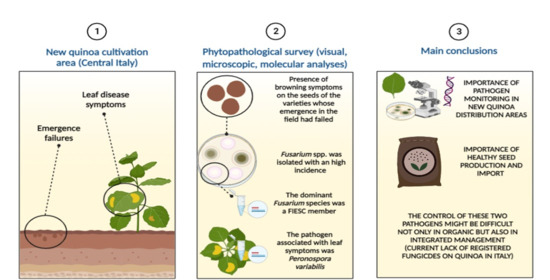
Phytopathological Threats Associated with Quinoa (Chenopodium quinoa Willd.) Cultivation and Seed Production in an Area of Central Italy
Abstract
1. Introduction
2. Results
2.1. Examination of Sampled Material
2.1.1. Seed Material
2.1.2. Plant Material
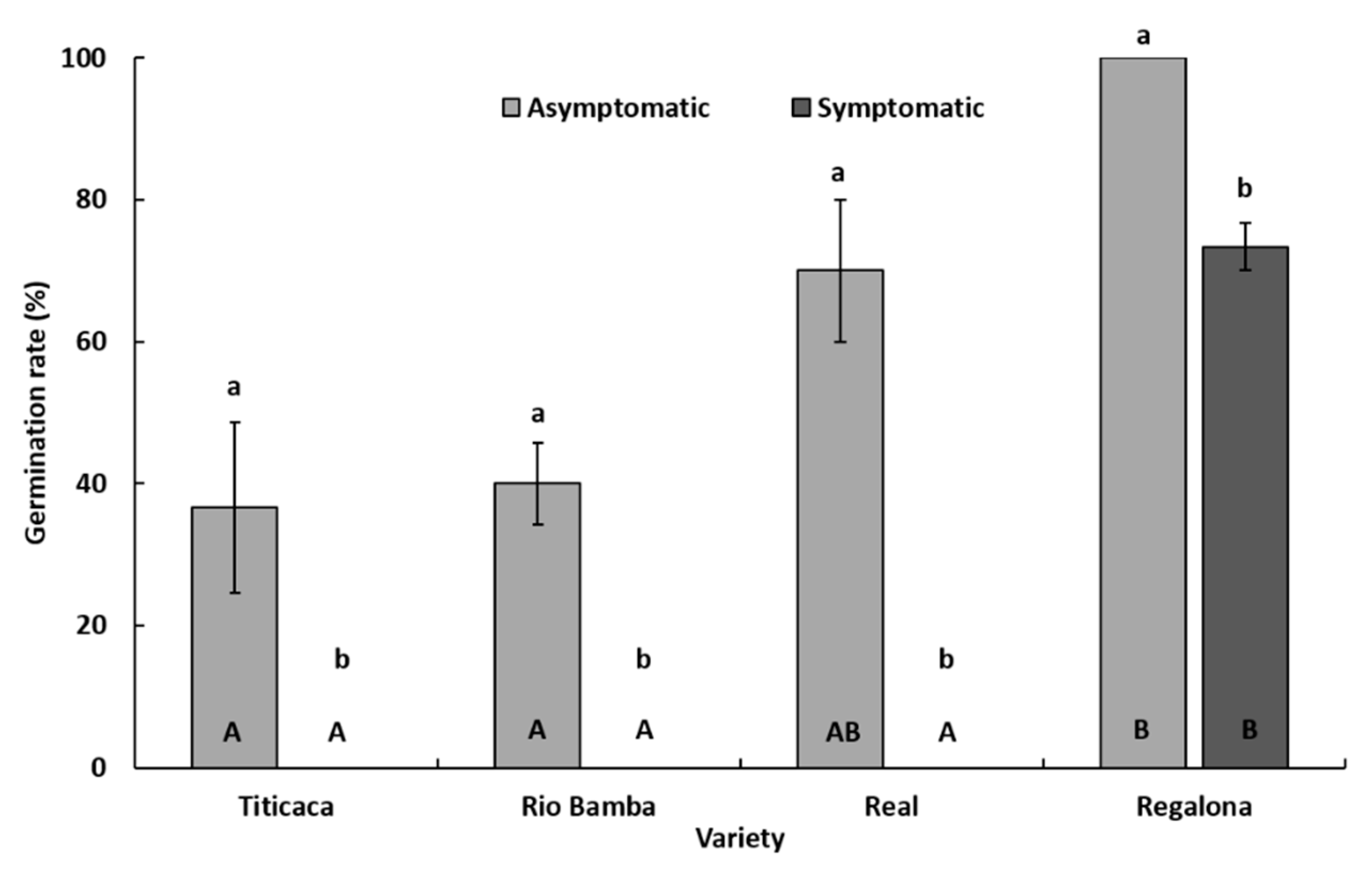
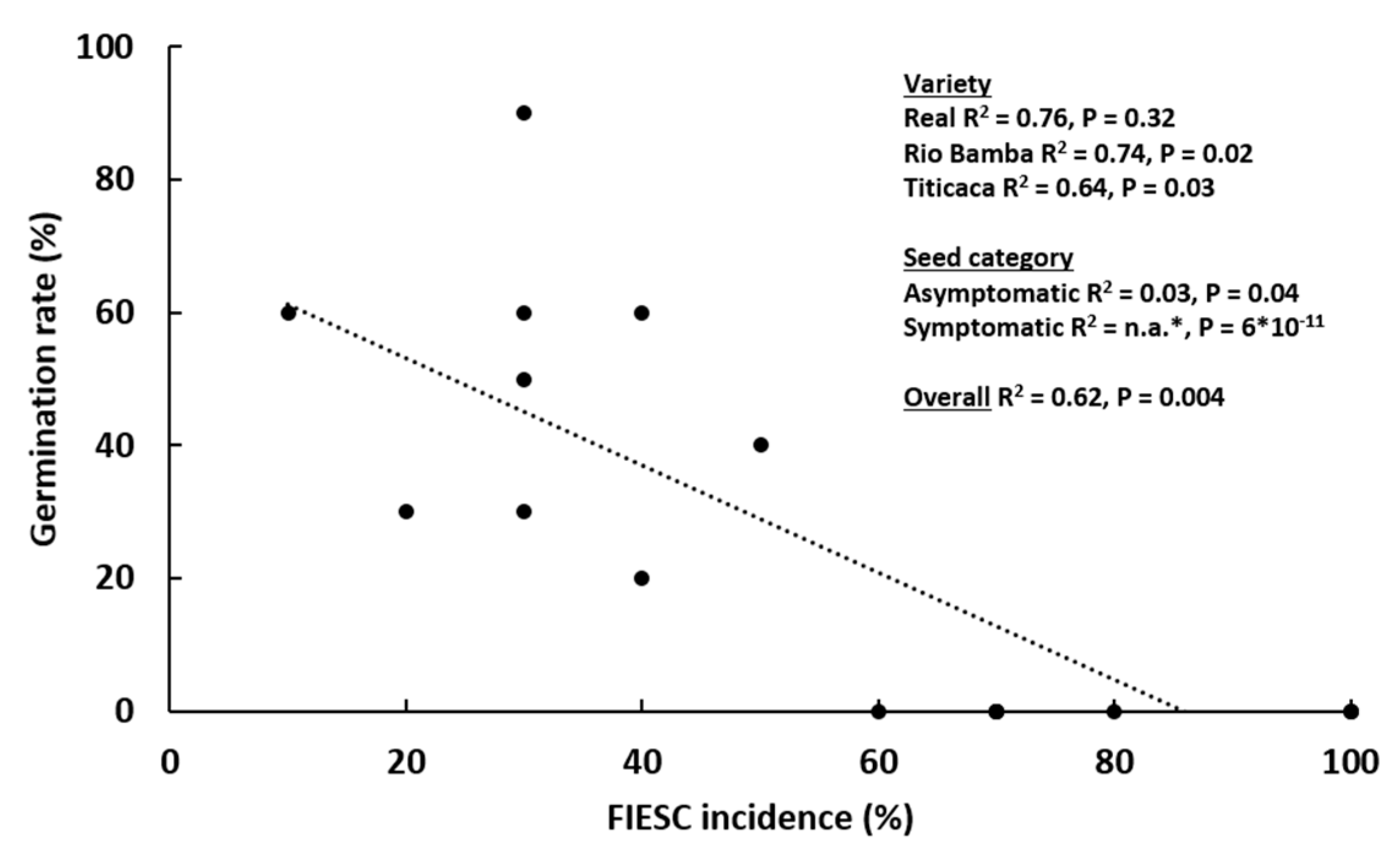
2.2. Seed Germination Rate
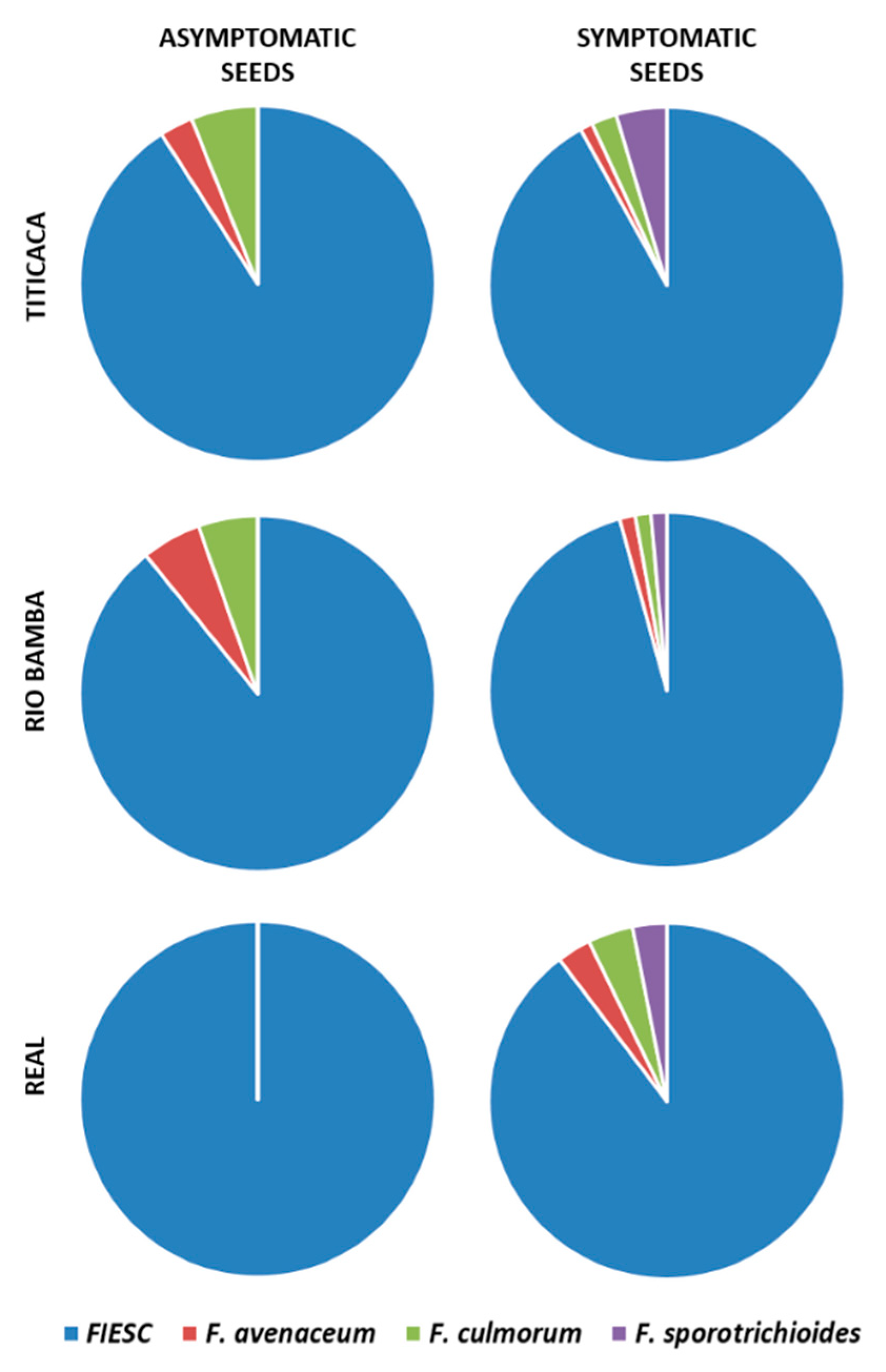
2.3. Seed Mycological Analysis
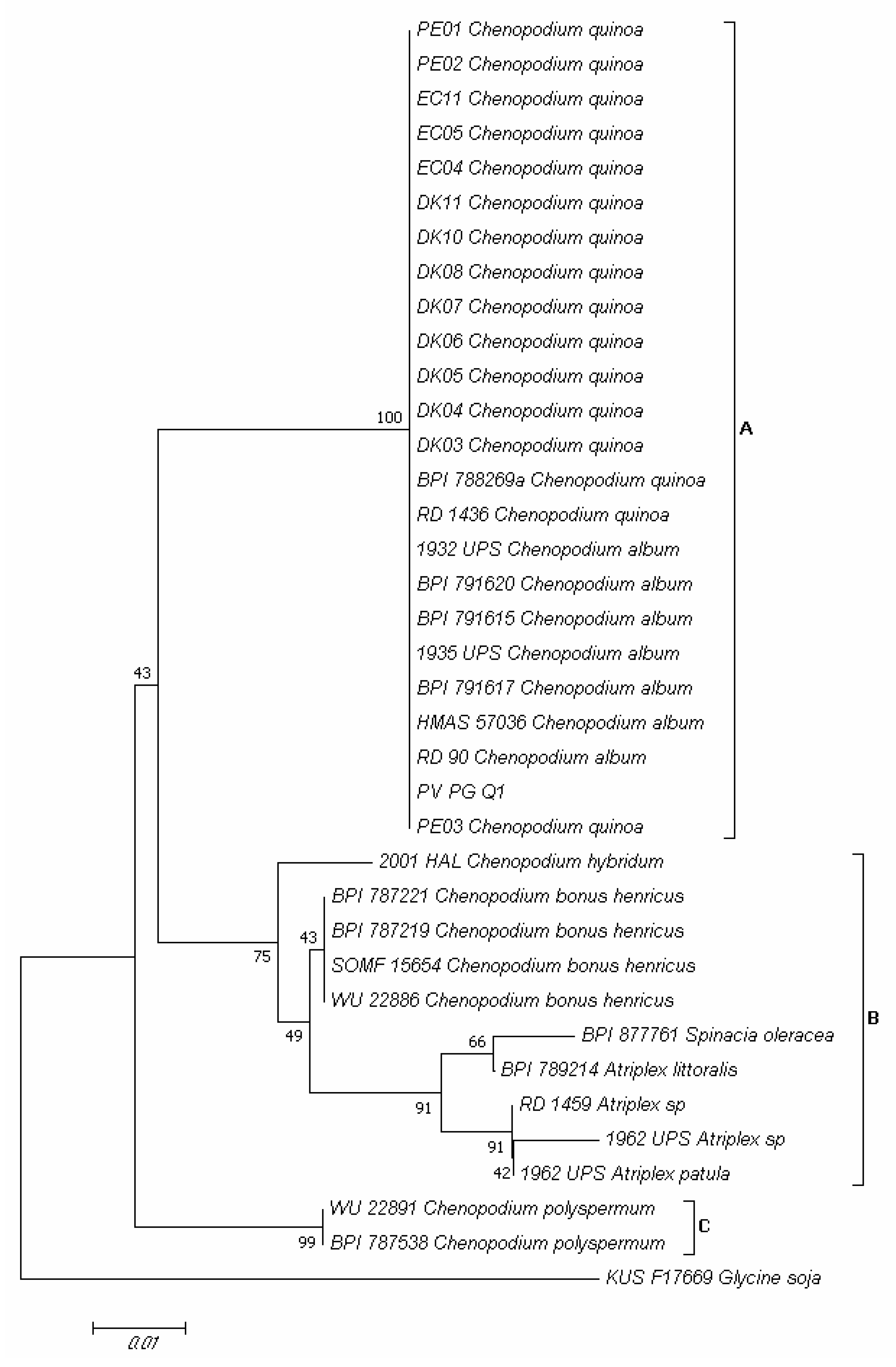
2.4. Molecular Identification of Fusarium spp. Associated to Quinoa Seeds
2.5. Molecular Identification of Pathogen Infecting Quinoa Leaves and Its Detection Seeds
3. Discussion
4. Materials and Methods
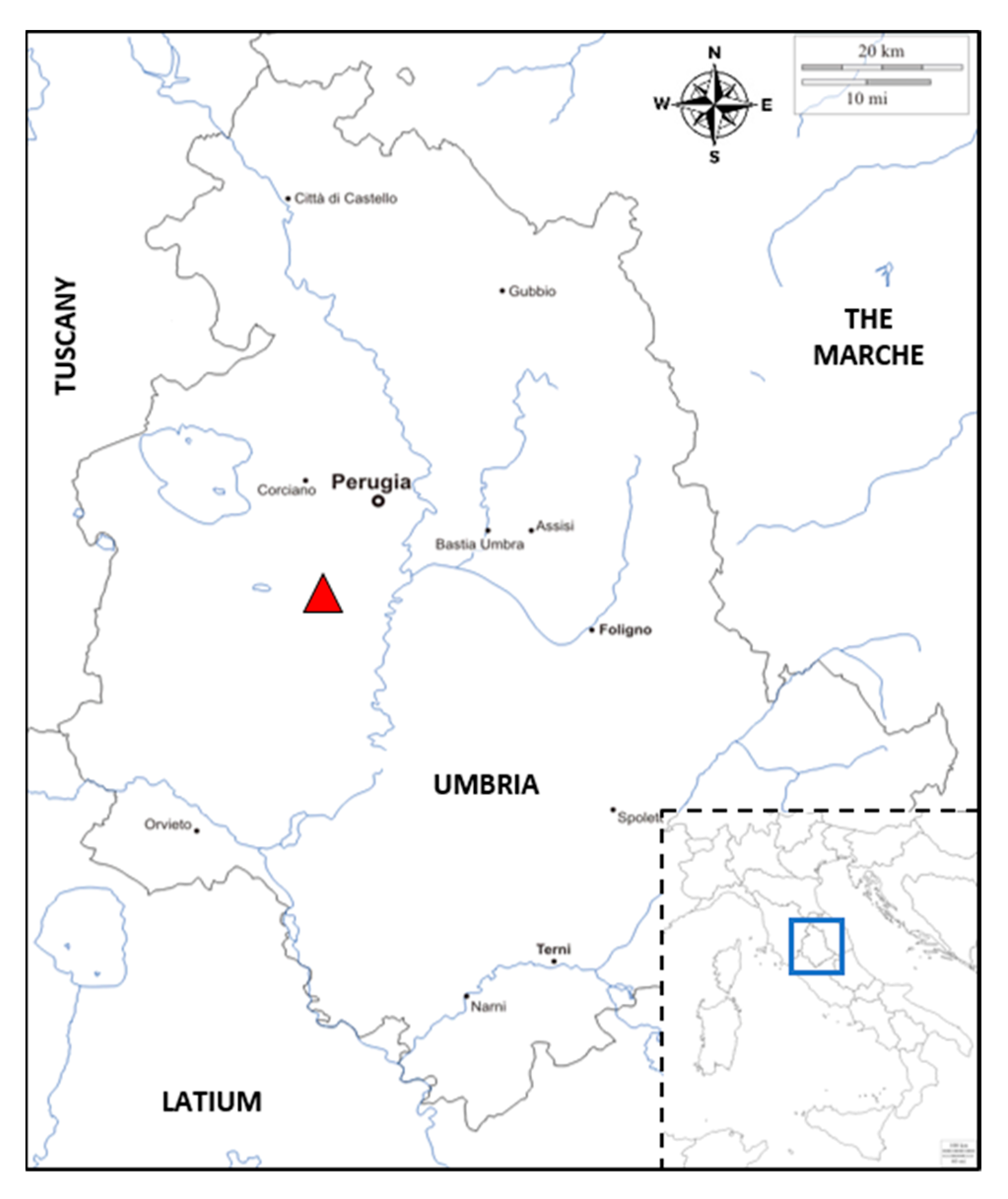
4.1. Field Observations, Samples Collection, and Examination of Sampled Materials
4.2. Seed Germination Test
4.3. Seed Mycological Analysis
4.4. Molecular Identification of Fusarium spp. Associated to Quinoa Seeds
4.5. Molecular Identification of the Pathogen Associated with Quinoa Leaves
4.6. Statistical Analysis
- Incidence (%) of asymptomatic and symptomatic seeds is indicated for each variety as the average (±standard error, SE) of three biological replicates. Data were subject to one-way analysis of variance by considering “variety” as a factor and “incidence” as a variable.
- The germination rate (%) was indicated for each variety as the average (± SE) of three biological replicates (Petri dishes), both for the asymptomatic and symptomatic selected seeds. Data were subject to one-way analysis of variance by considering, within a variety or between varieties, “seed category (asymptomatic, symptomatic)” as a factor and “germination rate” as a variable.
- The incidences (%) of each fungal genus recovered during the entire survey are expressed as the average (± SE) of three biological replicates (Petri dishes), both for the asymptomatic and symptomatic selected seeds. Data were subject to one-way analysis of variance by considering: within the variety–seed category combination, “fungal genera” as a factor and “incidence” as a variable; within the variety–fungal genera combination, “seed category (asymptomatic, symptomatic)” as a factor and “incidence” as a variable; and within the fungal genera–seed category combination, “variety” as a factor and “incidence” as a variable.
- The incidences (%) of each Fusarium species recovered during the entire survey were calculated as the incidence of isolates belonging to the morphotype from which the identified isolate was sub-sampled and are expressed as the average of three biological replicates (Petri dishes), both for the asymptomatic and symptomatic selected seeds. Data were subject to one-way analysis of variance by considering, within the variety–seed combination category, “Fusarium species” as a factor and “incidence” as a variable;
- The incidence (%) of FIESC recovered during the entire survey is expressed as the average (± SE) of three biological replicates (Petri dishes) both for the asymptomatic and symptomatic selected seeds. Data were subject to one-way analysis of variance by considering: within the variety–seed combination category, “fungal genera” as a factor and “incidence” as a variable; within the variety–fungal genera combination, “seed category (asymptomatic, symptomatic)” as a factor and “incidence” as a variable; within the fungal genera–seed combination category, “variety” as a factor and “incidence” as a variable.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vega-Galvez, A.; Miranda, M.; Vergara, J.; Uribe, E.; Puente, L.; Martinez, E.A. Nutrition facts and functional potential of quinoa (Chenopodium quinoa Willd.) an ancient Andean grain: A review. J. Sci. Food Agric. 2010, 90, 2541–2547. [Google Scholar] [CrossRef] [PubMed]
- Bazile, D.; Jacobsen, S.E.; Verniau, A. The global expansion of quinoa: Trends and limits. Front. Plant Sci. 2016, 7, 622. [Google Scholar] [CrossRef] [PubMed]
- FAO. Catalogue of Commercial Varieties of Quinoa in Peru. Available online: http://www.fao.org/documents/card/en/c/f351e07b-bd68-4724-a849-778dd44e6358/ (accessed on 10 March 2021).
- Ruiz, K.B.; Biondi, S.; Oses, R.; Acuña-Rodriguez, I.S.; Antognoni, F.; Martinez-Mosquiera, E.A.; Coulibaly, A.; Canahua-Murillo, A.; Pinto, M.; Zurita-Silva, A.; et al. Quinoa biodiversity and sustainability for food security under climate change. A review. Agric. Sustain. Dev. 2014, 34, 349–359. [Google Scholar] [CrossRef]
- Ruiz, K.B.; Biondi, S.; Martínez, E.A.; Orsini, F.; Antognoni, F.; Jacobsen, S.E. Quinoa—A model crop for understanding salt tolerance mechanisms in halophytes. Plant Biosyst. 2015, 150, 357–371. [Google Scholar] [CrossRef]
- Jarvis, D.E.; Ho, Y.S.; Lightfoot, D.J.; Schmöckel, S.M.; Li, B.; Borm, T.J.A.; Ohyanagi, H.; Mineta, K.; Michell, C.T.; Saber, N.; et al. The genome of Chenopodium quinoa. Nature 2017, 542, 307–312. [Google Scholar] [CrossRef]
- Gordillo-Bastidas, E.; Díaz-Rizzolo, D.A.; Roura, E.; Massanés, T.; Gomis, R. Quinoa (Chenopodium quinoa Willd.), from nutritional value to potential health benefits: An integrative review. J. Nutr. Food Sci. 2016, 6, 497. [Google Scholar]
- Wright, K.H.; Pike, O.A.; Fairbanks, D.J.; Huber, S.C. Composition of Atriplex hortensis, sweet and bitter Chenopodium quinoa seeds. J. Food Sci. 2002, 67, 1383–1385. [Google Scholar] [CrossRef]
- Repo-Carrasco, R.; Espinoza, C.; Jacobsen, S.E. Nutritional value and use of the Andean crops quinoa (Chenopodium quinoa) and kañiwa (Chenopodium pallidicaule). Food Rev. Int. 2003, 19, 179–189. [Google Scholar] [CrossRef]
- FAO. The International Year of Quinoa. Available online: http://www.fao.org/quinoa-2013/en/ (accessed on 10 March 2021).
- FAO. FAOSTAT. Statistic Division. Food and Agriculture Organization of the UN. Database 2018. Available online: http://faostat.fao.org (accessed on 29 October 2020).
- Basantes-Morales, E.R.; Alconada, M.M.; Pantoja, J.L. Quinoa (Chenopodium quinoa Willd.) production in the Andean region: Challenges and potentials. J. Exp. Agric. Int. 2019, 36, 1–18. [Google Scholar] [CrossRef]
- García-Parra, M.; Zurita-Silva, A.; Stechauner-Rohringer, R.; Roa-Acosta, D.; Jacobsen, S. Quinoa (Chenopodium quinoa Willd.) and its relationship with agroclimatic characteristics: A Colombian perspective. Chil. J. Agric. Res. 2020, 80, 290–302. [Google Scholar] [CrossRef]
- Alandia, G.; Rodriguez, J.P.; Jacobsen, S.E.; Bazile, D.; Condori, B. Global expansion of quinoa and challenges for Andean region. Glob. Food Secur. 2020, 26, 100429. [Google Scholar] [CrossRef]
- Lavini, A.; Pulvento, C.; d’Andria, R.; Riccardi, M.; Choukr-Allah, R.; Balhabib, O.; Yazar, A.; Incekaya, C.; MetinSezen, S.; Qadir, M.; et al. Quinoa’s potential in the Mediterranean region. J. Agron. Crop Sci. 2014, 200, 344–360. [Google Scholar] [CrossRef]
- Pulvento, C.; Riccardi, M.; Biondi, S.; Orsini, F.; Jacobsen, S.E.; Ragab, R.; D’Andria, R.; Lavini, A. Quinoa in Italy: Research and perspectives. In FAO & CIRAD; State of the Art Report on Quinoa in the World in 2013; Bazile, D., Bertero, D., Nieto, C., Eds.; Food and Agriculture Organization of the United Nations (FAO): Rome, Italy, 2015; Chapter 6.13; pp. 192–217. [Google Scholar]
- Damiani, F.; Brunetti, M.; Pannacci, E. Quinoa cultivation in Italy. In Food Science and Technology—Quinoa Cultivation, Nutritional Properties and Effect on Health; Peiretti, P.G., Gai, F., Eds.; Nova Science Publisher Inc.: New York, NY, USA, 2019; pp. 1–31. [Google Scholar]
- Bilalis, D.J.; Roussis, I.; Kakabouki, I.; Folina, A. Quinoa (Chenopodium quinoa Willd.) crop under Mediterranean conditions: A review. Int. J. Agric. Nat. Resour. 2019, 46, 51–68. [Google Scholar] [CrossRef]
- Danielsen, S.; Bonifacio, A.; Ames, T. Diseases of Quinoa (Chenopodium quinoa). Food Rev. Int. 2003, 19, 43–59. [Google Scholar] [CrossRef]
- Gandarillas, A.; Saravia, R.; Plata, G.; Quispe, R.; Ortiz-Romero, R. Principle quinoa pests and diseases. In FAO & CIRAD; State of the Art Report on Quinoa in the World in 2013; Bazile, D., Bertero, D., Nieto, C., Eds.; Food and Agriculture Organization of the United Nations (FAO): Rome, Italy, 2015; Chapter 2.6; pp. 192–217. [Google Scholar]
- Testen, A.L.; Jiménez-Gasco, M.M.; Ochoa, J.B.; Backman, P.A. Molecular detection of Peronospora variabilis in quinoa seed and phylogeny of the quinoa downy mildew pathogen in South America and the United States. Phytopathology 2014, 104, 379–386. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Colque-Little, C.; Amby, D.B.; Andreasen, C. A review of Chenopodium quinoa (Willd.) diseases—An updated perspective. Plants 2021, 10, 1228. [Google Scholar] [CrossRef]
- Li, J.; Zhou, X.; Huang, H.; Li, G. Diseases characteristic and control measurements for Chenopodium quinoa Willd. Adv. Eng. Res. 2017, 143, 305–308. [Google Scholar]
- Cavalier-Smith, T. Kingdom Chromista and its eight phyla: A new synthesis emphasizing periplastid protein targeting, cytoskeletal and periplastid evolution, and ancient divergences. Protoplasma 2018, 255, 297–357. [Google Scholar] [CrossRef] [PubMed]
- Castellá, G.; Cabañes, F.J. Phylogenetic diversity of Fusaium incarnatum-equiseti species complex isolated from Spanish wheat. Antoine van Leeuwenhoek 2014, 106, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger dataset. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Gilbert, J.; Tekauz, A. Effect of fusarium head blight and seed treatment on germination, emergence and seedling vigor of spring wheat. Can. J. Plant Pathol. 1995, 3, 252–259. [Google Scholar] [CrossRef]
- Suthar, R.; Bhatt, D.P.; Bhatt, P.N. Effect of culture filtrate of Fusarium equiseti on seed germination and seedling growth of cumin (Cuminum cyminum). Indian Phytopathol. 2014, 67, 193–194. [Google Scholar]
- Causin, H.F.; Bordón, D.A.E.; Burrieza, H. Salinity tolerance mechanisms during germination and early seedling growth in Chenopodium quinoa Wild. Genotypes with different sensitivity to saline stress. Environ. Exp. Bot. 2020, 172, 103995. [Google Scholar] [CrossRef]
- Jacobsen, S.E.; Bach, A.P. The influence of temperature on seed germination rate in quinoa (Chenopodium quinoa Willd.). SST 1998, 26, 515–523. [Google Scholar]
- Drimalkova, M.; Veverka, K. Seedlings damping-off of Chenopodium quinoa Willd. Plant Prot. Sci. 2004, 40, 5–10. [Google Scholar] [CrossRef]
- Villani, A.; Moretti, A.; De Saeger, S.; Han, Z.; Di Mavungu, J.D.; Soares, C.M.G.; Proctor, R.H.; Venâncio, A.; Lima, N.; Stea, G.; et al. A polyphasic approach for characterization of a collection of cereal isolates of the Fusarium incarnatum-equiseti species complex. Int. J. Food Microbiol. 2016, 234, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Matić, S.; Tabone, G.; Guarnaccia, V.; Gullino, M.L.; Garibaldi, A. Emerging leafy vegetable crop diseases caused by the Fusarium incarnatum-equiseti species complex. Phytopathol. Mediterr. 2020, 59, 303–317. [Google Scholar]
- Beccari, G.; Caproni, L.; Tini, F.; Uhlig, S.; Covarelli, L. Presence of Fusarium species and other toxigenic fungi in malting barley and multi-mycotoxin analysis by liquid chromatography-high-resolution mass spectrometry. J. Agric. Food Chem. 2016, 64, 4390–4399. [Google Scholar] [CrossRef]
- Beccari, G.; Senatore, M.T.; Tini, F.; Sulyok, M.; Covarelli, L. Fungal community, Fusarium head blight complex and secondary metabolites associated with malting barley grains harvested in Umbria, Central Italy. Int. J. Food Microbiol. 2018, 271, 33–42. [Google Scholar] [CrossRef]
- Beccari, G.; Prodi, A.; Senatore, M.T.; Balmas, V.; Tini, F.; Onofri, A.; Pedini, L.; Sulyok, M.; Brocca, L.; Covarelli, L. Cultivation area affects the presence of fungal communities and secondary metabolites in Italian durum what grains. Toxins 2020, 12, 97. [Google Scholar] [CrossRef]
- Villani, A.; Proctor, R.H.; Kim, H.; Brown, D.W.; Logrieco, A.F.; Amatulli, M.T.; Moretti, A.; Susca, A. Variation in secondary metabolite production potential in the Fusarium incarnatum-equiseti species complex revealed by comparative analysis of 13 genomes. BMC Genom. 2019, 20, 314. [Google Scholar] [CrossRef] [PubMed]
- Avila, C.F.; Moreira, G.M.; Nicolli, C.P.; Gomes, L.B.; Abreu, L.M.; Pfenning, L.H.; Haidukowski, M.; Moretti, A.; Logrieco, A.; Del Ponte, E.M. Fusarium incarnatum-equiseti species complex associated with brazilian rice: Phylogeny, morphology and toxigenic potential. Int. J. Food Microbiol. 2019, 306, 108267. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.W.; Sandoval-Denis, M.; Crous, P.W.; Zhang, X.G.; Lombard, L. Numbers to names—Restyling the Fusarium incarnatum-equiseti species complex. Persoonia 2019, 43, 186–221. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.A.; Hoekstra, E.S.; Frisvad, J.C. Introduction to Food- and Airborne Fungi, 7th ed.; Centraalbureau voor Schimmelcultures: Utrech, The Netherlands, 2004. [Google Scholar]
- Marín, P.; Moretti, A.; Ritieni, A.; Jurado, M.; Vázquez, C.; González-Jaén, M.T. Phylogenetic analysis and toxigenic profiles of Fusarium equiseti and Fusarium incarnatum isolates from Southern Europe. Food Microbiol. 2012, 31, 229–237. [Google Scholar] [CrossRef]
- Balmas, V.; Migheli, Q.; Schern, B.; Garau, P.; O’Donnel, K.; Ceccherelli, G.; Kang, S.; Geiser, D. Multilocus phylogenetics shows high levels of endemic fusaria inhabiting Sardinian soils (Tyrrhenian Islands). Mycologia 2010, 102, 803–812. [Google Scholar] [CrossRef] [PubMed]
- Mendes, M.A.S.; da Silva, V.L.; Dianese, J.C.; Ferreira, M.A.S.V.; dos Santos, C.E.N. Fungos em Plants no Brasil; Embrapa-SPI/Embrapa-Cenargen: Brasilia, Brazil, 1998; 555p. [Google Scholar]
- Gilardi, G.; Pintore, I.; Gullino, M.L.; Garibaldi, A. Occurrence of Fusarium equiseti as a contaminant of Diplotaxis tenuifolia seeds. J. Plant Pathol. 2017, 99, 245–248. [Google Scholar]
- Abd El Moity, T.H.; Badrawy, H.B.M.; Ali, A.M. Survey on diseases and pests attack quinoa in Egypt. In Proceedings of the Sixth International Scientific Agricultural Symposium, Jahorina, Bosnia and Herzegovina, 15–18 October 2015; pp. 868–876. [Google Scholar]
- El-Hadidy, A.E. Effect of biocontrol agents on damping-off and root-rot diseases of quinoa (Chenopodium quinoa Willd.) seedlings. Egypt. J. Desert Res. 2019, 69, 21–38. [Google Scholar] [CrossRef]
- Choi, Y.J.; Denchev, C.M.; Shin, H.D. Morphological and molecular analyses support the existence of host-specific Peronospora species infecting Chenopodium. Mycopathologia 2008, 165, 155–164. [Google Scholar] [CrossRef]
- Choi, Y.; Danielsen, S.; Lübeck, M.; Hong, S.; Delhey, R.; Shin, H. Morphological and molecular characterization of the causal agent of downy mildew on Quinoa (Chenopodium quinoa). Mycopathologia 2010, 169, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Farr, D.F.; Rossman, A.Y.; Fungal Databases, U.S. National Fungus Collections, ARS, USDA. Retrieved 20 July 2021. Available online: https://nt.ars-grin.gov/fungaldatabases/ (accessed on 20 July 2021).
- Jacobsen, S.E. The scope for adaptation of quinoa in Northern latitudes of Europe. J. Agric. Crop Sci. 2017, 203, 603–613. [Google Scholar] [CrossRef]
- Danielsen, S.; Jacobsen, S.E.; Echegaray, J.; Ames, T. Impact of Downy Mildew on the Yield of Quinoa; Program Report 1999–2000; International Potato Center (Centro Internacional de la Papa): Lima, Peru; pp. 397–401.
- Gul, E. First report of Peronospora variabilis on Chenopodium quinoa in Turkey. J. Plant Pathol. 2021, 103, 389–390. [Google Scholar] [CrossRef]
- Danielsen, S.; Ames, T. Mildew (Peronospora Farinosa) of Quinoa (Chenopodium Quinoa) in the Andean Region: Practical Manual for the Study of the Disease and Pathogen; International Potato Center (Centro Internacional de la Papa): Lima, Peru, 2004. [Google Scholar]
- El-Assiuty, E.M.; Taha, E.M.; Fahmy, Z.M.; Fahmy, G.M. Histological and molecular detections of Peronospora variabilis Gäum oospores in seeds of Quinoa (Chenopodium Quino L.). Egypt. J. Exp. Biol. 2019, 15, 197–203. [Google Scholar]
- El-Assiuty, E.M.; Fahmy, G.M.; Taha, E.M.; Fahmy, Z.M.; Ismael, A.S.M.; Abd-Elghany, W.R.; Kafsheer, D.A. Microscopic visualization of Peronospora variabilis Gäum., the cause of Quinoa downy mildew in plant tissues at different stages of plant growth. Int. J. Sci. Eng. Res. 2019, 10, 1022–1033. [Google Scholar]
- Danielsen, S.; Mercado, V.H.; Ames, T.; Munk, L. Seed transmission of downy mildew (Peronospora farinosa f. sp. chenopodii) in quinoa and effect of relative humidity on seedling infection. Seed Sci. Technol. 2004, 32, 91–98. [Google Scholar] [CrossRef]
- Pannacci, E.; Lattanzi, B.; Tei, F. Non-chemical weed management strategies in minor crops: A review. Crop Prot. 2017, 96, 44–58. [Google Scholar] [CrossRef]
- Pannacci, E.; Farneselli, M.; Ottavini, D.; Tei, F. Mechanical and chemical weed control in quinoa. In Proceedings of the XLVIII Conference of Italian Society for Agronomy, Perugia, Italy, 18–20 September 2019; Seddaiu, G., Benincasa, P., Eds.; 2019; pp. 100–101. [Google Scholar]
- Yin, H.; Zhou, J.; Lv, H.; Qin, N.; Chang, F.J.; Zhao, X. Identification, pathogenicity, and fungicide sensitivity to Ascochyta caulina (Teleomorph: Neocamarosporium calvescens) associated with black stem on quinoa in China. Plant Dis. 2020, 104, 2585–2597. [Google Scholar] [CrossRef]
- Brahmanage, R.S.; Liu, M.; Wanasinghe, D.N.; Dayarathne, M.C.; Mei, L.; Jeewon, R.; Li, X.; Hyde, K.D. Heterosporicola beijingense sp. nov. (Leptosphariaceae, Pleosporales) associated with Chenopodium quinoa leaf spots. Phytopathol. Mediterr. 2020, 59, 219–227. [Google Scholar]
- Covarelli, L.; Beccari, G.; Prodi, A.; Generotti, S.; Etruschi, F.; Juan, C.; Ferrer, E.; Mañes, J. Fusarium species, chemotype characterization and trichothecene contamination of durum and soft wheat in an area of Central Italy. J. Sci. Food Agric. 2015, 95, 540–551. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef]
- Geiser, D.M.; Jimenez-Gasco, M.D.; Kang, S.C.; Makalowska, I.; Veeraghavan, N.; Ward, T.J.; Zhang, N.; Kuldau, G.A.; O’Donnel, K. FUSARIUM-ID v. 1.0: A DNA sequence for identifying Fusarium. Eur. J. Plant Pathol. 2004, 110, 473–479. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Beccari, G.; Arellano, C.; Covarelli, L.; Tini, F.; Sulyok, M.; Cowger, C. Effect of wheat infection timing on Fusarium head blight causal agents and secondary metabolites in grain. Int. J. Food Microbiol. 2019, 290, 214–225. [Google Scholar] [CrossRef] [PubMed]
- Cooke, D.E.L.; Drenth, A.; Duncan, J.M.; Wagels, G.; Brasier, C.M. A molecular phylogeny of Phytophtora and related oomycetes. Fungal Gen. Biol. 2000, 30, 17–32. [Google Scholar] [CrossRef]
- Sapkota, R.; Nicolaisen, M. An improved high throughput sequencing method for studying oomycete communities. J. Microbiol. Methods 2015, 110, 33–39. [Google Scholar] [CrossRef]
- Onofri, A.; Pannacci, E. Spreadsheet tools for biometry classes in crop science programmes. Commun. Biometry Crop. Sci. 2014, 9, 43–45. [Google Scholar]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beccari, G.; Quaglia, M.; Tini, F.; Pannacci, E.; Covarelli, L. Phytopathological Threats Associated with Quinoa (Chenopodium quinoa Willd.) Cultivation and Seed Production in an Area of Central Italy. Plants 2021, 10, 1933. https://doi.org/10.3390/plants10091933
Beccari G, Quaglia M, Tini F, Pannacci E, Covarelli L. Phytopathological Threats Associated with Quinoa (Chenopodium quinoa Willd.) Cultivation and Seed Production in an Area of Central Italy. Plants. 2021; 10(9):1933. https://doi.org/10.3390/plants10091933
Chicago/Turabian StyleBeccari, Giovanni, Mara Quaglia, Francesco Tini, Euro Pannacci, and Lorenzo Covarelli. 2021. "Phytopathological Threats Associated with Quinoa (Chenopodium quinoa Willd.) Cultivation and Seed Production in an Area of Central Italy" Plants 10, no. 9: 1933. https://doi.org/10.3390/plants10091933
APA StyleBeccari, G., Quaglia, M., Tini, F., Pannacci, E., & Covarelli, L. (2021). Phytopathological Threats Associated with Quinoa (Chenopodium quinoa Willd.) Cultivation and Seed Production in an Area of Central Italy. Plants, 10(9), 1933. https://doi.org/10.3390/plants10091933