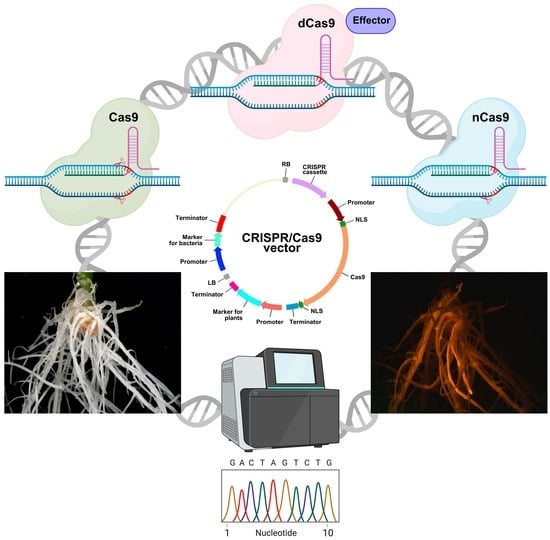
Hairy CRISPR: Genome Editing in Plants Using Hairy Root Transformation
Abstract
1. Introduction
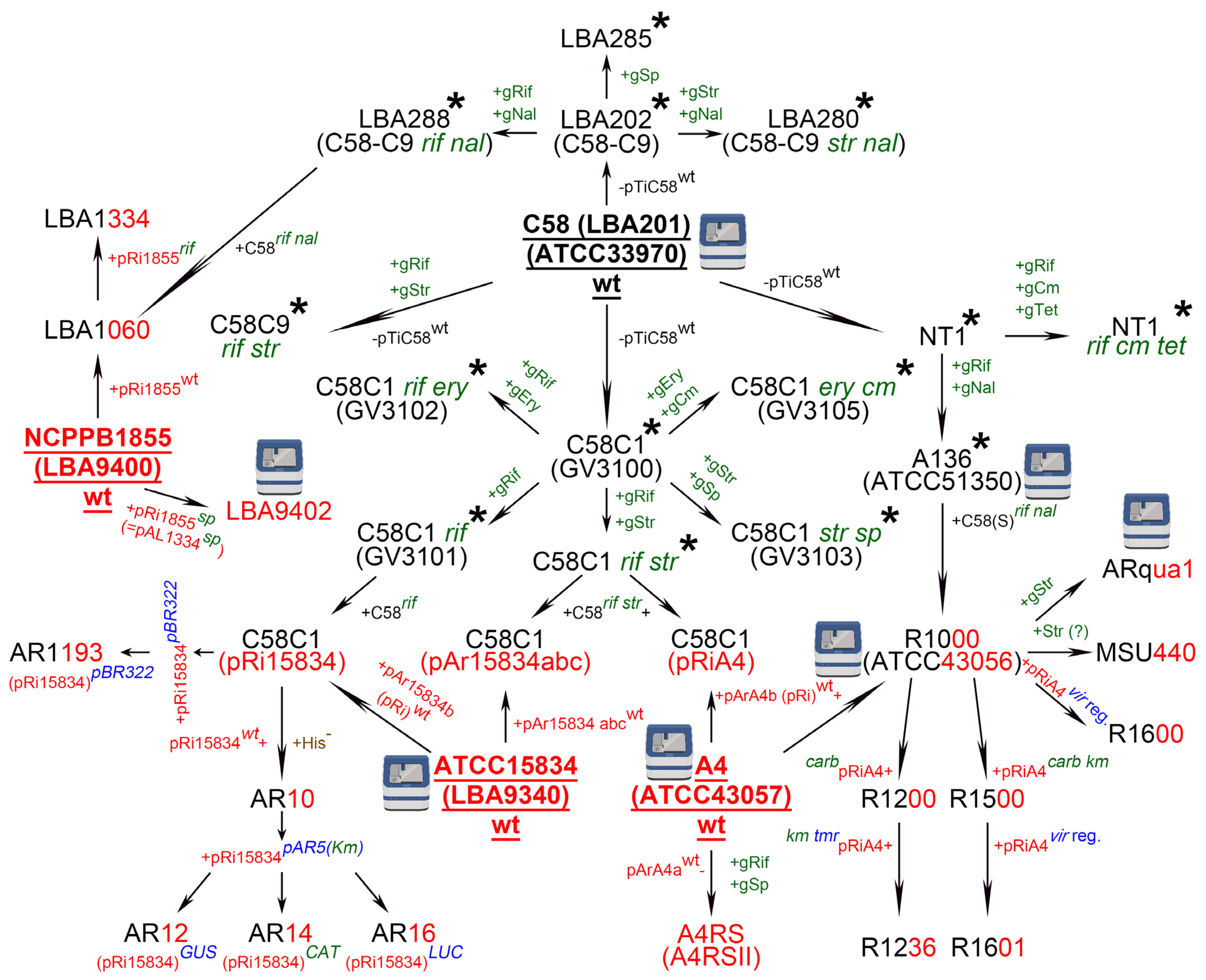
2. Agrobacterium rhizogenes: A Historical View of the Widely Used Strains and Their Nomenclature
3. Editing the Plant Genome in Transgenic Hairy Roots: Vector Components
3.1. Cassette for Cas Expression
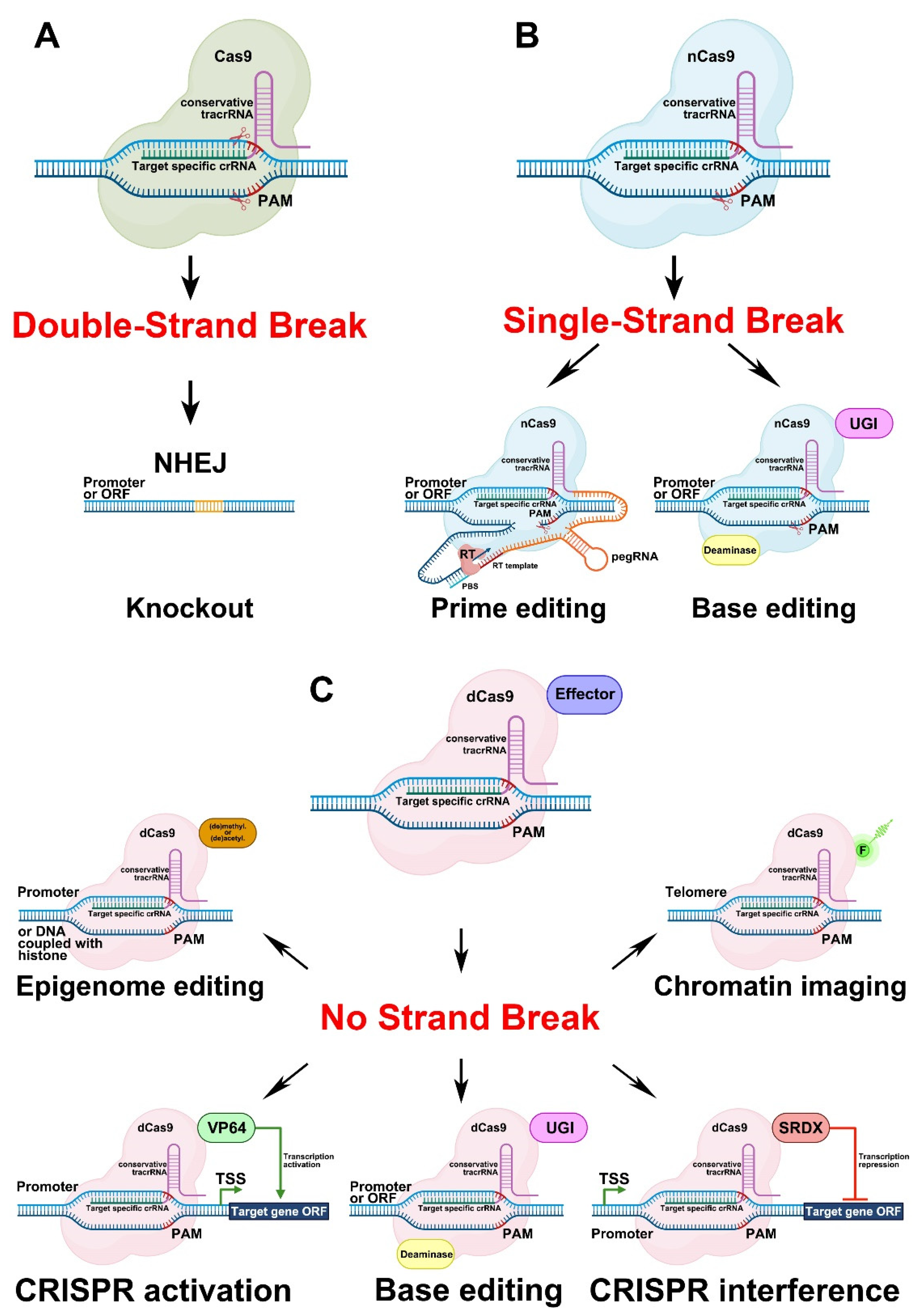
3.1.1. Genome Editing Based on Double-Strand Breaks in DNA Caused by Cas9 Activity
3.1.2. Genome Editing Based on Single-Strand Breaks in DNA Caused by Cas9 Activity
3.1.3. Genome Editing Based on a Version of Cas9 That Causes no Strand Breaks
3.2. gRNA: Design and Testing
3.3. Markers of Transgenicity Used in CRISPR/Cas Vectors: Old Players, New Tricks
3.3.1. Selectable Markers
3.3.2. β-Glucuronidase-Based Screenable Markers
3.3.3. Pigment Biosynthesis-Based Screenable Markers
3.3.4. Fluorescent Protein-Based Screenable Markers
3.3.5. New Strategies for the Insertion of a Marker Cassette into a CRISPR/Cas Vector
3.3.6. Construction of the CRISPR/Cas9 Vector with the DsRed1 Screenable Marker
4. Possible Applications of CRISPR/Cas9 Genome Editing Using Hairy Root Transformation
5. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CRISPR | clustered regularly interspaced short palindromic repeats |
| Cas | CRISPR associated nuclease |
| crRNA | crisprRNA |
| tracrRNA | trans-activating crisprRNA |
| PAM | protospacer adjacent motif |
| ORF | open reading frame |
| TF | transcription factor |
| TSS | transcription start site |
References
- Somssich, M. A short history of plant transformation. Peer J. 2019, 7, 1–28. [Google Scholar] [CrossRef]
- Young, J.M.; Kuykendall, L.D.; Martínez-Romero, E.; Kerr, A.; Sawada, H. A revision of Rhizobium Frank 1889, with an emended description of the genus, and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola de Lajudie et al. 1998 as new combinations: Rhizobium radiobacter, R. rhizogenes, R. rubi, R. undicola and R. vitis. Int. J. Syst. Evol. Microbiol. 2001, 51, 89–103. [Google Scholar] [CrossRef]
- Porter, J.R.; Flores, H. Host range and implications of plant infection by Agrobacterium rhizogenes. Crit. Rev. Plant. Sci. 1991, 10, 387–421. [Google Scholar] [CrossRef]
- Bahramnejad, B.; Naji, M.; Bose, R.; Jha, S. A critical review on use of Agrobacterium rhizogenes and their associated binary vectors for plant transformation. Biotechnol. Adv. 2019, 37, 1–14. [Google Scholar] [CrossRef]
- Desmet, S.; Dhooghe, E.; De Keyser, E.; Van Huylenbroeck, J.; Müller, R.; Geelen, D.; Lütken, H. Rhizogenic agrobacteria as an innovative tool for plant breeding: Current achievements and limitations. Appl. Microbiol. Biotechnol. 2020, 104, 2435–2451. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, Z.; Iqbal, M.S.; Ahmad, A.; Memon, A.G.; Ansari, M.I. New prospects on the horizon: Genome editing to engineer plants for desirable traits. Curr. Plant Biol. 2020, 24, 1–12. [Google Scholar] [CrossRef]
- Belhaj, K.; Chaparro-Garcia, A.; Kamoun, S.; Nekrasov, V. Plant genome editing made easy: Targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 2013, 9, 1–10. [Google Scholar] [CrossRef]
- Ron, M.; Kajala, K.; Pauluzzi, G.; Wang, D.; Reynoso, M.A.; Zumstein, K.; Garcha, J.; Winte, S.; Masson, H.; Inagaki, S.; et al. Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol. 2014, 166, 455–469. [Google Scholar] [CrossRef]
- Twyman, R.M.; Stöger, E.; Kohli, A.; Capell, T.; Christou, P. Selectable and screenable markers for rice transformation. In Testing for Genetic Manipulation in Plants, 1st ed.; Jackson, J.F., Linskens, H.F., Eds.; Springer: Berlin, Germany, 2002; pp. 1–17. [Google Scholar] [CrossRef]
- Dhiman, N.; Patial, V.; Bhattacharya, A. The current status and future applications of hairy root cultures. In Biotechnological Approaches for Medicinal and Aromatic Plants: Conservation, Genetic Improvement and Utilization, 1st ed.; Kumar, N., Ed.; Springer Science+Business Media LLC: Singapore, 2018; pp. 87–155. [Google Scholar] [CrossRef]
- Alok, A.; Kumar, J.; Upadhyay, S.K. Engineering in hairy roots using CRISPR/Cas9-mediated editing. In Hairy Roots: An Effective Tool of Plant Biotechnology; Srivastava, V., Mehrotra, S., Mishra, S., Eds.; Springer Science+Business Media LLC: Singapore, 2018; pp. 329–342. [Google Scholar] [CrossRef]
- De Saeger, J.; Park, J.; Chung, H.S.; Hernalsteens, J.-P.; Van Lijsebettens, M.; Inzé, D.; Van Montagu, M.; Depuydt, S. Agrobacterium strains and strain improvement: Present and outlook. Biotechnol. Adv. 2021, 107677, 1–25. [Google Scholar] [CrossRef]
- Agrobacterium rhizogenes (ATCC® 15834™). Available online: https://www.atcc.org/products/15834 (accessed on 15 December 2021).
- White, F.F.; Nester, E.W. Hairy root: Plasmid encodes virulence traits in Agrobacterium rhizogenes. J. Bacteriol. 1980, 141, 1134–1141. [Google Scholar] [CrossRef] [PubMed]
- White, F.F.; Nester, E.W. Relationship of plasmids responsible for hairy root and crown gall tumorigenicity. J. Bacteriol. 1980, 144, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, D.L.; Durbin, R.D. Induction of adventitious roots by culture filtrates of the hairy root bacterium, Agrobacterium rhizogenes. Cannadian J. Microbiol. 1971, 17, 1409–1412. [Google Scholar] [CrossRef]
- Kajala, K.; Coil, D.A.; Brady, S.M. Draft genome sequence of Rhizobium rhizogenes strain ATCC 15834. Genome Announc. 2014, 2, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Agrobacterium rhizogenes (ATCC® 43057™). Available online: https://www.atcc.org/products/43057 (accessed on 15 December 2021).
- Moore, L.; Warren, G.; Strobel, G. Involvement of a plasmid in the hairy root disease of plants caused by Agrobacterium rhizogenes. Plasmid 1979, 2, 617–626. [Google Scholar] [CrossRef]
- Jouanin, L.; Tourneur, J.; Tourneur, C.; Casse-Delbart, F. Restriction maps and homologies of the three plasmids of Agrobacterium rhizogenes strain A4. Plasmid 1986, 16, 124–134. [Google Scholar] [CrossRef]
- Cho, S.-T.; Lin, Y.-C.; Lai, E.-M.; Kuo, C.-H. Complete Genome Sequence of Agrobacterium rhizogenes A4 (EML492). Available online: https://www.ncbi.nlm.nih.gov/assembly/GCF_018138105.1 (accessed on 15 December 2021).
- White, F.F.; Sinkar, V.P. Molecular analysis of root induction by Agrobacterium rhizogenes. In Plant DNA Infectious Agents, 1st ed.; Hohn, T., Schell, J., Eds.; Springer: Vienna, Austria, 1987; pp. 149–177. [Google Scholar] [CrossRef]
- Alpizar, E.; Dechamp, E.; Espeout, S.; Royer, M.; Lecouls, A.C.; Nicole, M.; Bertrand, B.; Lashermes, P.; Etienne, H. Efficient production of Agrobacterium rhizogenes-transformed roots and composite plants for studying gene expression in coffee roots. Plant Cell. Rep. 2006, 25, 959–967. [Google Scholar] [CrossRef]
- Bonaldi, K.; Gherbi, H.; Franche, C.; Bastien, G.; Fardoux, J.; Barker, D.; Giraud, E.; Cartieaux, F. The Nod factor–independent symbiotic signaling pathway: Development of Agrobacterium rhizogenes–mediated transformation for the legume Aeschynomene indica. Mol. Plant-Microbe Interact. 2010, 23, 1537–1544. [Google Scholar] [CrossRef]
- Dai, Y.; Hu, G.; Dupas, A.; Medina, L.; Blandels, N.; San Clemente, H.; Ladouce, N.; Badawi, M.; Hernandez-Raquet, G.; Mounet, F.; et al. Implementing the CRISPR/Cas9 technology in Eucalyptus hairy roots using wood-related genes. Int. J. Mol. Sci. 2020, 21, 3408. [Google Scholar] [CrossRef] [PubMed]
- Pellegrineschi, A.; Damon, J.-P.; Valtorta, N.; Paillard, N.; Tapfer, D. Improvement of ornamental characters and fragrance production in lemon-scented Geranium through genetic transformation by Agrobacterium rhizogenes. Bio/Technology 1994, 12, 64–68. [Google Scholar] [CrossRef]
- Stiller, J.; Martirani, L.; Tuppale, S.; Chian, R.-J.; Chiurazzi, M.; Gresshoff, P.M. High frequency transformation and regeneration of transgenic plants in the model legume Lotus japonicus. J. Exp. Bot. 1997, 48, 1357–1365. [Google Scholar] [CrossRef]
- NCPPB No. 2659, Rhizobium radiobacter. Available online: http://ncppb.fera.defra.gov.uk/furtherinfo.cfm?ncppb_no=2659 (accessed on 15 December 2021).
- NCPPB No. 1855, Rhizobium rhizogenes. Available online: http://ncppb.fera.defra.gov.uk/furtherinfo.cfm?ncppb_no=1855 (accessed on 15 December 2021).
- Combard, A.; Brevet, J.; Borowski, D.; Cam, K.; Tempé, J. Physical map of the T-DNA region of Agrobacterium rhizogenes strain NCPPB2659. Plasmid 1987, 18, 70–75. [Google Scholar] [CrossRef]
- Mankin, S.L.; Hill, D.S.; Olhoft, P.M.; Toren, E.; Wenck, A.R.; Nea, L.; Xing, L.; Brown, J.A.; Fu, H.; Ireland, L.; et al. Disarming and sequencing of Agrobacterium rhizogenes strain K599 (NCPPB2659) plasmid pRi2659. In Vitro Cell. Dev. Biol. Plant 2007, 43, 521–535. [Google Scholar] [CrossRef]
- Valdes Franco, J.A.; Collier, R.; Wang, Y.; Huo, N.; Gu, Y.; Thilmony, R.; Thomson, J.G. Draft genome sequence of Agrobacterium rhizogenes strain NCPPB2659. Genome Announc. 2016, 4, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, S.D.; Karimi, M.; Impens, L.; Van Lerberge, E.; Coussens, G.; Aesaert, S.; Rombaut, D.; Holtappels, D.; Ibrahim, H.M.M.; Van Montagu, M.; et al. Efficient CRISPR-mediated base editing in Agrobacterium spp. Proc. Natl. Acad. Sci. USA 2021, 118, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hooykaas, M.J.G.; Hooykaas, P.J.J. The genome sequence of hairy root Rhizobium rhizogenes strain LBA9402: Bioinformatics analysis suggests the presence of a new opine system in the agropine Ri plasmid. MicrobiologyOpen 2021, 10, 1–18. [Google Scholar] [CrossRef]
- Costantino, P.; Hooykaas, P.J.J.; den Dulk-Ras, H.; Schilperoort, R.A. Tumor formation and rhizogenicity of Agrobacterium rhizogenes carrying Ti plasmids. Gene 1980, 11, 79–87. [Google Scholar] [CrossRef]
- Hooykaas, P.J.J.; Klapwijk, P.M.; Nuti, M.P.; Schilperoort, R.A.; Rörsch, A. Transfer of the Agrobacterium tumefaciens Ti plasmid to avirulent Agrobacteria and to Rhizobium ex planta. Microbiology 1977, 98, 477–484. [Google Scholar] [CrossRef]
- Wood, D.W.; Setubal, J.C.; Kaul, R.; Monks, D.E.; Kitajima, J.P.; Okura, V.K.; Zhou, Y.; Chen, L.; Wood, G.E.; Almeida, N.F.; et al. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 2001, 294, 2317–2323. [Google Scholar] [CrossRef]
- Goodner, B.; Hinkle, G.; Gattung, S.; Miller, N.; Blanchard, M.; Qurollo, B.; Goldman, B.S.; Cao, Y.; Askenazi, M.; Halling, C.; et al. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 2001, 294, 2323–2328. [Google Scholar] [CrossRef]
- Slater, S.; Setubal, J.C.; Goodner, B.; Houmiel, K.; Sun, J.; Kaul, R.; Goldman, B.S.; Farrand, S.K.; Almeida, N.; Burr, T.; et al. Reconciliation of sequence data and updated annotation of the genome of Agrobacterium tumefaciens C58, and distribution of a linear chromosome in the genus Agrobacterium. Appl. Environ. Microbiol. 2013, 79, 1414–1417. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, R.H.; Fall, M.Z. The loss of tumor-initiating ability in Agrobacterium tumefaciens by incubation at high temperature. Experientia 1971, 27, 229–230. [Google Scholar] [CrossRef]
- Van Larebeke, N.; Engler, G.; Holsters, M.; Den Elsacker, S.V.; Zaenen, I.; Schilperoort, R.A.; Schell, J. Large plasmid in Agrobacterium tumefaciens essential for crown gall-inducing ability. Nature 1974, 252, 169–170. [Google Scholar] [CrossRef] [PubMed]
- Watson, B.; Currier, T.C.; Gordon, M.P.; Chilton, M.D.; Nester, E.W. Plasmid required for virulence of Agrobacterium tumefaciens. J. Bacteriol. 1975, 123, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Rhizobium radiobacter (ATCC® 51350™). Available online: https://www.atcc.org/products/51350 (accessed on 15 December 2021).
- Van Larebeke, N.; Genetello, C.H.; Schell, J.; Schilperoort, R.A.; Hermans, A.K.; Hernalsteens, J.P.; Van Montagu, M. Acquisition of tumour-inducing ability by non-oncogenic agrobacteria as a result of plasmid transfer. Nature 1975, 255, 742–743. [Google Scholar] [CrossRef]
- Genetello, C.; Van Larebeke, N.; Holsters, M.; De Picker, A.; Van Montagu, M.; Schell, J. Ti plasmids of Agrobacterium as conjugative plasmids. Nature 1977, 265, 561–563. [Google Scholar] [CrossRef]
- Petit, A.; Tempé, J.; Kerr, A.; Holsters, M.; Van Montagu, M.; Schell, J. Substrate induction of conjugative activity of Agrobacterium tumefaciens Ti plasmids. Nature 1978, 271, 570–572. [Google Scholar] [CrossRef]
- Holsters, M.; Silva, B.; Van Vliet, F.; Genetello, C.; De Block, M.; Dhaese, P.; Depicker, A.; Inzé, D.; Engler, G.; Villarroel, R.; et al. The functional organization of the nopaline A. tumefaciens plasmid pTiC58. Plasmid 1980, 3, 212–230. [Google Scholar] [CrossRef]
- Holsters, M.; de Waele, D.; Depicker, A.; Messens, E.; Van Montagu, M.; Schell, J. Transfection and transformation of Agrobacterium tumefaciens. Mol. Gen. Genet. 1978, 163, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Tempé, J.; Petit, A.; Holsters, M.; Van Montagu, M.; Schell, J. Thermosensitive step associated with transfer of the Ti plasmid during conjugation: Possible relation to transformation in crown gall. Proc. Natl. Acad. Sci. USA 1977, 74, 2848–2849. [Google Scholar] [CrossRef]
- Bomhoff, G.; Klapwijk, P.M.; Kester, H.C.M.; Schilperoort, R.A.; Hernalsteens, J.P.; Schell, J. Octopine and nopaline synthesis and breakdown genetically controlled by a plasmid of Agrobacterium tumefaciens. Mol. Gen. Genet. 1976, 145, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Ledeboer, A.M.; Krol, A.J.M.; Dons, J.J.M.; Spier, F.; Schilperoort, R.A.; Zaenen, I.; Van Larebeke, N.; Schell, J. On the isolation of TI-plasmid from Agrobacterium tumefaciens. Nucleic Acids Res. 1976, 3, 449–464. [Google Scholar] [CrossRef][Green Version]
- Klapwijk, P.M.; de Jonge, A.J.R.; Schilperoort, R.A.; Rörsch, A. An enrichment technique for auxotrophs of Agrobacterium tumefaciens using a combination of carbenicillin and lysozyme. J. Gen. Microbiol. 1975, 91, 177–182. [Google Scholar] [CrossRef][Green Version]
- Hooykaas, P.J.J.; den Dulk-Ras, H.; Schilperoort, R.A. Molecular mechanism of Ti plasmid mobilization by R plasmids: Isolation of Ti plasmids with transposon-insertions in Agrobacterium tumefaciens. Plasmid 1980, 4, 64–75. [Google Scholar] [CrossRef]
- Hooykaas, P.J.; den Dulk-Ras, H.; Ooms, G.; Schilperoort, R.A. Interactions between octopine and nopaline plasmids in Agrobacterium tumefaciens. J. Bacteriol. 1980, 143, 1295–1306. [Google Scholar] [CrossRef] [PubMed]
- Fodor, A.; Makrai, L.; Vozik, D.; Olasz, F.; Kiss, J.; Klein, M.G.; Ghaffar, M.-A.B.-A.; Szabados, L.; El-Deen, A.N.; Fodor, L.; et al. Agrobacterium may be used as a suitable experimental system for genetic analysis of resistance to (at least Xenorhabdus budapestensis) antimicrobial peptide complexes. PeerJ 2018, 6, e26900v26901. [Google Scholar] [CrossRef]
- Owens, L.D.; Cress, D.E. Genotypic variability of soybean response to Agrobacterium strains harboring the Ti or Ri plasmids. Plant Physiol. 1985, 77, 87–94. [Google Scholar] [CrossRef] [PubMed]
- White, F.F.; Taylor, B.H.; Huffman, G.A.; Gordon, M.P.; Nester, E.W. Molecular and genetic analysis of the transferred DNA regions of the root-inducing plasmid of Agrobacterium rhizogenes. J. Bacteriol. 1985, 164, 33–44. [Google Scholar] [CrossRef]
- McInnes, E.; Davey, M.R.; Mulligan, B.J.; Davies, K.; Sargent, A.W.; Morgan, A.J. Use of a disarmed Ri plasmid vector in analysis of transformed root induction. J. Exp. Bot. 1989, 40, 1135–1144. [Google Scholar] [CrossRef]
- Dommisse, E.M.; Leung, D.W.M.; Shaw, M.L.; Conner, A.J. Onion is a monocotyledonous host for Agrobacterium. Plant Sci. 1990, 69, 249–257. [Google Scholar] [CrossRef]
- Thompson, M.G.; Cruz-Morales, P.; Moore, W.M.; Pearson, A.N.; Keasling, J.D.; Scheller, H.V.; Shih, P.M. Draft genome sequence of Agrobacterium fabrum ARqua1. Microbiol. Resour. Announc. 2020, 9, 1–3. [Google Scholar] [CrossRef]
- Zhou, Y.; Neuhäuser, B.; Neumann, G.; Ludewig, U. LaALMT1 mediates malate release from phosphorus-deficient white lupin root tips and metal root to shoot translocation. Plant Cell. Environ. 2020, 43, 1691–1706. [Google Scholar] [CrossRef] [PubMed]
- Sahu, L.; Jena, S.; Swain, S.S.; Sahoo, S.; Chand, P.K. Agrobacterium rhizogenes-mediated transformation of a multi-medicinal herb, Boerhaavia diffusa L.: Optimization of the process and anti-microbial activity against bacterial pathogens causing urinary tract infections. Front. Life Sci. 2013, 7, 197–209. [Google Scholar] [CrossRef]
- Rhizobium sp. (ATCC® 43056™). Available online: https://www.atcc.org/products/43056 (accessed on 15 December 2021).
- Irigoyen, S.; Ramasamy, M.; Pant, S.; Niraula, P.; Bedre, R.; Gurung, M.; Rossi, D.; Laughlin, C.; Gorman, Z.; Achor, D.; et al. Plant hairy roots enable high throughput identification of antimicrobials against Candidatus Liberibacter spp. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef]
- Genome of Rhizobium sp. (ATCC® 43056™). Available online: https://genomes.atcc.org/genomes/e86859d407384d1c?tab=related-genomes-tab (accessed on 15 December 2021).
- Yoshimatsu, K.; Jaziri, M.; Kamada, H.; Shimomura, K. Production of diploid and haploid transgenic Atropa belladonna plants: Morphological traits and tropane alkaloid production. Belg. J. Bot. 1997, 130, 38–46. [Google Scholar]
- Joseph Sahayarayan, J.; Udayakumar, R.; Arun, M.; Ganapathi, A.; Alwahibi, M.S.; Aldosari, N.S.; Morgan, A.M.A. Effect of different Agrobacterium rhizogenes strains for in-vitro hairy root induction, total phenolic, flavonoids contents, antibacterial and antioxidant activity of (Cucumis anguria L.). J. Biol. Sci. 2020, 27, 2972–2979. [Google Scholar] [CrossRef] [PubMed]
- Thwe, A.; Valan Arasu, M.; Li, X.; Park, C.H.; Kim, S.J.; Al-Dhabi, N.A.; Park, S.U. Effect of different Agrobacterium rhizogenes strains on hairy root induction and phenylpropanoid biosynthesis in tartary buckwheat (Fagopyrum tataricum Gaertn). Front. Microbiol. 2016, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sinkar, V.P.; Pythoud, F.; White, F.F.; Nester, E.W.; Gordon, M.P. rolA locus of the Ri plasmid directs developmental abnormalities in transgenic tobacco plants. Genes Dev. 1988, 2, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Pythoud, F.; Sinkar, V.P.; Nester, E.W.; Gordon, M.P. Increased virulence of Agrobacterium rhizogenes conferred by the vir region of pTiBo542: Application to genetic engineering of poplar. Nat. Biotechnol. 1987, 5, 1323–1327. [Google Scholar] [CrossRef]
- Quandt, H.-J.; Pühler, A.; Broer, I. Transgenic root nodules of Vicia hirsuta: A fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol. Plant-Microbe Interact. 1993, 6, 699–706. [Google Scholar] [CrossRef]
- Sonti, R.V.; Chiurazzi, M.; Wong, D.; Davies, C.S.; Harlow, G.R.; Mount, D.W.; Signer, E.R. Arabidopsis mutants deficient in T-DNA integration. Proc. Natl. Acad. Sci. USA 1995, 92, 11786–11790. [Google Scholar] [CrossRef]
- MSU440 Competent Cells. Available online: https://www.lifeasible.com/?s=MSU440 (accessed on 15 December 2021).
- Petit, A.; David, C.; Dahl, G.A.; Ellis, J.G.; Guyon, P.; Casse-Delbart, F.; Tempé, J. Further extension of the opine concept: Plasmids in Agrobacterium rhizogenes cooperate for opine degradation. Mol. Gen. Genet. 1983, 190, 204–214. [Google Scholar] [CrossRef]
- Hansen, J.; Jørgensen, J.-E.; Stougaard, J.; Marcker, K.A. Hairy roots—A short cut to transgenic root nodules. Plant Cell. Rep. 1989, 8, 12–15. [Google Scholar] [CrossRef]
- Petit, A.; Stougaard, J.; Kühle, A.; Marcker, K.A.; Tempé, J. Transformation and regeneration of the legume Lotus corniculatus: A system for molecular studies of symbiotic nitrogen fixation. Mol. Gen. Genet. 1987, 207, 245–250. [Google Scholar] [CrossRef]
- Stougaard, J.; Abildsten, D.; Marcker, K.A. The Agrobacterium rhizogenes pRi TL-DNA segment as a gene vector system for transformation of plants. Mol. Gen. Genet. 1987, 207, 251–255. [Google Scholar] [CrossRef]
- Bolivar, F.; Rodriguez, R.L.; Greene, P.J.; Betlach, M.C.; Heyneker, H.L.; Boyer, H.W.; Crosa, J.H.; Falkow, S. Construction and characterization of new cloning vehicle. II. A multipurpose cloning system. Gene 1977, 2, 95–113. [Google Scholar] [CrossRef]
- Scitable. Available online: https://www.nature.com/scitable/popular-students-page/149/ (accessed on 15 December 2021).
- Opabode, J.T. Agrobacterium-mediated transformation of plants: Emerging factors that influence efficiency. Biotechnol. Mol. Biol. Rev. 2006, 1, 12–20. [Google Scholar] [CrossRef]
- Vergauwe, A.; Van Geldre, E.; Inzé, D.; Van Montagu, M.; Van den Eeckhout, E. The use of amoxicillin and ticarcillin in combination with a β-lactamase inhibitor as decontaminating agents in the Agrobacterium tumefaciens-mediated transformation of Artemisia annua L. J. Biotechnol. 1996, 52, 89–95. [Google Scholar] [CrossRef]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Nielsen, A.M.; Stougaard, J. Agrobacterium rhizogenes pRi TL-DNA integration system: A gene vector for Lotus japonicus transformation. In Lotus Japonicus Handbook, 1st ed.; Márquez, A.J., Ed.; Springer Science+Business Media LLC: Dordrecht, The Netherlands, 2005; pp. 285–287. [Google Scholar] [CrossRef]
- Díaz, C.L.; Melchers, L.S.; Hooykaas, P.J.J.; Lugtenberg, B.J.J.; Kijne, J.W. Root lectin as a determinant of host–plant specificity in the Rhizobium–legume symbiosis. Nature 1989, 338, 579–581. [Google Scholar] [CrossRef]
- Offringa, I.A.; Melchers, L.S.; Regensburg-Tuink, A.J.G.; Costantino, P.; Schilperoort, R.A.; Hooykaas, P.J.J. Complementation of Agrobacterium tumefaciens tumor-inducing aux mutants by genes from the TR-region of the Ri plasmid of Agrobacterium rhizogenes. Proc. Natl. Acad. Sci. USA 1986, 83, 6935–6939. [Google Scholar] [CrossRef]
- Díaz, C.L.; Grønlund, M.; Schlaman, H.R.M.; Spaink, H.P. Induction of hairy roots for symbiotic gene expression studies. In Lotus Japonicus Handbook, 1st ed.; Márquez, A.J., Ed.; Springer Science+Business Media LLC: Dordrecht, The Netherlands, 2005; pp. 261–277. [Google Scholar] [CrossRef]
- Nonaka, S.; Someya, T.; Kadota, Y.; Nakamura, K.; Ezura, H. Super-Agrobacterium ver. 4: Improving the transformation frequencies and genetic engineering possibilities for crop plants. Front. Plant Sci. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Christey, M.C. Use of Ri-mediated transformation for production of transgenic plants. In Vitro Cell. Dev. Biol. Plant 2001, 37, 687–700. [Google Scholar] [CrossRef]
- Amack, S.C.; Antunes, M.S. CaMV35S promoter–a plant biology and biotechnology workhorse in the era of synthetic biology. Curr. Plant Biol. 2020, 24, 1–9. [Google Scholar] [CrossRef]
- Sheen, J. Metabolic repression of transcription in higher plants. Plant Cell 1990, 2, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; McHale, L.K.; Finer, J.J. Isolation and characterization of “GmScream” promoters that regulate highly expressing soybean (Glycine max Merr.) genes. Plant Sci. 2015, 241, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Decaestecker, W.; Andrade Buono, R.; Pfeiffer, M.L.; Vangheluwe, N.; Jourquin, J.; Karimi, M.; Van Isterdael, G.; Beeckman, T.; Nowack, M.K.; Jacobs, T.B.B. CRISPR-TSKO: A technique for efficient mutagenesis in specific cell types, tissues, or organs in Arabidopsis. Plant Cell 2019, 31, 2868–2887. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ye, L.; Ursache, R.; Mähönen, A.P. An inducible genome editing system for plants. Nat. Plants 2020, 6, 766–772. [Google Scholar] [CrossRef] [PubMed]
- Omary, M.; Gil-Yarom, N.; Yahav, C.; Steiner, E.; Efroni, I. A conserved superlocus regulates above- and belowground root initiation. bioRxiv 2020. [Google Scholar] [CrossRef]
- De Rybel, B.; Vassileva, V.; Parizot, B.; Demeulenaere, M.; Grunewald, W.; Audenaert, D.; Van Campenhout, J.; Overvoorde, P.; Jansen, L.; Vanneste, S.; et al. A novel Aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol. 2010, 20, 1697–1706. [Google Scholar] [CrossRef]
- Vangheluwe, N.; Beeckman, T. Lateral root initiation and the analysis of gene function using genome editing with CRISPR in Arabidopsis. Genes 2021, 12, 884. [Google Scholar] [CrossRef]
- Ilina, E.L.; Kiryushkin, A.S.; Semenova, V.A.; Demchenko, N.P.; Pawlowski, K.; Demchenko, K.N. Lateral root initiation and formation within the parental root meristem of Cucurbita pepo: Is auxin a key player? Ann. Bot. 2018, 122, 873–888. [Google Scholar] [CrossRef]
- Kiryushkin, A.S.; Ilina, E.L.; Puchkova, V.A.; Guseva, E.D.; Pawlowski, K.; Demchenko, K.N. Lateral root initiation in the parental root meristem of cucurbits: Old players in a new position. Front. Plant Sci. 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Aoki, H.; Teramura, H.; Schepetilnikov, M.; Ryabova, L.A.; Kusano, H.; Shimada, H. Enhanced translation of the downstream ORF attributed to a long 5’ untranslated region in the OsMac1 gene family members, OsMac2 and OsMac3. Plant Biotechnol. 2014, 31, 221–228. [Google Scholar] [CrossRef][Green Version]
- Onodera, H.; Shingu, S.; Ohnuma, M.; Horie, T.; Kihira, M.; Kusano, H.; Teramura, H.; Shimada, H. Establishment of a conditional TALEN system using the translational enhancer dMac3 and an inducible promoter activated by glucocorticoid treatment to increase the frequency of targeted mutagenesis in plants. PLoS ONE 2018, 13, 1–13. [Google Scholar] [CrossRef]
- Kusano, H.; Ohnuma, M.; Mutsuro-Aoki, H.; Asahi, T.; Ichinosawa, D.; Onodera, H.; Asano, K.; Noda, T.; Horie, T.; Fukumoto, K.; et al. Establishment of a modified CRISPR/Cas9 system with increased mutagenesis frequency using the translational enhancer dMac3 and multiple guide RNAs in potato. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef]
- Xing, H.-L.; Dong, L.; Wang, Z.-P.; Zhang, H.-Y.; Han, C.-Y.; Liu, B.; Wang, X.-C.; Chen, Q.-J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Huang, T.-K.; Puchta, H. Novel CRISPR/Cas applications in plants: From prime editing to chromosome engineering. Transgenic Res. 2021, 30, 529–549. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.M.; Zhang, Y.; Yuan, G.; De, K.; Chen, J.-G.; Muchero, W.; Tuskan, G.A.; Qi, Y.; Yang, X. Construct design for CRISPR/Cas-based genome editing in plants. Trends Plant Sci. 2021, 26, 1133–1152. [Google Scholar] [CrossRef] [PubMed]
- Molla, K.A.; Sretenovic, S.; Bansal, K.C.; Qi, Y. Precise plant genome editing using base editors and prime editors. Nat. Plants 2021, 7, 1166–1187. [Google Scholar] [CrossRef]
- Grützner, R.; Martin, P.; Horn, C.; Mortensen, S.; Cram, E.J.; Lee-Parsons, C.W.T.; Stuttmann, J.; Marillonnet, S. High-efficiency genome editing in plants mediated by a Cas9 gene containing multiple introns. Plant Commun. 2021, 2, 1–15. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Karlson, C.K.S.; Mohd-Noor, S.N.; Nolte, N.; Tan, B.C. CRISPR/dCas9-based systems: Mechanisms and applications in plant sciences. Plants 2021, 10, 2055. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.K.; Haber, J.E. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996, 16, 2164–2173. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Marzec, M.; Brąszewska-Zalewska, A.; Hensel, G. Prime editing: A new way for genome editing. Trends Cell Biol. 2020, 30, 257–259. [Google Scholar] [CrossRef]
- Marzec, M.; Hensel, G. Prime editing: Game changer for modifying plant genomes. Trends Plant Sci. 2020, 25, 722–724. [Google Scholar] [CrossRef]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Nishida, K.; Arazoe, T.; Yachie, N.; Banno, S.; Kakimoto, M.; Tabata, M.; Mochizuki, M.; Miyabe, A.; Araki, M.; Hara, K.Y.; et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 2016, 353, 1–10. [Google Scholar] [CrossRef]
- Kim, Y.B.; Komor, A.C.; Levy, J.M.; Packer, M.S.; Zhao, K.T.; Liu, D.R. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 2017, 35, 371–376. [Google Scholar] [CrossRef]
- Komor, A.C.; Zhao, K.T.; Packer, M.S.; Gaudelli, N.M.; Waterbury, A.L.; Koblan, L.W.; Kim, Y.B.; Badran, A.H.; Liu, D.R. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci. Adv. 2017, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Rees, H.A.; Komor, A.C.; Yeh, W.-H.; Caetano-Lopes, J.; Warman, M.; Edge, A.S.B.; Liu, D.R. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat. Commun. 2017, 8, 1–10. [Google Scholar] [CrossRef]
- Koblan, L.W.; Doman, J.L.; Wilson, C.; Levy, J.M.; Tay, T.; Newby, G.A.; Maianti, J.P.; Raguram, A.; Liu, D.R. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 2018, 36, 843–846. [Google Scholar] [CrossRef]
- Richter, M.F.; Zhao, K.T.; Eton, E.; Lapinaite, A.; Newby, G.A.; Thuronyi, B.W.; Wilson, C.; Koblan, L.W.; Zeng, J.; Bauer, D.E.; et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 2020, 38, 883–891. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Lam, D.K.; Rees, H.A.; Solá-Esteves, N.M.; Barrera, L.A.; Born, D.A.; Edwards, A.; Gehrke, J.M.; Lee, S.-J.; Liquori, A.J.; et al. Directed evolution of adenine base editors with increased activity and therapeutic application. Nat. Biotechnol. 2020, 38, 892–900. [Google Scholar] [CrossRef]
- Grünewald, J.; Zhou, R.; Lareau, C.A.; Garcia, S.P.; Iyer, S.; Miller, B.R.; Langner, L.M.; Hsu, J.Y.; Aryee, M.J.; Joung, J.K. A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing. Nat. Biotechnol. 2020, 38, 861–864. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhu, B.; Chen, L.; Xie, L.; Yu, W.; Wang, Y.; Li, L.; Yin, S.; Yang, L.; Hu, H.; et al. Dual base editor catalyzes both cytosine and adenine base conversions in human cells. Nat. Biotechnol. 2020, 38, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Sakata, R.C.; Ishiguro, S.; Mori, H.; Tanaka, M.; Tatsuno, K.; Ueda, H.; Yamamoto, S.; Seki, M.; Masuyama, N.; Nishida, K.; et al. Base editors for simultaneous introduction of C-to-T and A-to-G mutations. Nat. Biotechnol. 2020, 38, 865–869. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Huang, X.; Wang, X.; Gou, S.; Liang, Y.; Chen, F.; Li, N.; Ouyang, Z.; Zhang, Q.; Ge, W.; et al. ACBE, a new base editor for simultaneous C-to-T and A-to-G substitutions in mammalian systems. BMC Biol. 2020, 18, 1–14. [Google Scholar] [CrossRef]
- Li, C.; Zhang, R.; Meng, X.; Chen, S.; Zong, Y.; Lu, C.; Qiu, J.-L.; Chen, Y.-H.; Li, J.; Gao, C. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat. Biotechnol. 2020, 38, 875–882. [Google Scholar] [CrossRef]
- Xu, R.; Kong, F.; Qin, R.; Li, J.; Liu, X.; Wei, P. Development of an efficient plant dual cytosine and adenine editor. J. Integr. Plant Biol. 2021, 63, 1600–1605. [Google Scholar] [CrossRef]
- Bharat, S.S.; Li, S.; Li, J.; Yan, L.; Xia, L. Base editing in plants: Current status and challenges. Crop J. 2020, 8, 384–395. [Google Scholar] [CrossRef]
- Azameti, M.K.; Dauda, W.P. Base editing in plants: Applications, challenges, and future prospects. Front. Plant Sci. 2021, 12, 1–13. [Google Scholar] [CrossRef]
- Pant, S.R.; McNeece, B.T.; Sharma, K.; Nirula, P.M.; Jiang, J.; Harris, J.L.; Lawrence, G.W.; Klink, V.P. A plant transformation system designed for high throughput genomics in Gossypium hirsutum to study root–organism interactions. J. Plant Interact. 2015, 10, 11–20. [Google Scholar] [CrossRef]
- Sindarovska, Y.R.; Gerasymenko, I.M.; Sheludko, Y.V.; Komarnytskyy, I.K.; Bannikova, M.A.; Kuchuk, N.V. Transgenic plants regenerated from hairy roots of Nicotiana benthamiana: A promising host for transient expression of foreign proteins. Tsitologiya I Genet. 2005, 39, 9–14. [Google Scholar]
- Horn, P.; Santala, J.; Nielsen, S.L.; Hühns, M.; Broer, I.; Valkonen, J.P.T. Composite potato plants with transgenic roots on non-transgenic shoots: A model system for studying gene silencing in roots. Plant Cell Rep. 2014, 33, 1977–1992. [Google Scholar] [CrossRef]
- Menze, A.; Möllers, C. Transformation of Different Brassica napus Cultivars with Three Different Strains of Agrobacterium rhizogenes. In Proceedings of the 10th International Rapeseed Congress, Canberra, Australia, 26–29 September 1999. [Google Scholar]
- Kajikawa, M.; Morikawa, K.; Abe, Y.; Yokota, A.; Akashi, K. Establishment of a transgenic hairy root system in wild and domesticated watermelon (Citrullus lanatus) for studying root vigor under drought. Plant Cell Rep. 2010, 29, 771–778. [Google Scholar] [CrossRef] [PubMed]
- Moradpour, M.; Abdulah, S.N.A. CRISPR/dCas9 platforms in plants: Strategies and applications beyond genome editing. Plant Biotechnol. J. 2020, 18, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Wang, Y.; Li, C.; Zhang, R.; Chen, K.; Ran, Y.; Qiu, J.-L.; Wang, D.; Gao, C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 438–440. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Kuang, Y.; Ren, B.; Wang, J.; Zhang, D.; Lin, H.; Yang, B.; Zhou, X.; Zhou, H. Highly efficient A·T to G·C base editing by Cas9n-guided tRNA adenosine deaminase in rice. Mol. Plant 2018, 11, 631–634. [Google Scholar] [CrossRef]
- Triezenberg, S.J.; Kingsbury, R.C.; McKnight, S.L. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988, 2, 718–729. [Google Scholar] [CrossRef] [PubMed]
- Beerli, R.R.; Segal, D.J.; Dreier, B.; Barbas, C.F. Toward controlling gene expression at will: Specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl. Acad. Sci. USA 1998, 95, 14628–14633. [Google Scholar] [CrossRef]
- La Russa, M.F.; Qi, L.S. The new state of the art: Cas9 for gene activation and repression. Mol. Cell. Biol. 2015, 35, 3800–3809. [Google Scholar] [CrossRef] [PubMed]
- Hiratsu, K.; Ohta, M.; Matsui, K.; Ohme-Takagi, M. The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett. 2002, 514, 351–354. [Google Scholar] [CrossRef]
- Hiratsu, K.; Mitsuda, N.; Matsui, K.; Ohme-Takagi, M. Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochem. Biophys. Res. Commun. 2004, 321, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Piatek, A.; Ali, Z.; Baazim, H.; Li, L.; Abulfaraj, A.; Al-Shareef, S.; Aouida, M.; Mahfouz, M.M. RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol. J. 2015, 13, 578–589. [Google Scholar] [CrossRef]
- Lowder, L.G.; Zhang, D.; Baltes, N.J.; Paul, J.W.; Tang, X.; Zheng, X.; Voytas, D.F.; Hsieh, T.-F.; Zhang, Y.; Qi, Y. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 2015, 169, 971–985. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, D.; Xiong, X.; Yan, B.; Xie, W.; Sheen, J.; Li, J.-F. A potent Cas9-derived gene activator for plant and mammalian cells. Nat. Plants 2017, 3, 930–936. [Google Scholar] [CrossRef]
- Lowder, L.G.; Paul, J.W.; Qi, Y. Multiplexed transcriptional activation or repression in plants using CRISPR-dCas9-based systems. In Plant Gene Regulatory Networks: Methods and Protocols, 1st ed.; Kaufmann, K., Mueller-Roeber, B., Eds.; Springer Science+Business Media LLC: New York, NY, USA, 2017; pp. 167–184. [Google Scholar] [CrossRef]
- Lowder, L.G.; Zhou, J.; Zhang, Y.; Malzahn, A.; Zhong, Z.; Hsieh, T.-F.; Voytas, D.F.; Zhang, Y.; Qi, Y. Robust transcriptional activation in plants using multiplexed CRISPR-Act2.0 and mTALE-Act systems. Mol. Plant 2018, 11, 245–256. [Google Scholar] [CrossRef]
- Lee, J.E.; Neumann, M.; Duro, D.I.; Schmid, M. CRISPR-based tools for targeted transcriptional and epigenetic regulation in plants. PLoS ONE 2019, 14, 1–20. [Google Scholar] [CrossRef]
- Li, J.; Blue, R.; Zeitler, B.; Strange, T.L.; Pearl, J.R.; Huizinga, D.H.; Evans, S.; Gregory, P.D.; Urnov, F.D.; Petolino, J.F. Activation domains for controlling plant gene expression using designed transcription factors. Plant Biotechnol. J. 2013, 11, 671–680. [Google Scholar] [CrossRef]
- Ikeda, M.; Ohme-Takagi, M. A novel group of transcriptional repressors in Arabidopsis. Plant Cell. Physiol. 2009, 50, 970–975. [Google Scholar] [CrossRef]
- Selma, S.; Orzáez, D. Perspectives for epigenetic editing in crops. Transgenic Res. 2021, 30, 381–400. [Google Scholar] [CrossRef]
- Dreissig, S.; Schiml, S.; Schindele, P.; Weiss, O.; Rutten, T.; Schubert, V.; Gladilin, E.; Mette, M.F.; Puchta, H.; Houben, A. Live-cell CRISPR imaging in plants reveals dynamic telomere movements. Plant J. 2017, 91, 565–573. [Google Scholar] [CrossRef]
- Fujimoto, S.; Matsunaga, S. Visualization of chromatin loci with transiently expressed CRISPR/Cas9 in plants. Cytologia 2017, 82, 559–562. [Google Scholar] [CrossRef]
- Khosravi, S.; Schindele, P.; Gladilin, E.; Dunemann, F.; Rutten, T.; Puchta, H.; Houben, A. Application of aptamers improves CRISPR-based live imaging of plant telomeres. Front. Plant Sci. 2020, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Konermann, S.; Brigham, M.D.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.D.; Habib, N.; Gootenberg, J.S.; Nishimasu, H.; et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2015, 517, 583–588. [Google Scholar] [CrossRef]
- Tanenbaum, M.E.; Gilbert, L.A.; Qi, L.S.; Weissman, J.S.; Vale, R.D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 2014, 159, 635–646. [Google Scholar] [CrossRef]
- Wang, L.; Sun, Z.; Su, C.; Wang, Y.; Yan, Q.; Chen, J.; Ott, T.; Li, X. A GmNINa-miR172c-NNC1 regulatory network coordinates the nodulation and autoregulation of nodulation pathways in soybean. Mol. Plant 2019, 12, 1211–1226. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Xie, C.; Ruan, Q.; Zhang, X.; Wu, C.; Han, B.; Qian, J.; Zhou, W.; Nützmann, H.-W.; Kai, G. The transcription factor OpWRKY2 positively regulates the biosynthesis of the anticancer drug camptothecin in Ophiorrhiza pumila. Hortic. Res. 2021, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Limpens, E.; Ramos, J.; Franken, C.; Raz, V.; Compaan, B.; Franssen, H.; Bisseling, T.; Geurts, R. RNA interference in Agrobacterium rhizogenes–transformed roots of Arabidopsis and Medicago truncatula. J. Exp. Bot. 2004, 55, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Waibel, F.; Filipowicz, W. U6 snRNA genes of Arabidopsis are transcribed by RNA polymerase III but contain the same two upstream promoter elements as RNA polymerase ll-transcribed U-snRNA genes. Nucleic Acids Res. 1990, 18, 3451–3458. [Google Scholar] [CrossRef] [PubMed]
- Marshallsay, C.; Kiss, T.; Filipowicz, W. Amplification of plant U3 and U6 snRNA gene sequences using primers specific for an upstream promoter element and conserved intragenic regions. Nucleic Acids Res. 1990, 18, 3459–3466. [Google Scholar] [CrossRef]
- Xie, K.; Minkenberg, B.; Yang, Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA 2015, 112, 3570–3575. [Google Scholar] [CrossRef]
- Čermák, T.; Curtin, S.J.; Gil-Humanes, J.; Čegan, R.; Kono, T.J.Y.; Konečná, E.; Belanto, J.J.; Starker, C.G.; Mathre, J.W.; Greenstein, R.L.; et al. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 2017, 29, 1196–1217. [Google Scholar] [CrossRef]
- Marshallsay, C.; Connelly, S.; Filipowicz, W. Characterization of the U3 and U6 snRNA genes from wheat: U3 snRNA genes in monocot plants are transcribed by RNA polymerase III. Plant Mol. Biol. 1992, 19, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Qu, F.; Zhai, W.; Chen, H.; Zhu, L.-H.; Morris, T.J. Cloning, characterization and transient expression of the gene encoding a rice U3 small nuclear RNA. Gene 1996, 172, 217–220. [Google Scholar] [CrossRef]
- plaBiPD. Available online: https://www.plabipd.de/plant_genomes_pa.ep (accessed on 15 December 2021).
- Hu, B.; Li, D.; Liu, X.; Qi, J.; Gao, D.; Zhao, S.; Huang, S.; Sun, J.; Yang, L. Engineering non-transgenic gynoecious cucumber using an improved transformation protocol and optimized CRISPR/Cas9 system. Mol. Plant 2017, 10, 1575–1578. [Google Scholar] [CrossRef]
- Long, L.; Guo, D.-D.; Gao, W.; Yang, W.-W.; Hou, L.-P.; Ma, X.-N.; Miao, Y.-C.; Botella, J.R.; Song, C.-P. Optimization of CRISPR/Cas9 genome editing in cotton by improved sgRNA expression. Plant Methods 2018, 14, 1–9. [Google Scholar] [CrossRef]
- Ren, C.; Liu, Y.; Guo, Y.; Duan, W.; Fan, P.; Li, S.; Liang, Z. Optimizing the CRISPR/Cas9 system for genome editing in grape by using grape promoters. Hortic. Res. 2021, 8, 1–12. [Google Scholar] [CrossRef]
- Qi, X.; Dong, L.; Liu, C.; Mao, L.; Liu, F.; Zhang, X.; Cheng, B.; Xie, C. Systematic identification of endogenous RNA polymerase III promoters for efficient RNA guide-based genome editing technologies in maize. Crop. J. 2018, 6, 314–320. [Google Scholar] [CrossRef]
- Alja, S.; Catalin, V.; Jennifer, B.; Karin, L.; John, V.; Markus, P.; Christian, S.H. Broad spectrum developmental role of Brachypodium AUX1. New Phytol. 2018, 219, 1216–1223. [Google Scholar] [CrossRef]
- Charleson, P.; Lorelle, P.; Barbara, G.; Cathie, R.; Mathias, S.; Glenn, T. Genome editing with CRISPR/Cas9 in Pinus radiata (D. Don). BMC Plant Biol. 2021, 21, 1–9. [Google Scholar] [CrossRef]
- Sugano, S.S.; Shirakawa, M.; Takagi, J.; Matsuda, Y.; Shimada, T.; Hara-Nishimura, I.; Kohchi, T. CRISPR/Cas9-mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol. 2014, 55, 475–481. [Google Scholar] [CrossRef]
- Collonnier, C.; Epert, A.; Mara, K.; Maclot, F.; Guyon-Debast, A.; Charlot, F.; White, C.; Schaefer, D.G.; Nogué, F. CRISPR-Cas9-mediated efficient directed mutagenesis and RAD51-dependent and RAD51-independent gene targeting in the moss Physcomitrella patens. Plant Biotechnol. J. 2017, 15, 122–131. [Google Scholar] [CrossRef]
- Liang, G.; Zhang, H.; Lou, D.; Yu, D. Selection of highly efficient sgRNAs for CRISPR/Cas9-based plant genome editing. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Shan, S.; Soltis, P.S.; Soltis, D.E.; Yang, B. Considerations in adapting CRISPR/Cas9 in nongenetic model plant systems. Appl. Plant Sci. 2020, 8, 1–17. [Google Scholar] [CrossRef]
- Bradford, J.; Perrin, D. A benchmark of computational CRISPR-Cas9 guide design methods. PLOS Comput. Biol. 2019, 15, 1–18. [Google Scholar] [CrossRef]
- Sledzinski, P.; Nowaczyk, M.; Olejniczak, M. Computational tools and resources supporting CRISPR-Cas experiments. Cells 2020, 9, 1288. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zhang, Y.; Zhang, T. Computational approaches for effective CRISPR guide RNA design and evaluation. Comput. Struct. Biotechnol. J. 2020, 18, 35–44. [Google Scholar] [CrossRef]
- Torres-Perez, R.; Garcia-Martin, J.A.; Montoliu, L.; Oliveros, J.C.; Pazos, F. WeReview: CRISPR tools—Live repository of computational tools for assisting CRISPR/Cas experiments. Bioengineering 2019, 6, 63. [Google Scholar] [CrossRef]
- Haeussler, M.; Schönig, K.; Eckert, H.; Eschstruth, A.; Mianné, J.; Renaud, J.-B.; Schneider-Maunoury, S.; Shkumatava, A.; Teboul, L.; Kent, J.; et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Minkenberg, B.; Zhang, J.; Xie, K.; Yang, Y. CRISPR-PLANT v2: An online resource for highly specific guide RNA spacers based on improved off-target analysis. Plant Biotechnol. J. 2019, 17, 5–8. [Google Scholar] [CrossRef]
- Yuan, M.; Zhu, J.; Gong, L.; He, L.; Lee, C.; Han, S.; Chen, C.; He, G. Mutagenesis of FAD2 genes in peanut with CRISPR/Cas9 based gene editing. BMC Biotechnol. 2019, 19, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, L.; Tan, Q.; Fan, Q.; Zhu, H.; Hong, Z.; Zhang, Z.; Duanmu, D. Efficient inactivation of symbiotic nitrogen fixation related genes in Lotus japonicus using CRISPR/Cas9. Front. Plant Sci. 2016, 7, 1–22. [Google Scholar] [CrossRef]
- Nadakuduti, S.S.; Starker, C.G.; Ko, D.K.; Jayakody, T.B.; Buell, C.R.; Voytas, D.F.; Douches, D.S. Evaluation of methods to assess in vivo activity of engineered genome-editing nucleases in protoplasts. Front. Plant Sci. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Gerasimova, S.V.; Hertig, C.; Korotkova, A.M.; Kolosovskaya, E.V.; Otto, I.; Hiekel, S.; Kochetov, A.V.; Khlestkina, E.K.; Kumlehn, J. Conversion of hulled into naked barley by Cas endonuclease-mediated knockout of the NUD gene. BMC Plant Biol. 2020, 20, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Budhagatapalli, N.; Schedel, S.; Gurushidze, M.; Pencs, S.; Hiekel, S.; Rutten, T.; Kusch, S.; Morbitzer, R.; Lahaye, T.; Panstruga, R.; et al. A simple test for the cleavage activity of customized endonucleases in plants. Plant Methods 2016, 12, 1–10. [Google Scholar] [CrossRef]
- Yuan, G.; Hassan, M.M.; Yao, T.; Lu, H.; Vergara, M.M.; Labbé, J.L.; Muchero, W.; Pan, C.; Chen, J.-G.; Tuskan, G.A.; et al. Plant-based biosensors for detecting CRISPR-mediated genome engineering. ACS Synth. Biol. 2021, 10, 3600–3603. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Estrella, L.; De Block, M.; Messens, E.; Hernalsteens, J.-P.; Van Montagu, M.; Schell, J. Chimeric genes as dominant selectable markers in plant cells. EMBO J. 1983, 2, 987–995. [Google Scholar] [CrossRef]
- Sundar, I.K.; Sakthivel, N. Advances in selectable marker genes for plant transformation. J. Plant Physiol. 2008, 165, 1698–1716. [Google Scholar] [CrossRef]
- Rosellini, D. Selectable markers and reporter genes: A well furnished toolbox for plant science and genetic engineering. Crit. Rev. Plant Sci. 2012, 31, 401–453. [Google Scholar] [CrossRef]
- Breyer, D.; Kopertekh, L.; Reheul, D. Alternatives to antibiotic resistance marker genes for in vitro selection of genetically modified plants–scientific developments, current use, operational access and biosafety considerations. Crit. Rev. Plant Sci. 2014, 33, 286–330. [Google Scholar] [CrossRef]
- Lin, Y.; Huang, J. Characterization of an algal phosphomannose isomerase gene and its application as a selectable marker for genetic manipulation of tomato. Plant Divers. 2021, 43, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Gu, J.; Wang, X.; Zhang, S. A novel non-antibiotic selectable marker GASA6 for plant transformation. Plant Cell Tiss. Org. Cult. 2021, 148, 1–12. [Google Scholar] [CrossRef]
- Pena, L.; Cervera, M.; Juárez, J.; Ortega, C.; Pina, J.; Durán-Vila, N.; Navarro, L. High efficiency Agrobacterium-mediated transformation and regeneration of citrus. Plant Sci. 1995, 104, 183–191. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Burgess, S.M.; Hirsh, D. β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc. Natl. Acad. Sci. USA 1986, 83, 8447–8451. [Google Scholar] [CrossRef]
- Novel, G.; Novel, M. Mutants d’Escherichia coli K 12 affectés pour leur croissance sur méthyl-β-D-glucuronide: Localisation du gène de structure de la β-D-glucuronidase (uid A). Mol. Gen. Genet. 1973, 120, 319–335. [Google Scholar] [CrossRef]
- Cervera, M. Histochemical and fluorometric assays for uidA (GUS) gene detection. In Transgenic Plants: Methods and Protocols, 1st ed.; Peña, L., Ed.; Humana Press: Totowa, NJ, USA, 2004; pp. 203–213. [Google Scholar] [CrossRef]
- Jefferson, R.A. Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 1987, 5, 387–405. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Klein, T.M.; Gradziel, T.; Fromm, M.E.; Sanford, J.C. Factors influencing gene delivery into Zea mays cells by high–velocity microprojectiles. Bio/Technology 1988, 6, 559–563. [Google Scholar] [CrossRef]
- Berthomieu, P.; Jouanin, L. Transformation of rapid cycling cabbage (Brassica oleracea var. capitata) with Agrobacterium rhizogenes. Plant Cell Rep. 1992, 11, 334–338. [Google Scholar] [CrossRef]
- Graham, J.; McNicol, R.J.; Kumar, A. Use of the GUS gene as a selectable marker for Agrobacterium-mediated transformation of Rubus. Plant Cell Tiss. Org. Cult. 1990, 20, 35–39. [Google Scholar] [CrossRef]
- Hosoki, T.; Kigo, T. Transformation of Brussels sprouts (Brassica oleracea var. gemmifera Zenk.) by Agrobacterium rhizogenes harboring a reporter β-glucuronidase gene. J. Jpn. Soc. Hortic. Sci. 1994, 63, 589–592. [Google Scholar] [CrossRef]
- Lee, N.-G.; Stein, B.; Suzuki, H.; Verma, D.P.S. Expression of antisense nodulin-35 RNA in Vigna aconitifolia transgenic root nodules retards peroxisome development and affects nitrogen availability to the plant. Plant J. 1993, 3, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Boisson-Dernier, A.; Chabaud, M.; Garcia, F.; Bécard, G.; Rosenberg, C.; Barker, D.G. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant-Microbe Interact. 2001, 14, 695–700. [Google Scholar] [CrossRef]
- Puddephat, I.J.; Robinson, H.T.; Fenning, T.M.; Barbara, D.J.; Morton, A.; Pink, D.A.C. Recovery of phenotypically normal transgenic plants of Brassica oleracea upon Agrobacterium rhizogenes-mediated co-transformation and selection of transformed hairy roots by GUS assay. Mol. Breed. 2001, 7, 229–242. [Google Scholar] [CrossRef]
- Datla, R.S.S.; Hammerlindl, J.K.; Pelcher, L.E.; Crosby, W.L.; Selvaraj, G. A bifunctional fusion between β-glucuronidase and neomycin phosphotransferase: A broad-spectrum marker enzyme for plants. Gene 1991, 101, 239–246. [Google Scholar] [CrossRef]
- Kamo, K.; McElroy, D.; Chamberiain, D. Transforming embryogenic cell lines of Gladiolus with either a bar-uidA fusion gene or cobombardment. In Vitro Cell. Dev. Biol. Plant 2000, 36, 182–187. [Google Scholar] [CrossRef]
- Ludwig, S.R.; Bowen, B.; Beach, L.; Wessler, S.R. A regulatory gene as a novel visible marker for maize transformation. Science 1990, 247, 449–450. [Google Scholar] [CrossRef]
- Dhir, S.K.; Pajeau, M.E.; Frommn, M.E.; Fry, J.E. Anthocyanin genes as visual markers for wheat transformation. In Improvement of Cereal Quality by Genetic Engineering, 1st ed.; Henry, R.J., Ronalds, J.A., Eds.; Plenum Press: New York, NY, USA, 1994; pp. 71–75. [Google Scholar] [CrossRef]
- Chawla, H.S.; Cass, L.A.; Simmonds, J.A. Expression of anthocyanin pigmentation in wheat tissues transformed with anthocyanin regulatory genes. Curr. Sci. 1999, 76, 1365–1370. [Google Scholar]
- Lloyd, A.M.; Walbot, V.; Davis, R.W. Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 1992, 258, 1773–1775. [Google Scholar] [CrossRef]
- Quattrocchio, F.; Wing, J.F.; Leppen, H.; Mol, J.; Koes, R.E. Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell 1993, 5, 1497–1512. [Google Scholar] [CrossRef]
- Damiani, F.; Paolocci, F.; Consonni, G.; Crea, F.; Tonelli, C.; Arcioni, S. A maize anthocyanin transactivator induces pigmentation in hairy roots of dicotyledonous species. Plant Cell. Rep. 1998, 17, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Khusnutdinov, E.; Sukhareva, A.; Panfilova, M.; Mikhaylova, E. Anthocyanin biosynthesis genes as model genes for genome editing in plants. Int. J. Mol. Sci. 2021, 22, 8752. [Google Scholar] [CrossRef] [PubMed]
- Paz-Ares, J.; Wienand, U.; Peterson, P.A.; Saedler, H. Molecular cloning of the c locus of Zea mays: A locus regulating the anthocyanin pathway. EMBO J. 1986, 5, 829–833. [Google Scholar] [CrossRef]
- Chandler, V.L.; Radicella, J.P.; Robbins, T.P.; Chen, J.; Turks, D. Two regulatory genes of the maize anthocyanin pathway are homologous: Isolation of B utilizing R genomic sequences. Plant Cell 1989, 1, 1175–1183. [Google Scholar] [CrossRef]
- Ludwig, S.R.; Habera, L.F.; Dellaporta, S.L.; Wessler, S.R. Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc. Natl. Acad. Sci. USA 1989, 86, 7092–7096. [Google Scholar] [CrossRef] [PubMed]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.-L.; Zeng, H.-N.; Shi, M.-Z.; Xie, D.-Y. Development of tobacco callus cultures over expressing Arabidopsis PAP1/MYB75 transcription factor and characterization of anthocyanin biosynthesis. Planta 2008, 229, 37–51. [Google Scholar] [CrossRef]
- Kim, C.Y.; Ahn, Y.O.; Kim, S.H.; Kim, Y.-H.; Lee, H.-S.; Catanach, A.S.; Jacobs, J.M.E.; Conner, A.J.; Kwak, S.-S. The sweet potato IbMYB1 gene as a potential visible marker for sweet potato intragenic vector system. Physiol. Plant. 2010, 139, 229–240. [Google Scholar] [CrossRef]
- Kortstee, A.J.; Khan, S.A.; Helderman, C.; Trindade, L.M.; Wu, Y.; Visser, R.G.F.; Brendolise, C.; Allan, A.; Schouten, H.J.; Jacobsen, E. Anthocyanin production as a potential visual selection marker during plant transformation. Transgenic Res. 2011, 20, 1253–1264. [Google Scholar] [CrossRef]
- Naing, A.H.; Lim, K.B.; Kim, C.K. The usage of snapdragon Delila (Del) gene as a visible selection marker for the antibiotic-free transformation system. J. Plant Biol. 2015, 58, 110–116. [Google Scholar] [CrossRef]
- Kandel, R.; Bergey, D.R.; Dutt, M.; Sitther, V.; Li, Z.T.; Gray, D.J.; Dhekney, S.A. Evaluation of a grapevine-derived reporter gene system for precision breeding of Vitis. Plant Cell. Tiss. Org. Cult. 2016, 124, 599–609. [Google Scholar] [CrossRef]
- Zhang, S.; Kondorosi, É.; Kereszt, A. An anthocyanin marker for direct visualization of plant transformation and its use to study nitrogen-fixing nodule development. J. Plant Res. 2019, 132, 695–703. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhu, M.; Wu, J.; Ouyang, L.; Wang, R.; Sun, H.; Yan, L.; Wang, L.; Xu, M.; Zhan, H.; et al. Repurposing of anthocyanin biosynthesis for plant transformation and genome editing. Front. Plant Sci. 2020, 2, 1–10. [Google Scholar] [CrossRef]
- Huang, T.; Xin, S.; Fang, Y.; Chen, T.; Chang, J.; Ko, N.C.K.; Huang, H.; Hua, Y. Use of a novel R2R3-MYB transcriptional activator of anthocyanin biosynthesis as visual selection marker for rubber tree (Hevea brasiliensis) transformation. Ind. Crops Prod. 2021, 174, 1–11. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, X.; Li, H.; Liu, S.; Jin, L.; Lyu, Y.; Shi, M.; Liu, S.; Yang, X.; Lyu, S. Anthocyanin, a novel and user-friendly reporter for convenient, non-destructive, low cost, directly visual selection of transgenic hairy roots in the study of rhizobia-legume symbiosis. Plant Methods 2020, 16, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.; Chen, K.; Su, Y.; Jiang, S.; Xu, P.; Murray, J.D. A root tip-specific expressing anthocyanin marker for direct identification of transgenic tissues by the naked eye in symbiotic studies. Plants 2021, 10, 605. [Google Scholar] [CrossRef]
- He, Y.; Zhang, T.; Sun, H.; Zhan, H.; Zhao, Y. A reporter for noninvasively monitoring gene expression and plant transformation. Hortic. Res. 2020, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sunnadeniya, R.; Bean, A.; Brown, M.; Akhavan, N.; Hatlestad, G.; Gonzalez, A.; Symonds, V.V.; Lloyd, A. Tyrosine hydroxylation in betalain pigment biosynthesis is performed by cytochrome P450 enzymes in beets (Beta vulgaris). PLoS ONE 2016, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Halpin, C.; Cooke, S.E.; Barakate, A.; Amrani, A.E.; Ryan, M.D. Self-processing 2A-polyproteins–a system for co-ordinate expression of multiple proteins in transgenic plants. Plant J. 1999, 17, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, O.; Wall, J.B.J.; Zheng, M.; Zhou, Y.; Wang, L.; Ruth Vaseghi, H.; Qian, L.; Liu, J. Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef]
- Khosla, A.; Rodriguez-Furlan, C.; Kapoor, S.; Van Norman, J.M.; Nelson, D.C. A series of dual-reporter vectors for ratiometric analysis of protein abundance in plants. Plant Direct 2020, 4, 1–14. [Google Scholar] [CrossRef]
- Grützner, R.; Schubert, R.; Horn, C.; Yang, C.; Vogt, T.; Marillonnet, S. Engineering betalain biosynthesis in tomato for high level betanin production in fruits. Front. Plant Sci. 2021, 12, 1–17. [Google Scholar] [CrossRef]
- Timoneda, A.; Yunusov, T.; Quan, C.; Gavrin, A.; Brockington, S.F.; Schornack, S. MycoRed: Betalain pigments enable in vivo real-time visualisation of arbuscular mycorrhizal colonisation. PLOS Biol. 2021, 19, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; He, Y.; Gan, S. Bidirectionalization of polar promoters in plants. Nat. Biotechnol. 2001, 19, 677–679. [Google Scholar] [CrossRef]
- Dey, N.; Sarkar, S.; Acharya, S.; Maiti, I.B. Synthetic promoters in planta. Planta 2015, 242, 1077–1094. [Google Scholar] [CrossRef]
- Ren, Q.; Zhong, Z.; Wang, Y.; You, Q.; Li, Q.; Yuan, M.; He, Y.; Qi, C.; Tang, X.; Zheng, X.; et al. Bidirectional promoter-based CRISPR/Cas9 systems for plant genome editing. Front. Plant Sci. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Shimomura, K.; Sudo, H.; Saga, H.; Kamada, H. Shikonin production and secretion by hairy root cultures of Lithospermum erythrorhizon. Plant Cell. Rep. 1991, 10, 282–285. [Google Scholar] [CrossRef]
- Brigham, L.A.; Michaels, P.J.; Flores, H.E. Cell-specific production and antimicrobial activity of naphthoquinones in roots of Lithospermum erythrorhizon. Plant Physiol. 1999, 119, 417–428. [Google Scholar] [CrossRef]
- Chaudhury, A.; Pal, M. Induction of shikonin production in hairy root cultures of Arnebia hispidissima via Agrobacterium rhizogenes-mediated genetic transformation. J. Crop Sci. Biotechnol. 2010, 13, 99–106. [Google Scholar] [CrossRef]
- Moyano, E.; Jouhikainen, K.; Tammela, P.; Palazón, J.; Cusidó, R.M.; Piñol, M.T.; Teeri, T.H.; Oksman–Caldentey, K.M. Effect of pmt gene overexpression on tropane alkaloid production in transformed root cultures of Datura metel and Hyoscyamus muticus. J. Exp. Bot. 2003, 54, 203–211. [Google Scholar] [CrossRef]
- Abbasi, B.H.; Tian, C.-L.; Murch, S.J.; Saxena, P.K.; Liu, C.-Z. Light-enhanced caffeic acid derivatives biosynthesis in hairy root cultures of Echinacea purpurea. Plant Cell. Rep. 2007, 26, 1367–1372. [Google Scholar] [CrossRef]
- Thimmaraju, R.; Venkatachalam, L.; Bhagyalakshmi, N. Morphometric and biochemical characterization of red beet (Beta vulgaris L.) hairy roots obtained after single and double transformations. Plant Cell. Rep. 2008, 27, 1039–1052. [Google Scholar] [CrossRef] [PubMed]
- Neelwarne, B. Red beet hairy root cultures. In Red Beet Biotechnology: Food and Pharmaceutical Applications, 1st ed.; Neelwarne, B., Ed.; Springer Science+Business Media LLC: Boston, MA, USA, 2013; pp. 199–249. [Google Scholar] [CrossRef]
- Thwe, A.A.; Kim, J.K.; Li, X.; Bok Kim, Y.; Romij Uddin, M.; Kim, S.J.; Suzuki, T.; Park, N.I.; Park, S.U. Metabolomic analysis and phenylpropanoid biosynthesis in hairy root culture of tartary buckwheat cultivars. PLoS ONE 2013, 8, 1–13. [Google Scholar] [CrossRef]
- Barba-Espín, G.; Chen, S.-T.; Agnolet, S.; Hegelund, J.N.; Stanstrup, J.; Christensen, J.H.; Müller, R.; Lütken, H. Ethephon-induced changes in antioxidants and phenolic compounds in anthocyanin-producing black carrot hairy root cultures. J. Exp. Bot. 2020, 71, 7030–7045. [Google Scholar] [CrossRef] [PubMed]
- Prasher, D.C.; Eckenrode, V.K.; Ward, W.W.; Prendergast, F.G.; Cormier, M.J. Primary structure of the Aequorea victoria green-fluorescent protein. Gene 1992, 111, 229–233. [Google Scholar] [CrossRef]
- Chiu, W.-l.; Niwa, Y.; Zeng, W.; Hirano, T.; Kobayashi, H.; Sheen, J. Engineered GFP as a vital reporter in plants. Curr. Biol. 1996, 6, 325–330. [Google Scholar] [CrossRef]
- Haseloff, J.; Siemering, K.R.; Prasher, D.C.; Hodge, S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 1997, 94, 2122–2127. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.J.; Vierstra, R.D. Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol. Biol. 1998, 36, 521–528. [Google Scholar] [CrossRef]
- Niedz, R.P.; Sussman, M.R.; Satterlee, J.S. Green fluorescent protein: An in vivo reporter of plant gene expression. Plant Cell Rep. 1995, 14, 403–406. [Google Scholar] [CrossRef]
- Kumagai, H.; Kouchi, H. Gene silencing by expression of hairpin RNA in Lotus japonicus roots and root nodules. Mol. Plant-Microbe Interact. 2003, 16, 663–668. [Google Scholar] [CrossRef] [PubMed]
- Matz, M.V.; Fradkov, A.F.; Labas, Y.A.; Savitsky, A.P.; Zaraisky, A.G.; Markelov, M.L.; Lukyanov, S.A. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 1999, 17, 969–973. [Google Scholar] [CrossRef] [PubMed]
- Jach, G.; Binot, E.; Frings, S.; Luxa, K.; Schell, J. Use of red fluorescent protein from Discosoma sp. (dsRED) as a reporter for plant gene expression. Plant J. 2001, 28, 483–491. [Google Scholar] [CrossRef]
- Stuitje, A.R.; Verbree, E.C.; Van Der Linden, K.H.; Mietkiewska, E.M.; Nap, J.-P.; Kneppers, T.J.A. Seed-expressed fluorescent proteins as versatile tools for easy (co)transformation and high-throughput functional genomics in Arabidopsis. Plant Biotechnol. J. 2003, 1, 301–309. [Google Scholar] [CrossRef]
- Lin, M.-H.; Gresshoff, P.M.; Indrasumunar, A.; Ferguson, B.J. pHairyRed: A novel binary vector containing the DsRed2 reporter gene for visual selection of transgenic hairy roots. Mol. Plant 2011, 4, 537–545. [Google Scholar] [CrossRef]
- Markmann, K.; Giczey, G.; Parniske, M. Functional adaptation of a plant receptor- kinase paved the way for the evolution of intracellular root symbioses with bacteria. PLoS Biol. 2008, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.; Maliga, P. Fluorescent antibiotic resistance marker for tracking plastid transformation in higher plants. Nat. Biotechnol. 1999, 17, 910–915. [Google Scholar] [CrossRef]
- Li, Z.; Jayasankar, S.; Gray, D.J. Expression of a bifunctional green fluorescent protein (GFP) fusion marker under the control of three constitutive promoters and enhanced derivatives in transgenic grape (Vitis vinifera). Plant Sci. 2001, 160, 877–887. [Google Scholar] [CrossRef]
- Ochiai-Fukuda, T.; Takahashi-Ando, N.; Ohsato, S.; Igawa, T.; Kadokura, K.; Hamamoto, H.; Nakasako, M.; Kudo, T.; Shibata, T.; Yamaguchi, I.; et al. A fluorescent antibiotic resistance marker for rapid production of transgenic rice plants. J. Biotechnol. 2006, 122, 521–527. [Google Scholar] [CrossRef]
- Dutt, M.; Lee, D.H.; Grosser, J.W. Bifunctional selection–reporter systems for genetic transformation of citrus: Mannose- and kanamycin-based systems. In Vitro Cell. Dev. Biol. Plant 2010, 46, 467–476. [Google Scholar] [CrossRef]
- Quaedvlieg, N.E.M.; Schlaman, H.R.M.; Admiraal, P.C.; Wijting, S.E.; Stougaard, J.; Spaink, H.P. Fusions between green fluorescent protein and β-glucuronidase as sensitive and vital bifunctional reporters in plants. Plant Mol. Biol. 1998, 38, 861–873. [Google Scholar] [CrossRef]
- Gao, X.; Chen, J.; Dai, X.; Zhang, D.; Zhao, Y. An effective strategy for reliably isolating heritable and Cas9-free Arabidopsis mutants generated by CRISPR/Cas9-mediated genome editing. Plant Physiol. 2016, 171, 1794–1800. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Yu, X.; Yang, H.; Gao, Q.; Ji, H.; Wang, Y.; Yan, G.; Peng, Y.; Luo, H.; Liu, K.; et al. Development and validation of an effective CRISPR/Cas9 vector for efficiently isolating positive transformants and transgene-free mutants in a wide range of plant species. Front. Plant Sci. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Aliaga, F.N.; Zhang, C.; Presa, S.; Granell, A.; Alabadi, D.; Sadanandom, A.; Blazquez, M.A.; Minguet, E.G. Identification of transgene-free CRISPR edited plants of rice, tomato and Arabidopsis by monitoring DsRED fluorescence in dry seeds. Front. Plant Sci. 2019, 10, 1–9. [Google Scholar] [CrossRef]
- Okada, A.; Arndell, T.; Borisjuk, N.; Sharma, N.; Watson-Haigh, N.S.; Tucker, E.J.; Baumann, U.; Langridge, P.; Whitford, R. CRISPR/Cas9-mediated knockout of Ms1 enables the rapid generation of male-sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnol. J. 2019, 17, 1905–1913. [Google Scholar] [CrossRef]
- Ursache, R.; Fujita, S.; Tendon, V.D.; Geldner, N. Combined fluorescent seed selection and multiplex CRISPR/Cas9 assembly for fast generation of multiple Arabidopsis mutants. Plant Methods 2021, 17, 1–14. [Google Scholar] [CrossRef]
- Yan, Y.; Zhu, J.; Qi, X.; Cheng, B.; Liu, C.; Xie, C. Establishment of an efficient seed fluorescence reporter-assisted CRISPR/Cas9 gene editing in maize. J. Integr. Plant Biol. 2021, 63, 1671–1680. [Google Scholar] [CrossRef] [PubMed]
- Lambert, T.J. FPbase: A community-editable fluorescent protein database. Nat. Methods 2019, 16, 277–278. [Google Scholar] [CrossRef] [PubMed]
- Nekrasov, V.; Staskawicz, B.; Weigel, D.; Jones, J.D.G.; Kamoun, S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013, 31, 691–693. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, K.; Osakabe, Y.; Toki, S. Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. Proc. Natl. Acad. Sci. USA 2010, 107, 12034–12039. [Google Scholar] [CrossRef]
- Osakabe, Y.; Watanabe, T.; Sugano, S.S.; Ueta, R.; Ishihara, R.; Shinozaki, K.; Osakabe, K. Optimization of CRISPR-Cas9 genome editing to modify abiotic stress responses in plants. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Ueta, R.; Abe, C.; Watanabe, T.; Sugano, S.S.; Ishihara, R.; Ezura, H.; Osakabe, Y.; Osakabe, K. Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef]
- Nishitani, C.; Hirai, N.; Komori, S.; Wada, M.; Okada, K.; Osakabe, K.; Yamamoto, T.; Osakabe, Y. Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Liang, Z.; Ren, C.; Nishitani, C.; Osakabe, K.; Wada, M.; Komori, S.; Malnoy, M.; Velasco, R.; Poli, M.; et al. CRISPR–Cas9-mediated genome editing in apple and grapevine. Nat. Protoc. 2018, 13, 2844–2863. [Google Scholar] [CrossRef]
- Hashimoto, R.; Ueta, R.; Abe, C.; Osakabe, Y.; Osakabe, K. Efficient multiplex genome editing induces precise, and self-ligated type mutations in tomato plants. Front. Plant Sci. 2018, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, F.; Suzuki, T.; Osakabe, Y.; Betsuyaku, S.; Kondo, Y.; Dohmae, N.; Fukuda, H.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 2018, 556, 235–238. [Google Scholar] [CrossRef]
- Petersen, B.L.; Möller, S.R.; Mravec, J.; Jørgensen, B.; Christensen, M.; Liu, Y.; Wandall, H.H.; Bennett, E.P.; Yang, Z. Improved CRISPR/Cas9 gene editing by fluorescence activated cell sorting of green fluorescence protein tagged protoplasts. BMC Biotechnol. 2019, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Hua, L.; Gupta, A.; Tricoli, D.; Edwards, K.J.; Yang, B.; Li, W. Development of an Agrobacterium-delivered CRISPR/Cas9 system for wheat genome editing. Plant Biotechnol. J. 2019, 17, 1623–1635. [Google Scholar] [CrossRef]
- Wang, Q.; Yang, S.; Liu, J.; Terecskei, K.; Ábrahám, E.; Gombár, A.; Domonkos, Á.; Szűcs, A.; Körmöczi, P.; Wang, T.; et al. Host-secreted antimicrobial peptide enforces symbiotic selectivity in Medicago truncatula. Proc. Natl. Acad. Sci. USA 2017, 114, 6854–6859. [Google Scholar] [CrossRef]
- Yang, S.; Wang, Q.; Fedorova, E.; Liu, J.; Qin, Q.; Zheng, Q.; Price, P.A.; Pan, H.; Wang, D.; Griffitts, J.S.; et al. Microsymbiont discrimination mediated by a host-secreted peptide in Medicago truncatula. Proc. Natl. Acad. Sci. USA 2017, 114, 6848–6853. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, J.; Li, H.; Yang, S.; Körmöczi, P.; Kereszt, A.; Zhu, H. Nodule-specific cysteine-rich peptides negatively regulate nitrogen-fixing symbiosis in a strain-specific manner in Medicago truncatula. Mol. Plant-Microbe Interact. 2018, 31, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Yang, S.; Liu, J.; Zhu, H. Rj4, a gene controlling nodulation specificity in soybeans, encodes a thaumatin-like protein but not the one previously reported. Plant Physiol. 2016, 170, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Limpens, E.; Mirabella, R.; Fedorova, E.; Franken, C.; Franssen, H.; Bisseling, T.; Geurts, R. Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc. Natl. Acad. Sci. USA 2005, 102, 10375–10380. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, S.; Harrison, M.J. A set of fluorescent protein-based markers expressed from constitutive and arbuscular mycorrhiza-inducible promoters to label organelles, membranes and cytoskeletal elements in Medicago truncatula. Plant J. 2014, 80, 1151–1163. [Google Scholar] [CrossRef]
- Ilina, E.L.; Logachov, A.A.; Laplaze, L.; Demchenko, N.P.; Pawlowski, K.; Demchenko, K.N. Composite Cucurbita pepo plants with transgenic roots as a tool to study root development. Ann. Bot. 2012, 110, 479–489. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, F.; Song, Y.; Li, X.; Chen, J.; Mo, L.; Zhang, X.; Lin, Z.; Zhang, L. Genome sequences of horticultural plants: Past, present, and future. Hortic. Res. 2019, 6, 11–23. [Google Scholar] [CrossRef]
- van Dijk, A.D.J. Plant Genomics Databases. Methods and Protocols, 1st ed.; Springer Science+Business Media LLC: New York, NY, USA, 2017; pp. 1–336. [Google Scholar]
- Chen, F.; Dong, W.; Zhang, J.; Guo, X.; Chen, J.; Wang, Z.; Lin, Z.; Tang, H.; Zhang, L. The sequenced angiosperm genomes and genome databases. Front. Plant Sci. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Shang, L.; Zhu, Q.-H.; Fan, L.; Guo, L. Twenty years of plant genome sequencing: Achievements and challenges. Trends Plant Sci. 2021, 2207, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Li, F.-W.; Harkess, A. A guide to sequence your favorite plant genomes. Appl. Plant Sci. 2018, 6, 1–7. [Google Scholar] [CrossRef]
- Paajanen, P.; Kettleborough, G.; López-Girona, E.; Giolai, M.; Heavens, D.; Baker, D.; Lister, A.; Cugliandolo, F.; Wilde, G.; Hein, I.; et al. A critical comparison of technologies for a plant genome sequencing project. GigaScience 2019, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Wang, X.; Cao, L.; Ji, J.; Liu, T.; Duan, K. Highly efficient Agrobacterium rhizogenes-mediated hairy root transformation for gene functional and gene editing analysis in soybean. Plant Methods 2021, 17, 1–12. [Google Scholar] [CrossRef]
- Cai, Y.; Chen, L.; Liu, X.; Sun, S.; Wu, C.; Jiang, B.; Han, T.; Hou, W. CRISPR/Cas9-mediated genome editing in soybean hairy roots. PLoS ONE 2015, 10, 1–13. [Google Scholar] [CrossRef]
- Du, H.; Zeng, X.; Zhao, M.; Cui, X.; Wang, Q.; Yang, H.; Cheng, H.; Yu, D. Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J. Biotechnol. 2016, 217, 90–97. [Google Scholar] [CrossRef]
- Pan, C.; Ye, L.; Qin, L.; Liu, X.; He, Y.; Wang, J.; Chen, L.; Lu, G. CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Chen, Y.; Ma, Y.; Wang, H.; Wei, J. Effective reduction in chimeric mutants of poplar trees produced by CRISPR/Cas9 through a second round of shoot regeneration. Plant Biotechnol. Rep. 2020, 14, 549–558. [Google Scholar] [CrossRef]
- Triozzi, P.; Schmidt, H.W.; Dervinis, C.; Kirst, M.; Conde, D. Simple, efficient and open-source CRISPR/Cas9 strategy for multi-site genome editing in Populus tremula × alba. Tree Physiol. 2021, 41, 2216–2227. [Google Scholar] [CrossRef] [PubMed]
- Hasebe, F.; Yuba, H.; Hashimoto, T.; Saito, K.; Funa, N.; Shoji, T. CRISPR/Cas9-mediated disruption of the PYRROLIDINE KETIDE SYNTHASE gene reduces the accumulation of tropane alkaloids in Atropa belladonna hairy roots. Biosci. Biotechnol. Biochem. 2021, 85, 2404–2409. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiryushkin, A.S.; Ilina, E.L.; Guseva, E.D.; Pawlowski, K.; Demchenko, K.N. Hairy CRISPR: Genome Editing in Plants Using Hairy Root Transformation. Plants 2022, 11, 51. https://doi.org/10.3390/plants11010051
Kiryushkin AS, Ilina EL, Guseva ED, Pawlowski K, Demchenko KN. Hairy CRISPR: Genome Editing in Plants Using Hairy Root Transformation. Plants. 2022; 11(1):51. https://doi.org/10.3390/plants11010051
Chicago/Turabian StyleKiryushkin, Alexey S., Elena L. Ilina, Elizaveta D. Guseva, Katharina Pawlowski, and Kirill N. Demchenko. 2022. "Hairy CRISPR: Genome Editing in Plants Using Hairy Root Transformation" Plants 11, no. 1: 51. https://doi.org/10.3390/plants11010051
APA StyleKiryushkin, A. S., Ilina, E. L., Guseva, E. D., Pawlowski, K., & Demchenko, K. N. (2022). Hairy CRISPR: Genome Editing in Plants Using Hairy Root Transformation. Plants, 11(1), 51. https://doi.org/10.3390/plants11010051