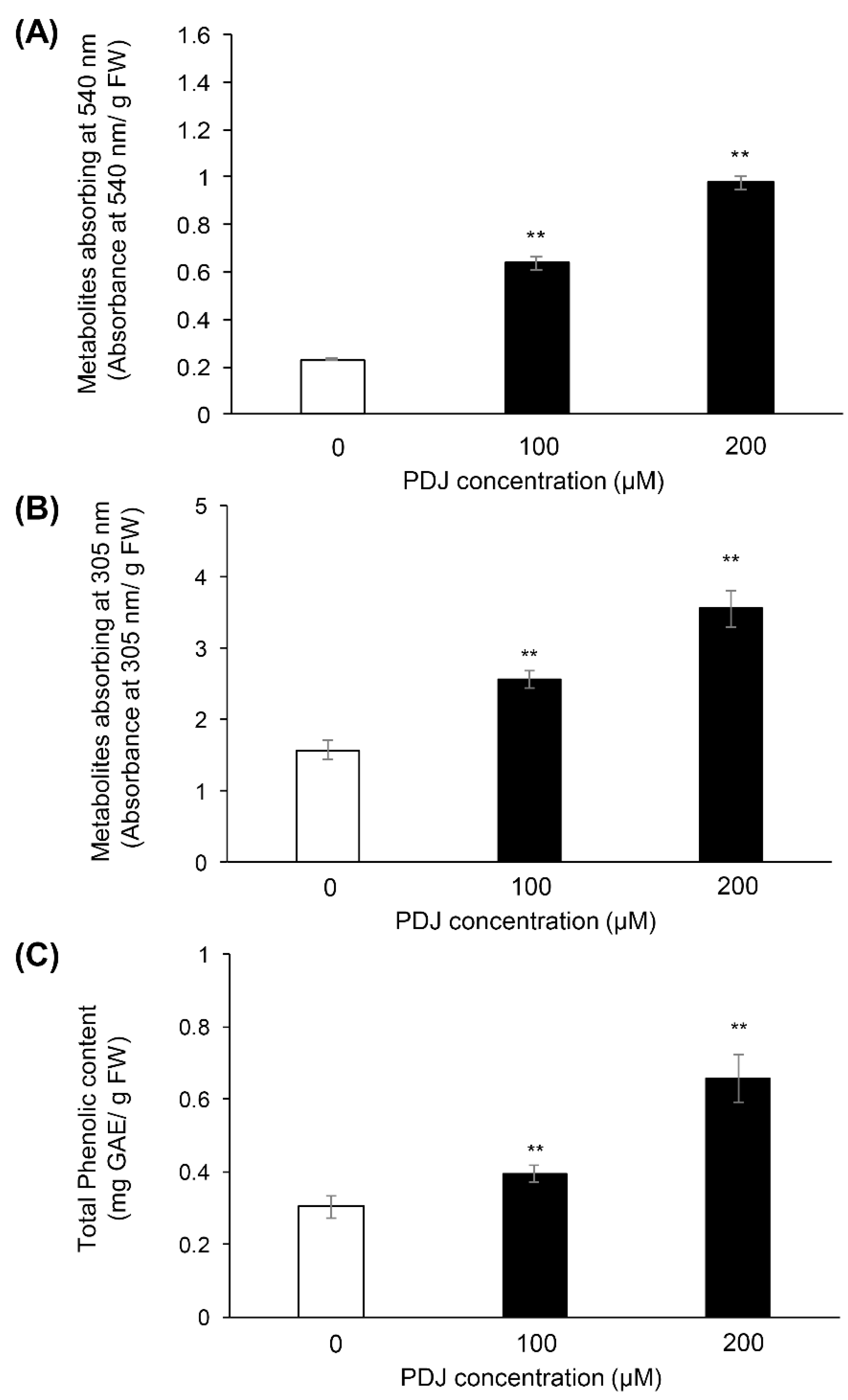


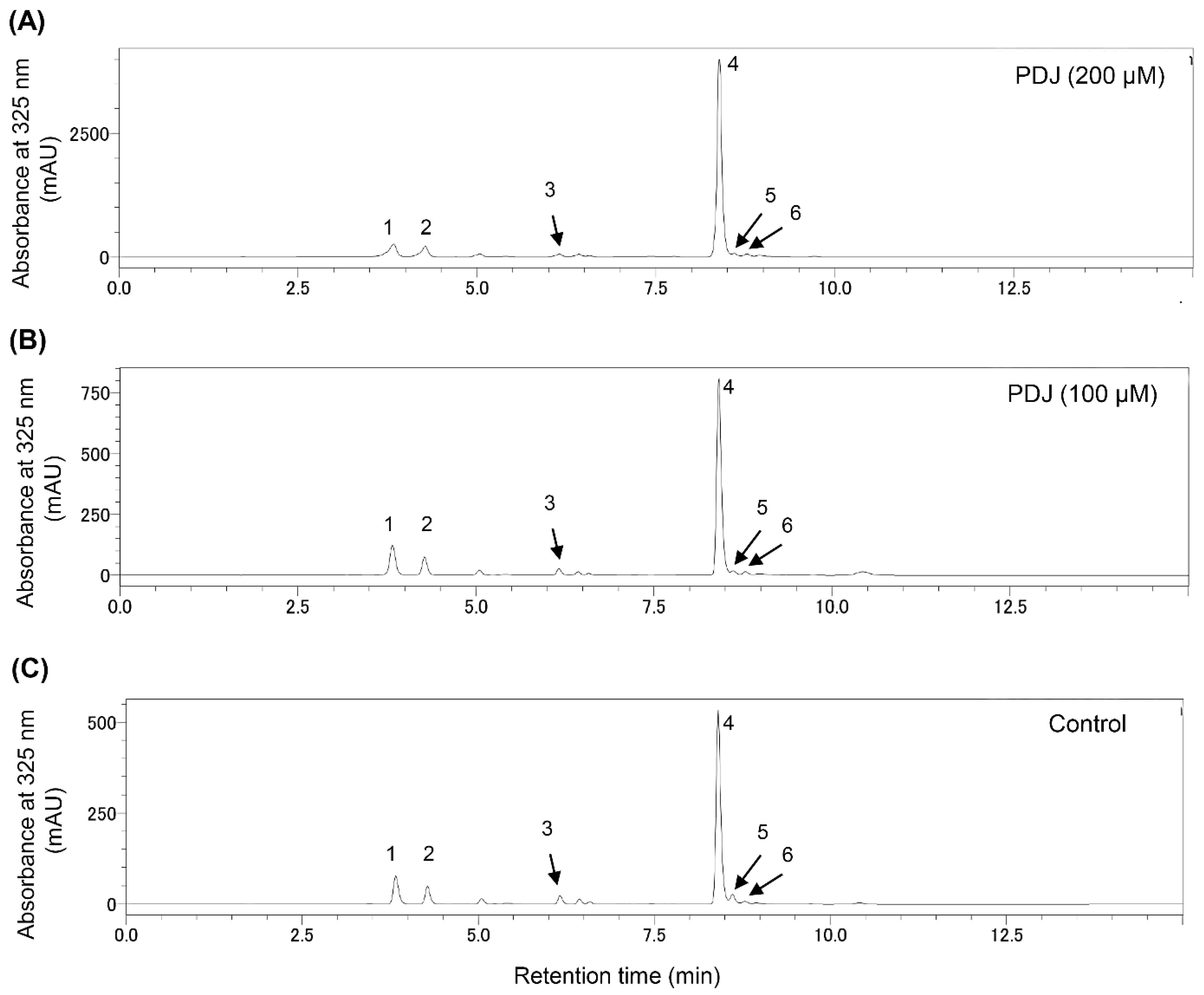
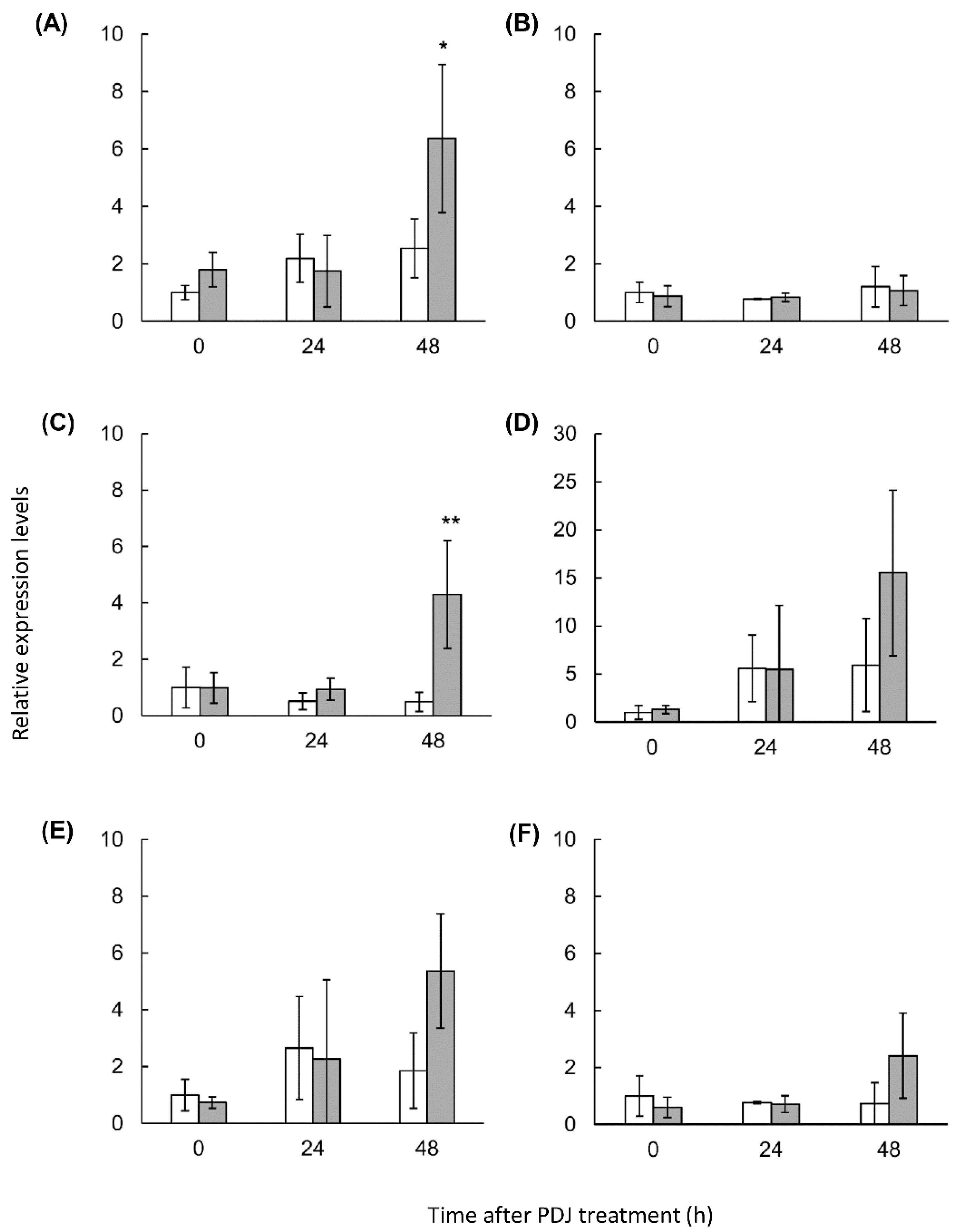
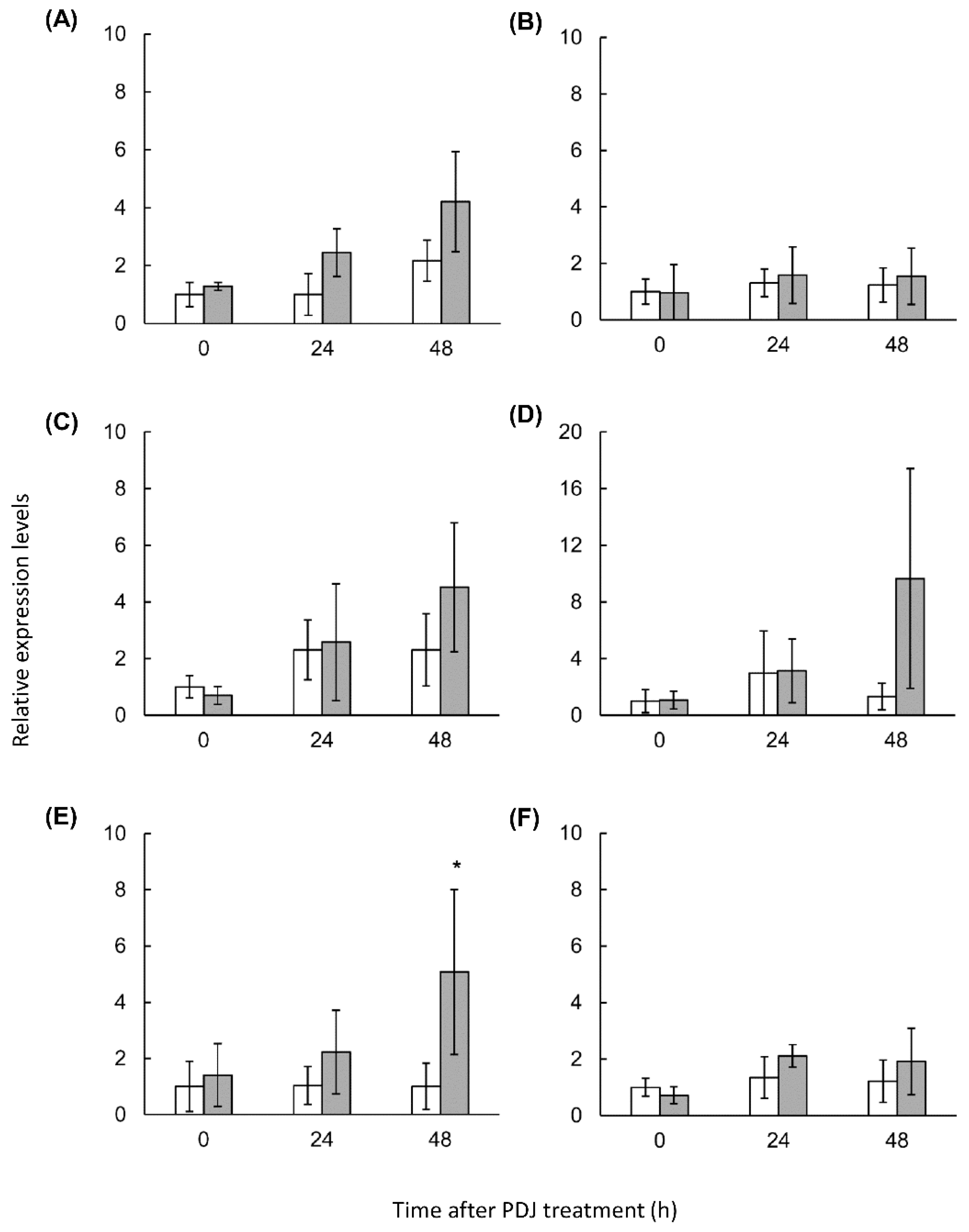
| Compound | Rt (min) | λmax (nm) | M+ m/z | Content [µg Cy3G eq./g Fresh Weight] (Mean ± SD) | ||
|---|---|---|---|---|---|---|
| 0 µM | 100 µM | 200 µM | ||||
| Cy3G | 5.97 | 273, 520 | 449 | 1.66 ± 1.54 c | 4.19 ± 0.66 b | 10.6 ± 0.2 a |
| Cy3MG | 7.70 | 277, 519 | 535 | 2.43 ± 0.04 c | 5.62 ± 0.02 b | 25.6 ± 0.4 a |
| Cy3MG-Me | 8.72 | 270, 523 | 549 | 0.64 ± 0.02 c | 1.25 ± 0.08 b | 5.15 ± 0.08 a |
| Compound | Rt (min) | UV λmax (nm) | [M − H]− m/z | Content [mg Chicoric Acid eq./g Fresh Weight] (Mean ± SD) | ||
|---|---|---|---|---|---|---|
| 0 µM | 100 µM | 200 µM | ||||
| CTA | 3.73 | 329 | 311 | 0.114 ± 0.001 c | 0.137 ± 0.002 b | 0.428 ± 0.005 a |
| Chlorogenic acid | 4.41 | 326 | 353 | 0.059 ± 0.0003 c | 0.069 ± 0.001 b | 0.269 ± 0.006 a |
| CMA | 6.12 | 328 | 295 | 0.038 ± 0.001 c | 0.033 ± 0.0001 b | 0.121 ± 0.002 a |
| Chicoric acid | 8.34 | 329 | 473 | 0.731 ± 0.028 c | 0.795 ± 0.015 b | 2.824 ± 0.025 a |
| diCQA | 8.78 | 327 | 515 | 0.030 ± 0.001 c | 0.023 ± 0.001 b | 0.077 ± 0.001 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takahashi, S.; Namioka, Y.; Azis, H.R.; Sano, T.; Aono, M.; Koshiyama, M.; Fujisawa, H.; Isoda, H. Prohydrojasmon Promotes the Accumulation of Phenolic Compounds in Red Leaf Lettuce. Plants 2021, 10, 1920. https://doi.org/10.3390/plants10091920
Takahashi S, Namioka Y, Azis HR, Sano T, Aono M, Koshiyama M, Fujisawa H, Isoda H. Prohydrojasmon Promotes the Accumulation of Phenolic Compounds in Red Leaf Lettuce. Plants. 2021; 10(9):1920. https://doi.org/10.3390/plants10091920
Chicago/Turabian StyleTakahashi, Shinya, Yui Namioka, Haidar Rafid Azis, Tomoharu Sano, Mitsuko Aono, Masami Koshiyama, Hiroshi Fujisawa, and Hiroko Isoda. 2021. "Prohydrojasmon Promotes the Accumulation of Phenolic Compounds in Red Leaf Lettuce" Plants 10, no. 9: 1920. https://doi.org/10.3390/plants10091920
APA StyleTakahashi, S., Namioka, Y., Azis, H. R., Sano, T., Aono, M., Koshiyama, M., Fujisawa, H., & Isoda, H. (2021). Prohydrojasmon Promotes the Accumulation of Phenolic Compounds in Red Leaf Lettuce. Plants, 10(9), 1920. https://doi.org/10.3390/plants10091920








