Profiling of N6-Methyladenosine (m6A) Modification Landscape in Response to Drought Stress in Apple (Malus prunifolia (Willd.) Borkh)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Stress Treatment
2.2. RNA-Seq Analysis
2.3. m6A-Seq Analysis
3. Results
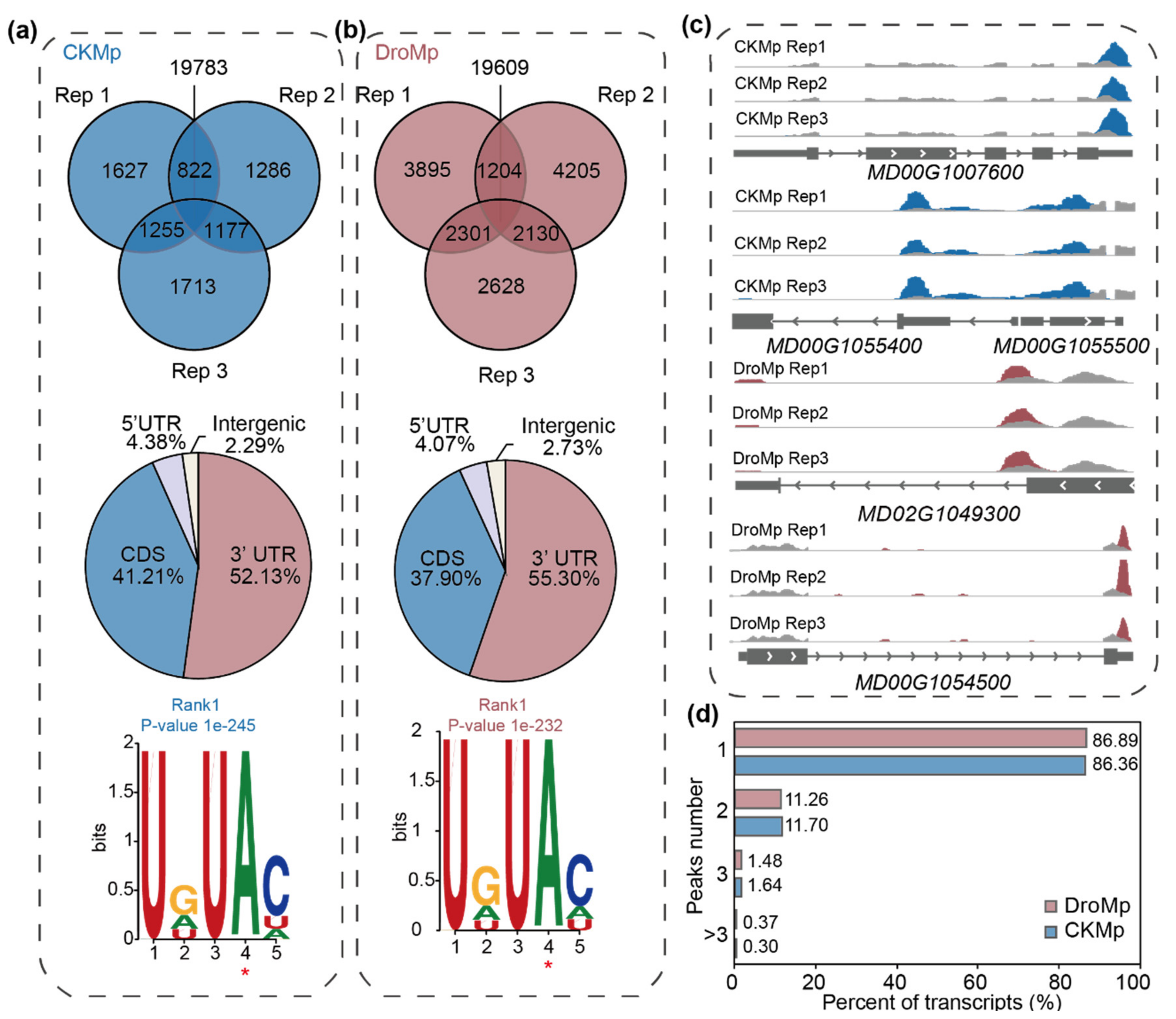
3.1. Transcriptome-Wide Mapping of m6A in Malus prunifolia Seedlings
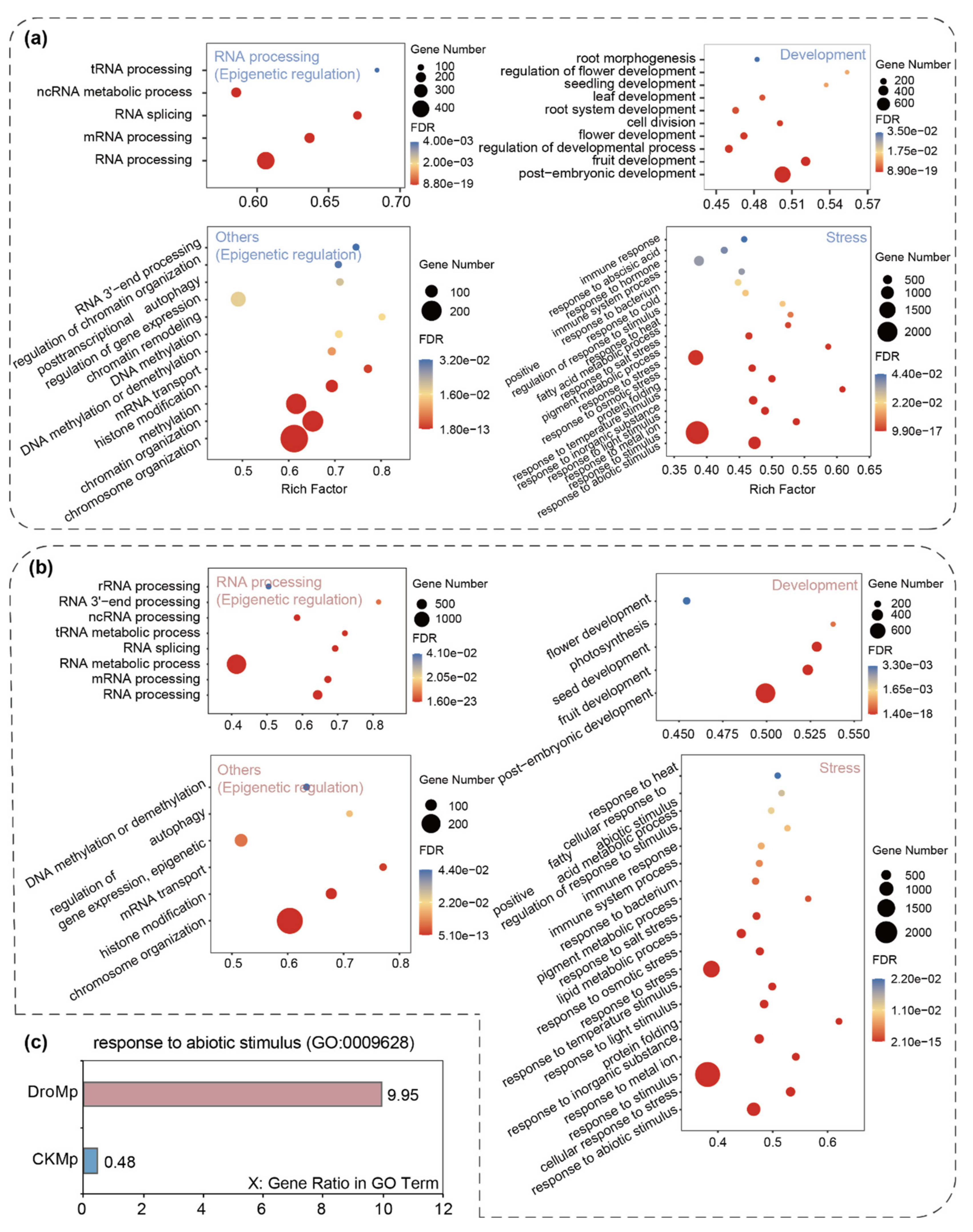
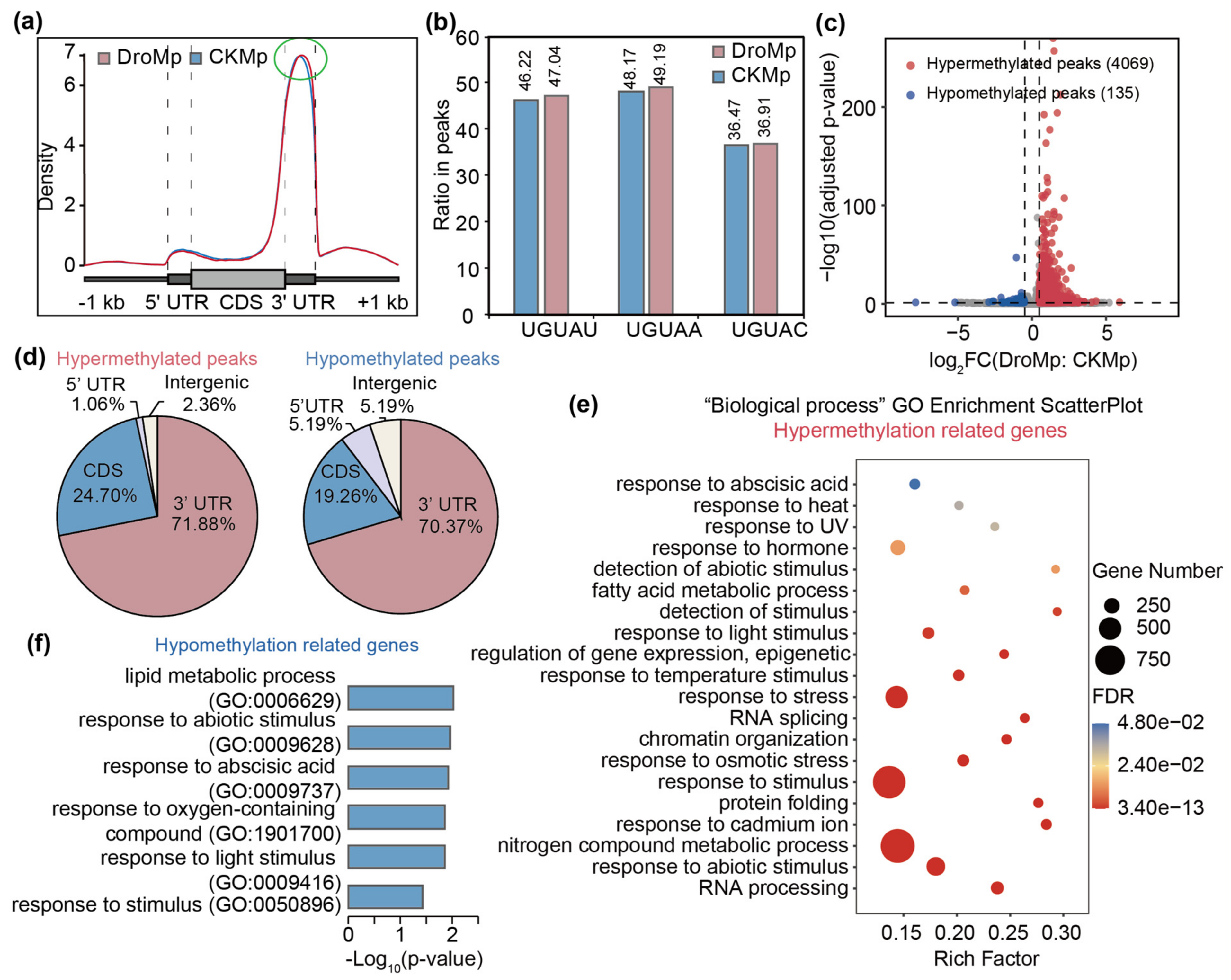
3.2. Differential m6A Methylation between Control and Drought-Treated M. prunifolia Seedlings
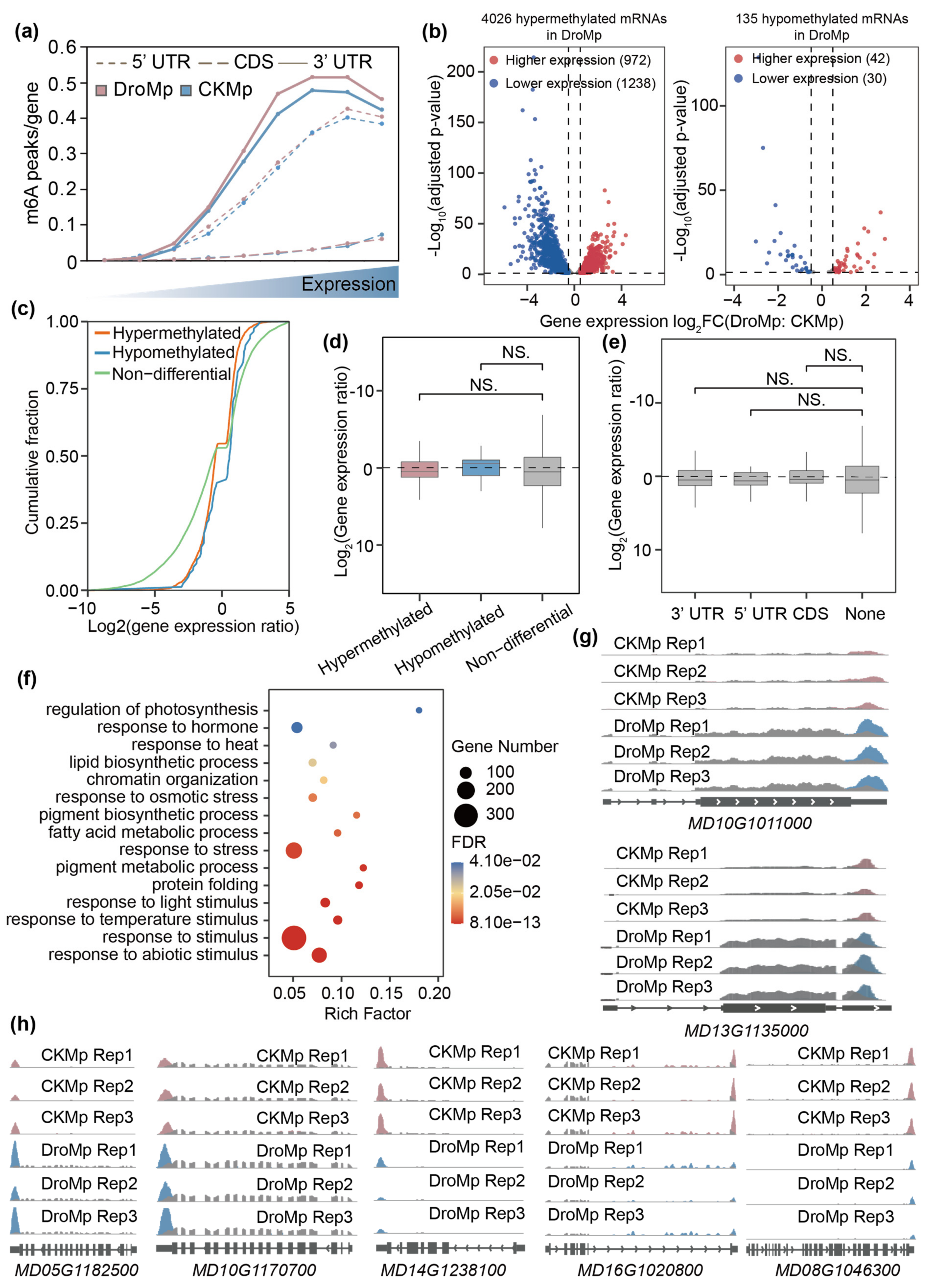
3.3. Differential Gene Expression Analysis
3.4. Association Analysis of m6A Levels with Gene Expressions Involved in Apple Drought Tolerance
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piątkowski, P.; Bagiński, B.; Wirecki, T.K.; de Crécy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A Database of RNA Modification Pathways. 2017 Update. Nucleic Acids Res. 2017, 46, D303–D307. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.-M.; Gershowitz, A.; Moss, B. Methylated Nucleotides Block 5′ Terminus of HeLa Cell Messenger RNA. Cell 1975, 4, 379–386. [Google Scholar] [CrossRef]
- Wei, W.; Ji, X.; Guo, X.; Ji, S. Regulatory Role of N6-Methyladenosine (M6A) Methylation in RNA Processing and Human Diseases. J. Cell. Biochem. 2017, 118, 2534–2543. [Google Scholar] [CrossRef]
- Cantara, W.A.; Crain, P.F.; Rozenski, J.; McCloskey, J.A.; Harris, K.A.; Zhang, X.; Vendeix, F.A.P.; Fabris, D.; Agris, P.F. The RNA Modification Database, RNAMDB: 2011 Update. Nucleic Acids Res. 2011, 39, D195–D201. [Google Scholar] [CrossRef] [Green Version]
- Yue, H.; Nie, X.; Yan, Z.; Weining, S. N6-methyladenosine Regulatory Machinery in Plants: Composition, Function and Evolution. Plant Biotechnol. J. 2019, 17, 1194–1208. [Google Scholar] [CrossRef]
- Kierzek, E. The Thermodynamic Stability of RNA Duplexes and Hairpins Containing N6-Alkyladenosines and 2-Methylthio-N6-Alkyladenosines. Nucleic Acids Res. 2003, 31, 4472–4480. [Google Scholar] [CrossRef]
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of Methylated Nucleosides in Messenger RNA from Novikoff Hepatoma Cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975. [Google Scholar] [CrossRef] [Green Version]
- Nichols, J.L.; Welder, L. Nucleotides Adjacent to N6-Methyladenosine in Maize Poly(A)-Containing RNA. Plant Sci. Lett. 1981, 21, 75–81. [Google Scholar] [CrossRef]
- Kennedy, T.D.; Lane, B.G. Wheat Embryo Ribonucleates. XIII. Methyl-Substituted Nucleoside Constituents and 5′-Terminal Dinucleotide Sequences in Bulk Poly(A)-Rich RNA from Imbibing Wheat Embryos. Can. J. Biochem. 1979, 57, 927–931. [Google Scholar] [CrossRef]
- Haugland, R.A.; Cline, M.G. Post-Transcriptional Modifications of Oat Coleoptile Ribonucleic Acids. 5′-Terminal Capping and Methylation of Internal Nucleosides in Poly(A)-Rich RNA. Eur. J. Biochem. 1980, 104, 271–277. [Google Scholar] [CrossRef]
- Saneyoshi, M.; Harada, F.; Nishimura, S. Isolation and Characterization of N6-Methyladenosine from Escherichia Coli Valine Transfer RNA. Biochim. Et Biophys. Acta (BBA) Nucleic Acids Protein Synth. 1969, 190, 264–273. [Google Scholar] [CrossRef]
- Meyer, K.D.; Jaffrey, S.R. Rethinking m6A Readers, Writers, and Erasers. Annu. Rev. Cell Dev. Biol. 2017, 33, 319–342. [Google Scholar] [CrossRef] [Green Version]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.-M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.-L.; Song, S.-H.; et al. ALKBH5 Is a Mammalian RNA Demethylase That Impacts RNA Metabolism and Mouse Fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [Green Version]
- Ping, X.-L.; Sun, B.-F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.-J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.-S.; et al. Mammalian WTAP Is a Regulatory Subunit of the RNA N6-Methyladenosine Methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G.; et al. N6-Methyladenosine in Nuclear RNA Is a Major Substrate of the Obesity-Associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Patil, D.P.; Pickering, B.F.; Jaffrey, S.R. Reading M6A in the Transcriptome: M6A-Binding Proteins. Trends Cell Biol. 2018, 28, 113–127. [Google Scholar] [CrossRef]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.-B.; Jaffrey, S.R. 5′ UTR M6A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Yang, Y.; Sun, B.-F.; Shi, Y.; Yang, X.; Xiao, W.; Hao, Y.-J.; Ping, X.-L.; Chen, Y.-S.; Wang, W.-J.; et al. FTO-Dependent Demethylation of N6-Methyladenosine Regulates MRNA Splicing and Is Required for Adipogenesis. Cell Res. 2014, 24, 1403–1419. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-Methyladenosine-Dependent Regulation of Messenger RNA Stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.-S.; Hao, Y.-J.; Sun, B.-F.; Sun, H.-Y.; Li, A.; Ping, X.-L.; Lai, W.-Y.; et al. Nuclear m6A Reader YTHDC1 Regulates MRNA Splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef] [Green Version]
- Kasowitz, S.D.; Ma, J.; Anderson, S.J.; Leu, N.A.; Xu, Y.; Gregory, B.D.; Schultz, R.M.; Wang, P.J. Nuclear M6A Reader YTHDC1 Regulates Alternative Polyadenylation and Splicing during Mouse Oocyte Development. PLoS Genet. 2018, 14, e1007412. [Google Scholar] [CrossRef]
- Fustin, J.-M.; Doi, M.; Yamaguchi, Y.; Hida, H.; Nishimura, S.; Yoshida, M.; Isagawa, T.; Morioka, M.S.; Kakeya, H.; Manabe, I.; et al. RNA-Methylation-Dependent RNA Processing Controls the Speed of the Circadian Clock. Cell 2013, 155, 793–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arribas-Hernández, L.; Bressendorff, S.; Hansen, M.H.; Poulsen, C.; Erdmann, S.; Brodersen, P. An M6A-YTH Module Controls Developmental Timing and Morphogenesis in Arabidopsis. Plant Cell 2018, 30, 952–967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, L.-H.; Song, P.; Wang, Y.; Lu, Z.; Tang, Q.; Yu, Q.; Xiao, Y.; Zhang, X.; Duan, H.-C.; Jia, G. The m6A Reader ECT2 Controls Trichome Morphology by Affecting MRNA Stability in Arabidopsis. Plant Cell 2018, 30, 968–985. [Google Scholar] [CrossRef] [Green Version]
- Duan, H.-C.; Wei, L.-H.; Zhang, C.; Wang, Y.; Chen, L.; Lu, Z.; Chen, P.R.; He, C.; Jia, G. ALKBH10B Is an RNA N6-Methyladenosine Demethylase Affecting Arabidopsis Floral Transition. Plant Cell 2017, 29, 2995–3011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, F.; Zhang, Y.-C.; Liao, J.-Y.; Yu, Y.; Zhou, Y.-F.; Feng, Y.-Z.; Yang, Y.-W.; Lei, M.-Q.; Bai, M.; Wu, H.; et al. The Subunit of RNA N6-Methyladenosine Methyltransferase OsFIP Regulates Early Degeneration of Microspores in Rice. PLoS Genet. 2019, 15, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Tian, S.; Qin, G. RNA Methylomes Reveal the M6A-Mediated Regulation of DNA Demethylase Gene SlDML2 in Tomato Fruit Ripening. Genome Biol. 2019, 20, 156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.; Tang, R.; Li, X.; Tian, S.; Li, B.; Qin, G. N6-Methyladenosine RNA Modification Regulates Strawberry Fruit Ripening in an ABA-Dependent Manner. Genome Biol. 2021, 22, 168. [Google Scholar] [CrossRef] [PubMed]
- Ok, S.H.; Jeong, H.J.; Bae, J.M.; Shin, J.-S.; Luan, S.; Kim, K.-N. Novel CIPK1-Associated Proteins in Arabidopsis Contain an Evolutionarily Conserved C-Terminal Region That Mediates Nuclear Localization. Plant Physiol. 2005, 139, 138–150. [Google Scholar] [CrossRef] [Green Version]
- Huong, T.T.; Ngoc, L.N.T.; Kang, H. Functional Characterization of a Putative RNA Demethylase ALKBH6 in Arabidopsis Growth and Abiotic Stress Responses. Int. J. Mol. Sci. 2020, 21, 6707. [Google Scholar] [CrossRef]
- Zhang, K.; Zhuang, X.; Dong, Z.; Xu, K.; Chen, X.; Liu, F.; He, Z. The Dynamics of N6-Methyladenine RNA Modification in Interactions between Rice and Plant Viruses. Genome Biol. 2021, 22, 189. [Google Scholar] [CrossRef]
- Miao, Z.; Zhang, T.; Qi, Y.; Song, J.; Han, Z.; Ma, C. Evolution of the RNA N6-Methyladenosine Methylome Mediated by Genomic Duplication. Plant Physiol. 2020, 182, 345–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.; Wang, J.; Hou, X. Transcriptome-Wide N6-Methyladenosine (M6A) Methylome Profiling of Heat Stress in Pak-Choi (Brassica Rapa ssp. Chinensis). Plants 2020, 9, 1080. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Liu, C.; Meng, F.; Hu, L.; Fu, X.; Yang, Z.; Wang, N.; Jiang, Q.; Ma, F. The M6A Reader MhYTP2 Regulates MdMLO19 MRNA Stability and Antioxidant Genes Translation Efficiency Conferring Powdery Mildew Resistance in Apple. Plant Biotechnol. J. 2021. [Google Scholar] [CrossRef]
- Nawaz, H.; Hussain, N.; Jamil, M.; Yasmeen, A.; Bukhari, S.A.H.; Aurangzaib, M.; Usman, M. Seed Biopriming Mitigates Terminal Drought Stress at Reproductive Stage of Maize by Enhancing Gas Exchange Attributes and Nutrient Uptake. Turk. J. Agric. For. 2020, 44, 250–261. [Google Scholar] [CrossRef]
- Li, X.; Chen, P.; Xie, Y.; Yan, Y.; Wang, L.; Dang, H.; Zhang, J.; Xu, L.; Ma, F.; Guan, Q. Apple SERRATE Negatively Mediates Drought Resistance by Regulating MdMYB88 and MdMYB124 and MicroRNA Biogenesis. Hortic. Res. 2020, 7, 98. [Google Scholar] [CrossRef]
- Radivojevic, D.; Milivojevic, J.; Pavlovic, M.; Stopar, M. Comparison of Metamitron Efficiency for Postbloom Thinning of Young ‘Gala’ and ‘Golden Delicious’ Apple Trees. Turk. J. Agric. For. 2020, 44, 83–94. [Google Scholar] [CrossRef]
- Tan, Y.; Li, M.; Yang, Y.; Sun, X.; Wang, N.; Liang, B.; Ma, F. Overexpression of MpCYS4, A Phytocystatin Gene from Malus Prunifolia (Willd.) Borkh., Enhances Stomatal Closure to Confer Drought Tolerance in Transgenic Arabidopsis and Apple. Front. Plant Sci. 2017, 8, 33. [Google Scholar] [CrossRef] [Green Version]
- Sokolov, V.V.; Savel’ev, N.I.; Goncharov, N.P.I.V. Michurin’S Work on Expansion of the Plant Horticulture Assortment and Improvement of Food Quality. Proc. Latv. Acad. Sci. Sect. B. Nat. Exact Appl. Sci. 2015, 69, 190–197. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.; Yan, M.; Li, L.; He, J.; Zhou, S.; Li, Z.; Niu, C.; Bao, C.; Zhi, F.; Ma, F.; et al. The Apple DNA-Binding One Zinc-Finger Protein MdDof54 Promotes Drought Resistance. Hortic. Res. 2020, 7, 195. [Google Scholar] [CrossRef]
- Xie, Y.; Chen, P.; Yan, Y.; Bao, C.; Li, X.; Wang, L.; Shen, X.; Li, H.; Liu, X.; Niu, C.; et al. An Atypical R2R3 MYB Transcription Factor Increases Cold Hardiness by CBF-Dependent and CBF-Independent Pathways in Apple. New Phytol. 2018, 218, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Daccord, N.; Celton, J.-M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; van de Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-Quality de Novo Assembly of the Apple Genome and Methylome Dynamics of Early Fruit Development. Nat. Genet. 2017, 49, 1099–1106. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, M.; Huber, W.; Pagès, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.T.; Carey, V.J. Software for Computing and Annotating Genomic Ranges. PLoS Comput. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Molecular Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Kolde, R. Pheatmap: Pretty Heatmaps. R Package Version 2012, 1, 726. [Google Scholar]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. AgriGO v2.0: A GO Analysis Toolkit for the Agricultural Community, 2017 Update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Fiancette, R.; Finlay, C.M.; Willis, C.; Bevington, S.L.; Soley, J.; Ng, S.T.H.; Baker, S.M.; Andrews, S.; Hepworth, M.R.; Withers, D.R. Reciprocal Transcription Factor Networks Govern Tissue-Resident ILC3 Subset Function and Identity. Nat. Immunol. 2021, 22, 1245–1255. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Song, B.; Ma, J.; Song, Y.; Zhang, S.-Y.; Tang, Y.; Wu, X.; Wei, Z.; Chen, K.; Su, J.; et al. Bioinformatics Approaches for Deciphering the Epitranscriptome: Recent Progress and Emerging Topics. Comput. Struct. Biotechnol. J. 2020, 18, 1587–1604. [Google Scholar] [CrossRef]
- Khan, A.; Mathelier, A. Intervene: A Tool for Intersection and Visualization of Multiple Gene or Genomic Region Sets. BMC Bioinform. 2017, 18, 287. [Google Scholar] [CrossRef] [Green Version]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple Combinations of Lineage-Determining Transcription Factors Prime Cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinlan, A.R.; Hall, I.M. BEDTools: A Flexible Suite of Utilities for Comparing Genomic Features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [Green Version]
- Zhai, J.; Song, J.; Cheng, Q.; Tang, Y.; Ma, C. PEA: An Integrated R Toolkit for Plant Epitranscriptome Analysis. Bioinformatics 2018, 34, 3747–3749. [Google Scholar] [CrossRef] [Green Version]
- Robinson, J.T.; Thorvaldsdóttir, H.; Wenger, A.M.; Zehir, A.; Mesirov, J.P. Variant Review with the Integrative Genomics Viewer. Cancer Res. 2017, 77, e31–e34. [Google Scholar] [CrossRef] [Green Version]
- Luo, G.-Z.; MacQueen, A.; Zheng, G.; Duan, H.; Dore, L.C.; Lu, Z.; Liu, J.; Chen, K.; Jia, G.; Bergelson, J.; et al. Unique Features of the M6A Methylome in Arabidopsis Thaliana. Nat. Commun. 2014, 5, 5630. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Tang, K.; Zhang, D.; Wan, Y.; Wen, Y.; Lu, Q.; Wang, L. High-Throughput M6A-Seq Reveals RNA M6A Methylation Patterns in the Chloroplast and Mitochondria Transcriptomes of Arabidopsis Thaliana. PLoS ONE 2017, 12, e0185612. [Google Scholar] [CrossRef]
- Bhat, S.S.; Bielewicz, D.; Gulanicz, T.; Bodi, Z.; Yu, X.; Anderson, S.J.; Szewc, L.; Bajczyk, M.; Dolata, J.; Grzelak, N.; et al. MRNA Adenosine Methylase (MTA) Deposits m6A on Pri-MiRNAs to Modulate MiRNA Biogenesis in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2020, 117, 21785–21795. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Wei, Z.; Zhang, L.; Liu, H.; Sun, L.; Zhang, S.-W.; Huang, Y.; Meng, J. Guitar: An R/Bioconductor Package for Gene Annotation Guided Transcriptomic Analysis of RNA-Related Genomic Features. BioMed Res. Int. 2016, 2016, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, Writing and Erasing MRNA Methylation. Nat. Rev. Mol. Cell. Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-M.; Sasaki, T.; Ueda, M.; Sako, K.; Seki, M. Chromatin Changes in Response to Drought, Salinity, Heat, and Cold Stresses in Plants. Front. Plant Sci. 2015, 6, 114. [Google Scholar] [CrossRef] [Green Version]
- Grigorova, B.; Vaseva, I.; Demirevska, K.; Feller, U. Combined Drought and Heat Stress in Wheat: Changes in Some Heat Shock Proteins. Biologia Plant. 2011, 55, 105–111. [Google Scholar] [CrossRef]
- Jia, J.; Zhou, J.; Shi, W.; Cao, X.; Luo, J.; Polle, A.; Luo, Z.-B. Comparative Transcriptomic Analysis Reveals the Roles of Overlapping Heat-/Drought-Responsive Genes in Poplars Exposed to High Temperature and Drought. Sci. Rep. 2017, 7, 43215. [Google Scholar] [CrossRef] [Green Version]
- Boyko, A.; Blevins, T.; Yao, Y.; Golubov, A.; Bilichak, A.; Ilnytskyy, Y.; Hollander, J.; Meins, F.; Kovalchuk, I. Transgenerational Adaptation of Arabidopsis to Stress Requires DNA Methylation and the Function of Dicer-Like Proteins. PLoS ONE 2010, 5, e9514. [Google Scholar] [CrossRef]
- Yao, Y.; Bilichak, A.; Golubov, A.; Kovalchuk, I. Arabidopsis Thaliana SiRNA Biogenesis Mutants Have the Lower Frequency of Homologous Recombination. Plant Signal. Behav. 2016, 11, e1151599. [Google Scholar] [CrossRef] [Green Version]
- Cheng, M.-C.; Liao, P.-M.; Kuo, W.-W.; Lin, T.-P. The Arabidopsis ETHYLENE RESPONSE FACTOR1 Regulates Abiotic Stress-Responsive Gene Expression by Binding to Different Cis-Acting Elements in Response to Different Stress Signals. Plant Physiol. 2013, 162, 1566–1582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manohara Reddy, B.; Kumari, V.; Anthony Johnson, A.; Jagadeesh Kumar, N.; Venkatesh, B.; Jayamma, N.; Sudhakar, C. Scarecrow like Protein 1,(Ct-SCL1) Involved in Drought Stress Tolerance by Interacting with SWI3B Component of Chromatin Modelling Complex in Cluster Bean, Cyamopsistetragonaloba (L.) Taub. Int. J. Res. Anal. Rev. 2018, 5. [Google Scholar]
- Luo, J.-H.; Wang, Y.; Wang, M.; Zhang, L.-Y.; Peng, H.-R.; Zhou, Y.-Y.; Jia, G.-F.; He, Y. Natural Variation in RNA m6A Methylation and Its Relationship with Translational Status. Plant Physiol. 2020, 182, 332–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, L.; Liang, Z.; Gu, X.; Chen, Y.; Teo, Z.W.N.; Hou, X.; Cai, W.M.; Dedon, P.C.; Liu, L.; Yu, H. N6-Methyladenosine RNA Modification Regulates Shoot Stem Cell Fate in Arabidopsis. Dev. Cell 2016, 38, 186–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, Z.; Zhang, T.; Xie, B.; Qi, Y.; Ma, C. Evolutionary Implications of the RNA N 6-Methyladenosine Methylome in Plants. Mol. Biol. Evol. 2021, msab299. [Google Scholar] [CrossRef]
- Wang, K.; Peng, J.; Yi, C. The m6A Consensus Motif Provides a Paradigm of Epitranscriptomic Studies. Biochemistry 2021, 60, 3410–3412. [Google Scholar] [CrossRef] [PubMed]
- Adams, H.D.; Guardiola-Claramonte, M.; Barron-Gafford, G.A.; Villegas, J.C.; Breshears, D.D.; Zou, C.B.; Troch, P.A.; Huxman, T.E. Temperature Sensitivity of Drought-Induced Tree Mortality Portends Increased Regional Die-off under Global-Change-Type Drought. Proc. Natl. Acad. Sci. USA 2009, 106, 7063–7066. [Google Scholar] [CrossRef] [Green Version]
- Šircelj, H.; Tausz, M.; Grill, D.; Batič, F. Biochemical Responses in Leaves of Two Apple Tree Cultivars Subjected to Progressing Drought. J. Plant Physiol. 2005, 162, 1308–1318. [Google Scholar] [CrossRef]
- Xu, Y.; Zhan, C.; Huang, B. Heat Shock Proteins in Association with Heat Tolerance in Grasses. Int. J. Proteom. 2011, 2011, 529648. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.-J.; Cheng, G.-X.; Khan, A.; Wei, A.-M.; Yu, Q.-H.; Yang, S.-B.; Luo, D.-X.; Gong, Z.-H. CaHSP16.4, a Small Heat Shock Protein Gene in Pepper, Is Involved in Heat and Drought Tolerance. Protoplasma 2019, 256, 39–51. [Google Scholar] [CrossRef]
- Cho, E.K.; Hong, C.B. Over-Expression of Tobacco NtHSP70-1 Contributes to Drought-Stress Tolerance in Plants. Plant Cell Rep. 2006, 25, 349–358. [Google Scholar] [CrossRef]
- Song, H.; Zhao, R.; Fan, P.; Wang, X.; Chen, X.; Li, Y. Overexpression of AtHsp90.2, AtHsp90.5 and AtHsp90.7 in Arabidopsis Thaliana Enhances Plant Sensitivity to Salt and Drought Stresses. Planta 2009, 229, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Tan, D.-X.; Liang, D.; Chang, C.; Jia, D.; Ma, F. Melatonin Mediates the Regulation of ABA Metabolism, Free-Radical Scavenging, and Stomatal Behaviour in Two Malus Species under Drought Stress. J. Exp. Bot. 2015, 66, 669–680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, F.; Kuromori, T.; Urano, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Drought Stress Responses and Resistance in Plants: From Cellular Responses to Long-Distance Intercellular Communication. Front. Plant Sci. 2020, 11, 556972. [Google Scholar] [CrossRef] [PubMed]
- Soma, F. Plant Raf-like Kinases Regulate the MRNA Population Upstream of ABA-Unresponsive SnRK2 Kinases under Drought Stress. Nat. Commun. 2020, 11, 1373. [Google Scholar] [CrossRef] [Green Version]
- Mizoguchi, M.; Umezawa, T.; Nakashima, K.; Kidokoro, S.; Takasaki, H.; Fujita, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two Closely Related Subclass II SnRK2 Protein Kinases Cooperatively Regulate Drought-Inducible Gene Expression. Plant Cell Physiol. 2010, 51, 842–847. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mao, X.; Hou, N.; Liu, Z.; He, J. Profiling of N6-Methyladenosine (m6A) Modification Landscape in Response to Drought Stress in Apple (Malus prunifolia (Willd.) Borkh). Plants 2022, 11, 103. https://doi.org/10.3390/plants11010103
Mao X, Hou N, Liu Z, He J. Profiling of N6-Methyladenosine (m6A) Modification Landscape in Response to Drought Stress in Apple (Malus prunifolia (Willd.) Borkh). Plants. 2022; 11(1):103. https://doi.org/10.3390/plants11010103
Chicago/Turabian StyleMao, Xiushan, Nan Hou, Zhenzhong Liu, and Jieqiang He. 2022. "Profiling of N6-Methyladenosine (m6A) Modification Landscape in Response to Drought Stress in Apple (Malus prunifolia (Willd.) Borkh)" Plants 11, no. 1: 103. https://doi.org/10.3390/plants11010103
APA StyleMao, X., Hou, N., Liu, Z., & He, J. (2022). Profiling of N6-Methyladenosine (m6A) Modification Landscape in Response to Drought Stress in Apple (Malus prunifolia (Willd.) Borkh). Plants, 11(1), 103. https://doi.org/10.3390/plants11010103




