Regulation of the Later Stages of Nodulation Stimulated by IPD3/CYCLOPS Transcription Factor and Cytokinin in Pea Pisum sativum L.
Abstract
1. Introduction
2. Results
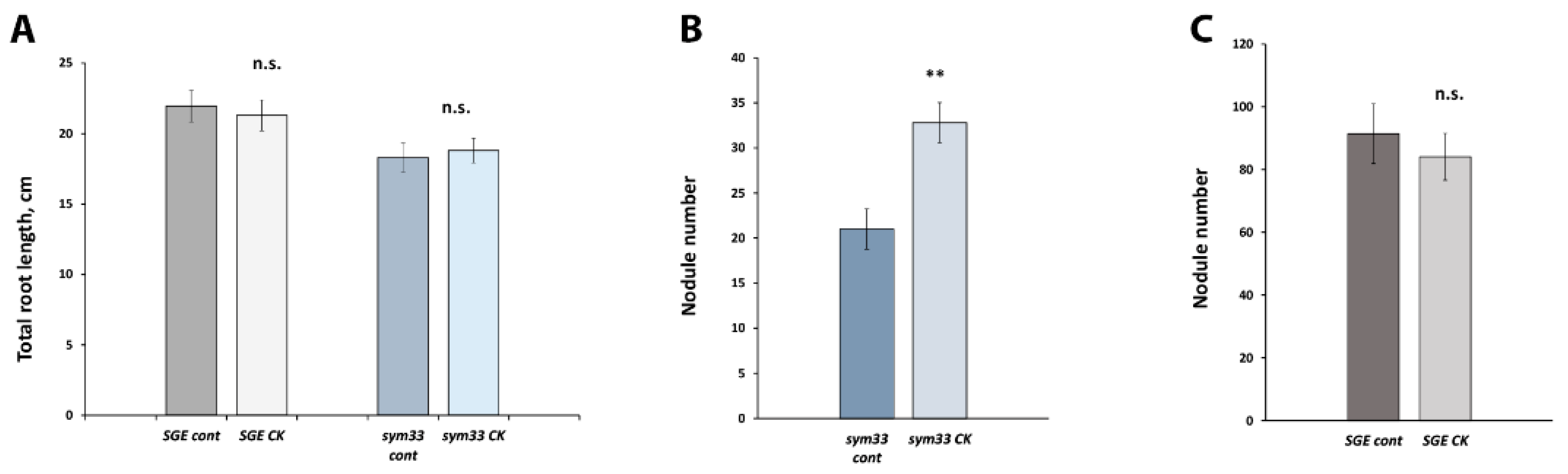
2.1. Effect of Exogenously Applied Cytokinin on the Morphology and Structural Characteristics of SGEFix--2 (sym33) Mutant Nodules
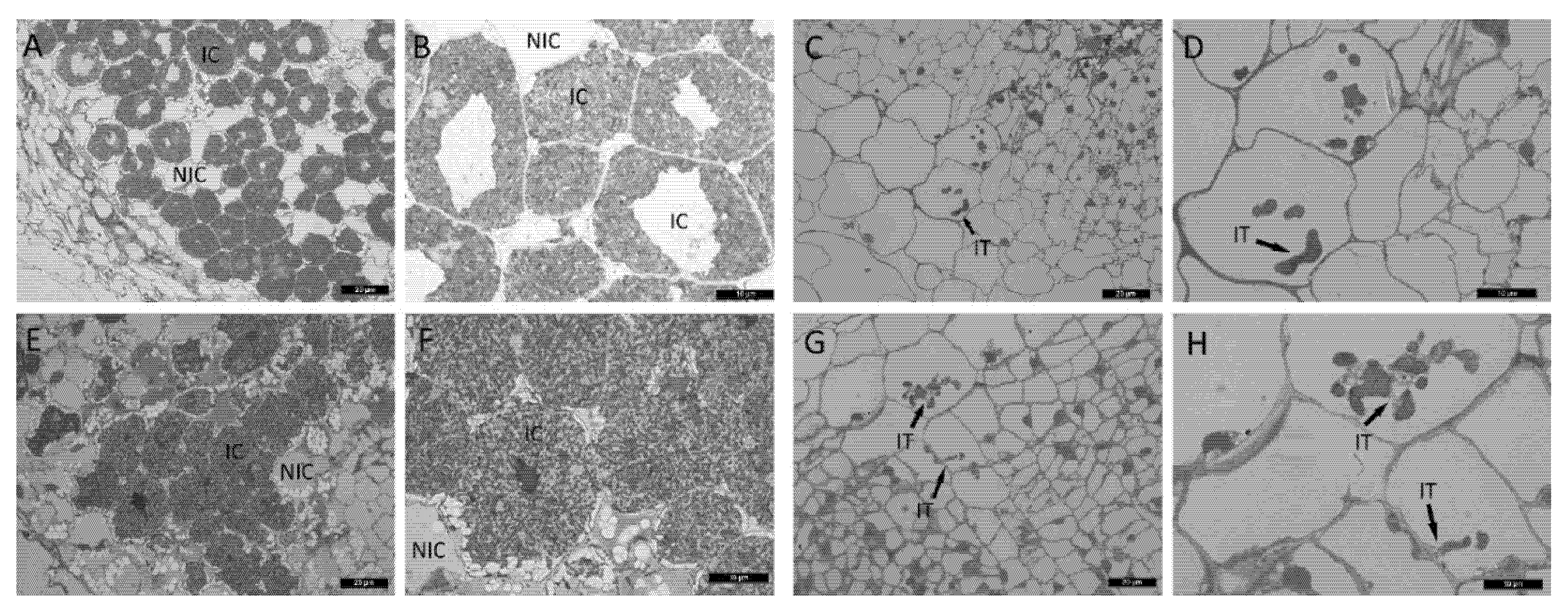
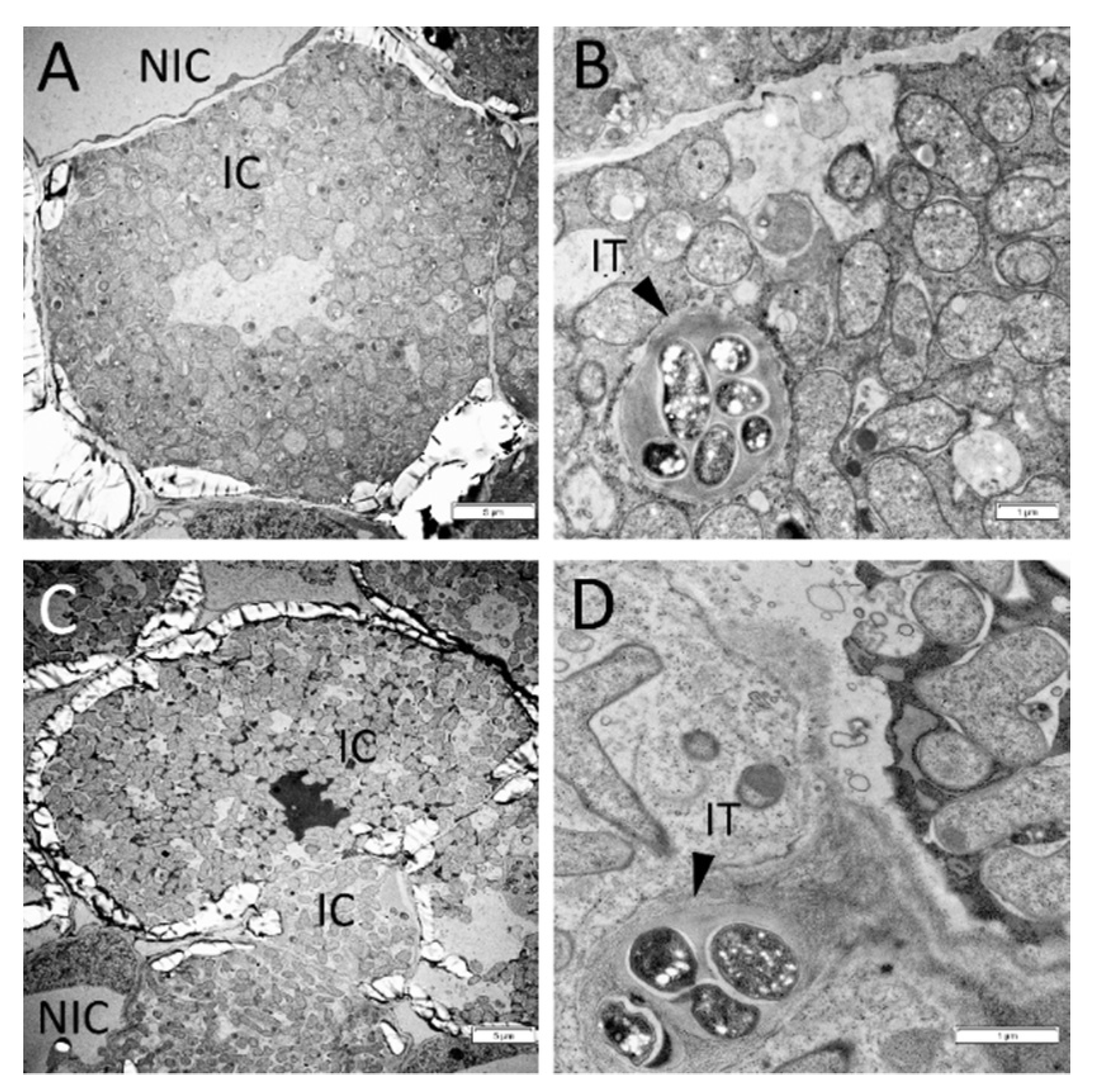
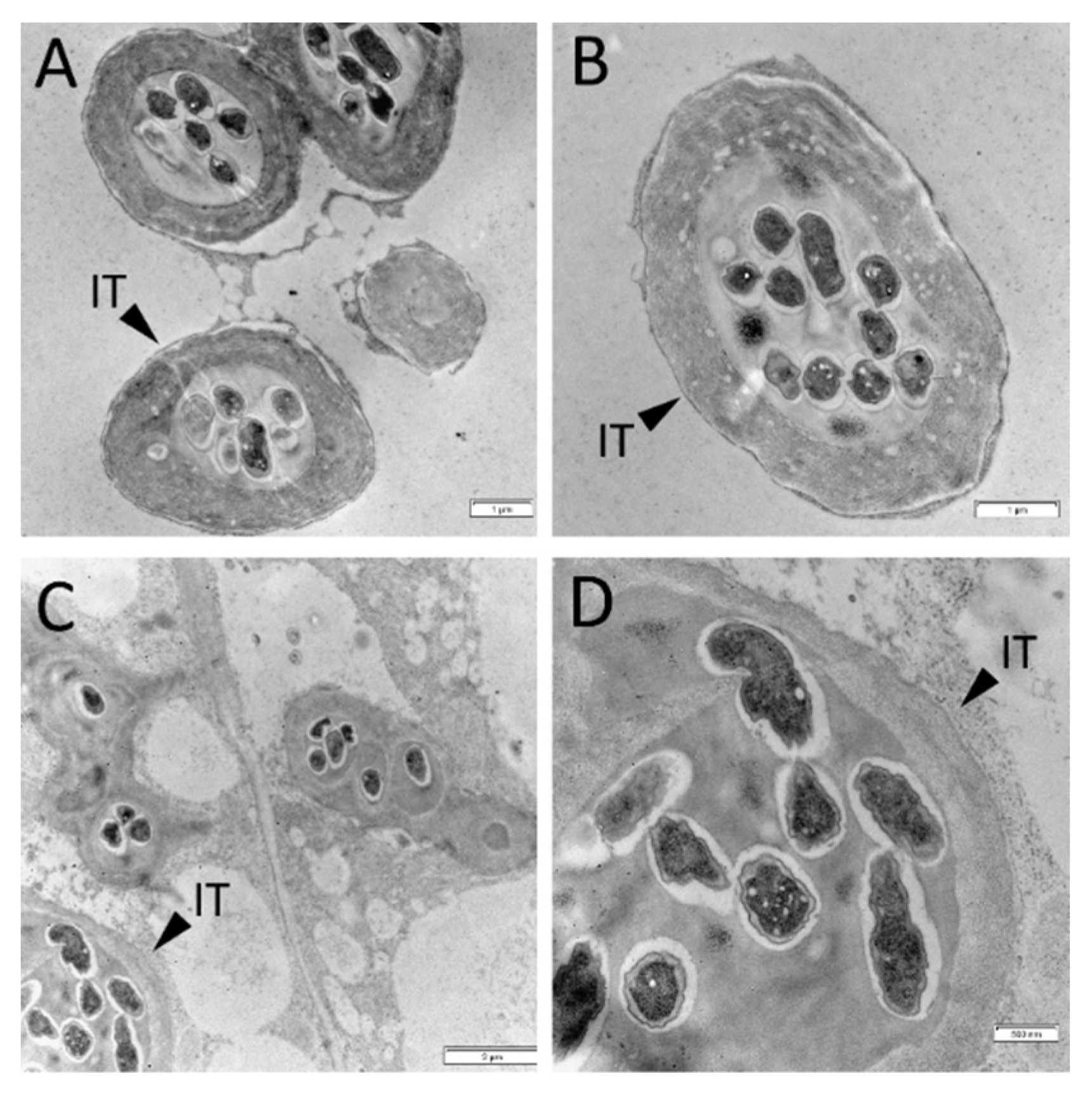
2.2. Histological Analysis of Nodules in Wild Type and sym33 Mutant Plants Treated with Cytokinin
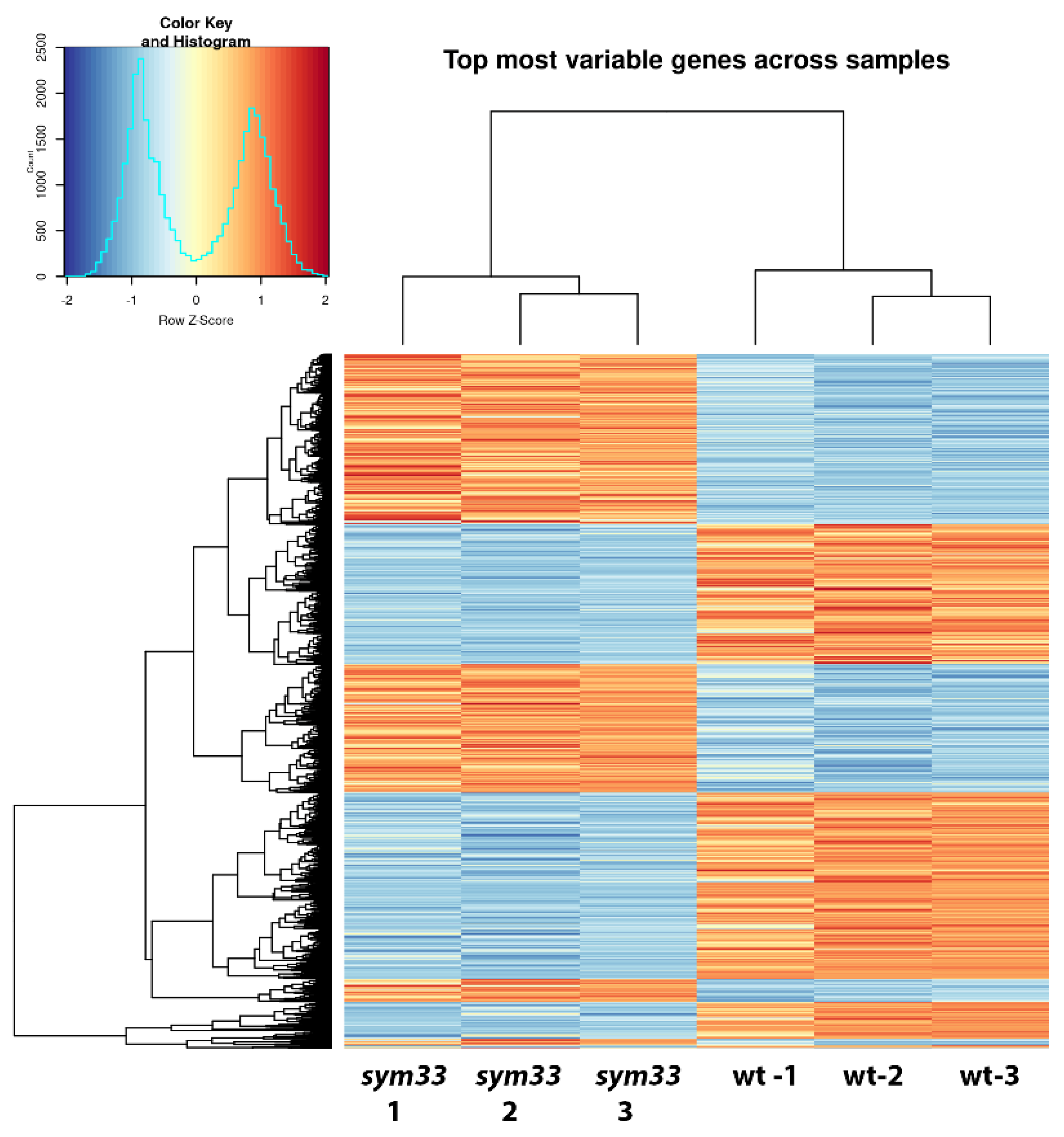
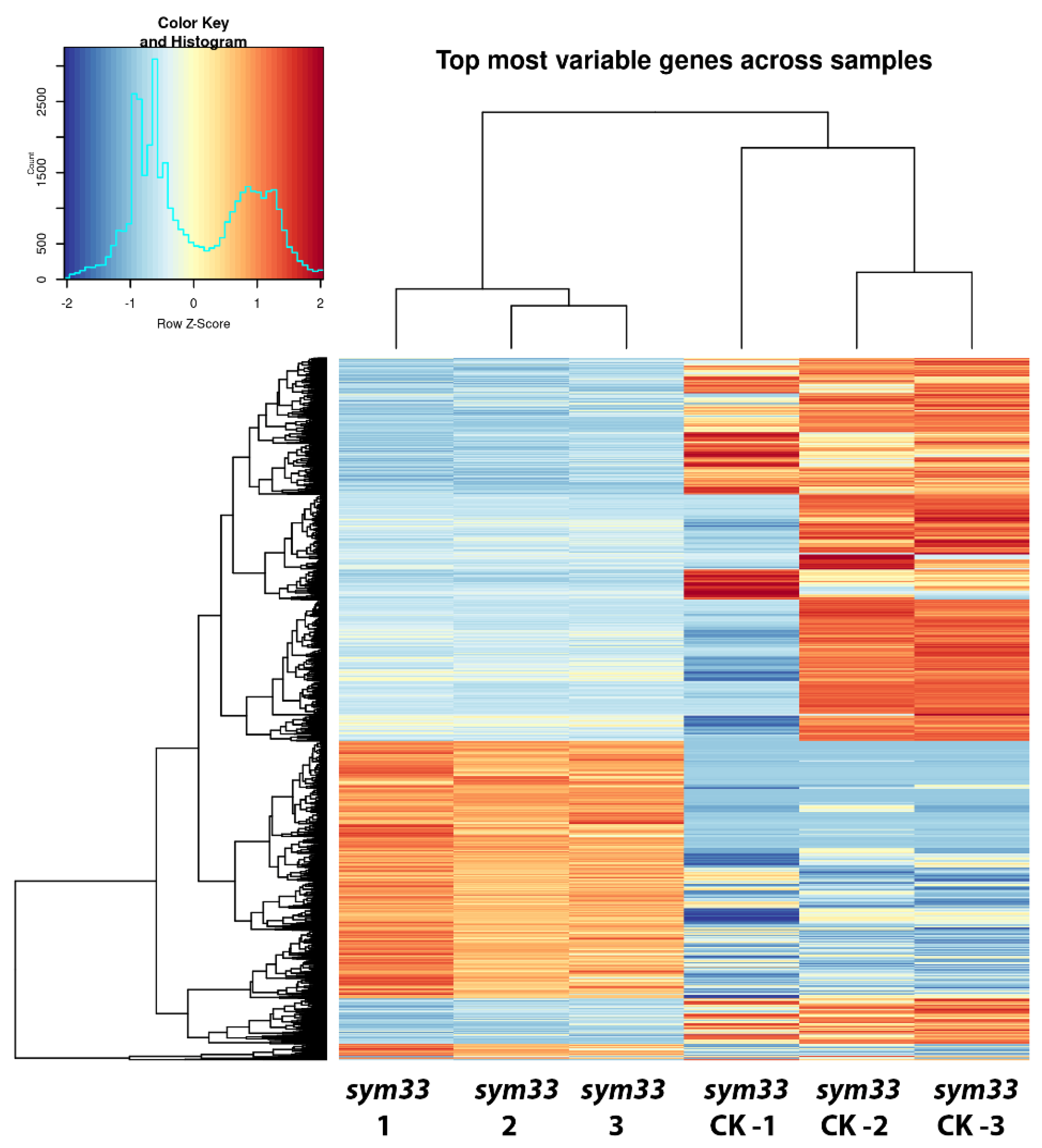
2.3. Comparative Analysis of Expression Patterns between Cv. SGE Wild Type and SGEFix--2 (Sym33) Mutant Nodules Using a Transcriptomic Approach
2.4. Functional Activity of Differentially Expressed Genes between cv. SGE Wild Type and SGEFix--2 (sym33) Mutant Based on Gene Ontology Analysis
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Inoculation
4.2. Plant Material and Growth Conditions
4.3. Material Fixation
4.4. Light and Transmission Electron Microscopy
4.5. Isolation of RNA and Preparation of Libraries
4.6. Illumina Sequencing and Data Analysis
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Madsen, E.B.; Madsen, L.H.; Radutoiu, S.; Olbryt, M.; Rakwalska, M.; Szczyglowski, K.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; et al. A receptor kinase gene of the LysM type is involved in legumeperception of rhizobial signals. Nature 2003, 425, 637–640. [Google Scholar] [CrossRef]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Jurkiewicz, A.; Fukai, E.; Quistgaard, E.M.H.; Albrektsen, A.S.; James, E.K.; Thirup, S.; Stougaard, J. LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 2007, 26, 3923–3935. [Google Scholar] [CrossRef] [PubMed]
- Broghammer, A.; Krusell, L.; Blaise, M.; Sauer, J.; Sullivan, J.T.; Maolanon, N.; Vinther, M.; Lorentzen, A.; Madsen, E.B.; Jensen, K.J.; et al. Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl. Acad. Sci. USA 2012, 109, 13859–13864. [Google Scholar] [CrossRef] [PubMed]
- Amor, B.B.; Shaw, S.L.; Oldroyd, G.E.D.; Maillet, F.; Penmetsa, R.V.; Cook, D.; Long, S.R.; Denarie, J.; Gough, C. The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 2003, 34, 495–506. [Google Scholar] [CrossRef]
- Limpens, E.; Franken, C.; Smit, P.; Willemse, J.; Bisseling, T.; Geurts, R. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 2003, 302, 630–633. [Google Scholar] [CrossRef] [PubMed]
- Smit, P.; Limpens, E.; Geurts, R.; Fedorova, E.; Dolgikh, E.; Gough, C.; Bisseling, T. Medicago LYK3, an Entry Receptor in Rhizobial Nodulation Factor Signaling. Plant Physiol. 2007, 145, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Bensmihen, S.; de Billy, F.; Gough, C. Contribution of NFP LysM domains to the recognition of Nod factors during the Medicago truncatula/Sinorhizobium meliloti symbiosis. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Zhukov, V.; Radutoiu, S.; Madsen, L.H.; Rychagova, T.; Ovchinnikova, E.; Borisov, A.; Tikhonovich, I.; Stougaard, J. The Pea Sym37 Receptor Kinase Gene Controls Infection-Thread Initiation and Nodule Development. Mol. Plant-Microbe Interact. 2008, 21, 1600–1608. [Google Scholar] [CrossRef] [PubMed]
- Kirienko, A.N.; Porozov, Y.B.; Malkov, N.V.; Akhtemova, G.A.; Le Signor, C.; Thompson, R.; Saffray, C.; Dalmais, M.; Bendahmane, A.; Tikhonovich, I.A.; et al. Role of a receptor-like kinase K1 in pea Rhizobium symbiosis development. Planta 2018, 248, 1101–1120. [Google Scholar] [CrossRef] [PubMed]
- Sulima, A.S.; Zhukov, V.A.; Afonin, A.A.; Zhernakov, A.I.; Tikhonovich, I.A.; Lutova, L.A. Selection Signatures in the First Exon of Paralogous Receptor Kinase Genes from the Sym2 Region of the Pisum sativum L. Genome. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Geurts, R.; Heidstra, R.; Hadri, A.E.; Downie, J.A.; Franssen, H.; Van Kammen, A.; Bisseling, T. SYM2 of pea is involved in a nodulation factor-perception mechanism that controls the infection process in the epidermis. Plant Physiol. 1997, 115, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Kirienko, A.N.; Vishnevskaya, N.A.; Kitaeva, A.B.; Shtark, O.Y.; Kozyulina, P.Y.; Thompson, R.; Dalmais, M.; Bendahmane, A.; Tikhonovich, I.A.; Dolgikh, E.A. Structural variations in LysM domains of LysM-RLK psK1 may result in a different effect on Pea–Rhizobial symbiosis development. Int. J. Mol. Sci. 2019, 20, 1624. [Google Scholar] [CrossRef] [PubMed]
- Endre, G.; Kereszt, A.; Kevei, Z.; Mihacea, S.; Kaló, P.; Kiss, G.B. A receptor kinase gene regulating symbiotic nodule development. Nature 2002, 417, 962–966. [Google Scholar] [CrossRef] [PubMed]
- Stracke, S.; Kistner, C.; Yoshida, S.; Mulder, L.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; Stougaard, J.; Szczyglowski, K.; et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 2002, 417, 959–962. [Google Scholar] [CrossRef]
- Ané, J.M.; Kiss, G.B.; Riely, B.K.; Penmetsa, R.V.; Oldroyd, G.E.D.; Ayax, C.; Lévy, J.; Debellé, F.; Baek, J.M.; Kalo, P.; et al. Medicago truncatula DMI1 Required for Bacterial and Fungal Symbioses in Legumes. Science 2004, 303, 1364–1367. [Google Scholar] [CrossRef]
- Charpentier, M.; Martins, T.V.; Granqvist, E.; Oldroyd, G.E.D.; Morris, R.J. The role of DMI1 in establishing Ca2+ oscillations in legume symbioses. Plant Signal. Behav. 2013, 8, e2289. [Google Scholar] [CrossRef] [PubMed]
- Kanamori, N.; Madsen, L.H.; Radutoiu, S.; Frantescu, M.; Quistgaard, E.M.H.; Miwa, H.; Downie, J.A.; James, E.K.; Felle, H.H.; Haaning, L.L.; et al. A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl. Acad. Sci. USA 2006, 103, 359–364. [Google Scholar] [CrossRef]
- Saito, K.; Yoshikawa, M.; Yano, K.; Miwa, H.; Uchida, H.; Asamizu, E.; Sato, S.; Tabata, S.; Imaizumi-Anraku, H.; Umehara, Y.; et al. Nucleoporin85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 2007, 19, 610–624. [Google Scholar] [CrossRef] [PubMed]
- Groth, M.; Takeda, N.; Perry, J.; Uchid, H.; Dräxl, S.; Brachmann, A.; Sato, S.; Tabata, S.; Kawaguchi, M.; Wang, T.L.; et al. NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 2010, 22, 2509–2526. [Google Scholar] [CrossRef] [PubMed]
- Chiasson, D.M.; Haage, K.; Sollweck, K.; Brachmann, A.; Dietrich, P.; Parniske, M. A quantitative hypermorphic CNGC allele confers ectopic calcium flux and impairs cellular development. Elife 2017, 6, e25012. [Google Scholar] [CrossRef]
- Capoen, W.; Sun, J.; Wysham, D.; Otegui, M.S.; Venkateshwaran, M.; Hirsch, S.; Miwa, H.; Downie, J.A.; Morris, R.J.; Ane, J.-M.; et al. Nuclear membranes control symbiotic calcium signaling of legumes. Proc. Natl. Acad. Sci. USA 2011, 108, 14348–14353. [Google Scholar] [CrossRef] [PubMed]
- Lévy, J.; Bres, C.; Geurts, R.; Chalhoub, B.; Kulikova, O.; Duc, G.; Journet, E.-P.; Ané, J.-M.; Lauber, E.; Bisseling, T.; et al. A Putative Ca2+ and Calmodulin-Dependent Protein Kinase Required for Bacterial and Fungal Symbioses. Science 2004, 303, 1361–1364. [Google Scholar] [CrossRef] [PubMed]
- Mitra, R.M.; Shaw, S.L.; Long, S.R. Six nonnodulating plant mutants defective for Nod factor-induced transcriptional changes associated with the legume-rhizobia symbiosis. Proc. Natl. Acad. Sci. USA 2004, 101, 10217–10222. [Google Scholar] [CrossRef]
- Gleason, C.; Chaudhuri, S.; Yang, T.; Muñoz, A.; Poovaiah, B.W.; Oldroyd, G.E.D. Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 2006, 441, 1149–1152. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D.; Downie, J.A. Coordinating Nodule Morphogenesis with Rhizobial Infection in Legumes. Annu. Rev. Plant Biol. 2008, 59, 519–546. [Google Scholar] [CrossRef]
- Messinese, E.; Mun, J.-H.; Yeun, L.H.; Jayaraman, D.; Rougé, P.; Barre, A.; Lougnon, G.; Schornack, S.; Bono, J.-J.; Cook, D.R.; et al. A Novel Nuclear Protein Interacts With the Symbiotic DMI3 Calcium- and Calmodulin-Dependent Protein Kinase of Medicago truncatula. Mol. Plant-Microbe Interact. 2007, 20, 912–921. [Google Scholar] [CrossRef] [PubMed]
- Yano, K.; Yoshida, S.; Muller, J.; Singh, S.; Banba, M.; Vickers, K.; Markmann, K.; White, C.; Schuller, B.; Sato, S.; et al. CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. USA 2008, 105, 20540–20545. [Google Scholar] [CrossRef]
- Limpens, E.; Bisseling, T. CYCLOPS: A new vision on rhizobium-induced nodule organogenesis. Cell Host Microbe 2014, 15, 127–129. [Google Scholar] [CrossRef]
- Singh, S.; Katzer, K.; Lambert, J.; Cerri, M.; Parniske, M. CYCLOPS, A DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 2014, 15, 139–152. [Google Scholar] [CrossRef]
- Liu, C.W.; Breakspear, A.; Guan, D.; Cerri, M.R.; Jackson, K.; Jiang, S.; Robson, F.; Radhakrishnan, G.V.; Roy, S.; Bone, C.; et al. NIN acts as a network hub controlling a growth module required for rhizobial infection. Plant Physiol. 2019, 179, 1704–1722. [Google Scholar] [CrossRef]
- Schauser, L.; Roussis, A.; Stiller, J.; Stougaard, J. A plant regulator controlling development of symbiotic root nodules. Nature 1999, 402, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Rutten, L.; Limpens, E.; Van Der Molen, T.; Van Velzen, R.; Chen, R.; Chen, Y.; Geurts, R.; Kohlen, W.; Kulikova, O.; et al. A remote cis-regulatory region is required for nin expression in the pericycle to initiate nodule primordium formation in medicago truncatula. Plant Cell 2019, 31, 68–83. [Google Scholar] [CrossRef] [PubMed]
- Soyano, T.; Shimoda, Y.; Kawaguchi, M.; Hayashi, M. A shared gene drives lateral root development and root nodule symbiosis pathways in Lotus. Science 2019, 366, 1021–1023. [Google Scholar] [CrossRef] [PubMed]
- Marsh, J.F.; Rakocevic, A.; Mitra, R.M.; Brocard, L.; Sun, J.; Eschstruth, A.; Long, S.R.; Schultze, M.; Ratet, P.; Oldroyd, G.E.D. Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 2007, 144, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Kosuta, S.; Held, M.; Hossain, M.S.; Morieri, G.; MacGillivary, A.; Johansen, C.; Antolín-Llovera, M.; Parniske, M.; Oldroyd, G.E.D.; Downie, A.J.; et al. Lotus japonicus symRK-14 uncouples the cortical and epidermal symbiotic program. Plant J. 2011, 67, 929–940. [Google Scholar] [CrossRef]
- Kaló, P.; Gleason, C.; Edwards, A.; Marsh, J.; Mitra, R.M.; Hirsch, S.; Jakab, J.; Sims, S.; Long, S.R.; Rogers, J.; et al. Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 2005, 308, 1786–1789. [Google Scholar] [CrossRef]
- Smit, P.; Raedts, J.; Portyanko, V.; Debellé, F.; Gough, C.; Bisseling, T.; Geurts, R. NSP1 of the GRAS protein family is essential for rhizobial nod factor-induced transcription. Science 2005, 308, 1789–1791. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, S.; Oldroyd, G.E.D. GRAS-domain transcription factors SCR/SHR—GRAS Proteins Involved in Root Radial Patterning NSP1/NSP2—Formation of GRAS Domain Complex at the DNA. Plant Signal. Behav. 2009, 4, 698–700. [Google Scholar] [CrossRef]
- Dolgikh, E.A.; Leppyanen, I.V.; Osipova, M.A.; Savelyeva, N.V.; Borisov, A.Y.; Tsyganov, V.E.; Geurts, R.; Tikhonovich, I.A. Genetic dissection of Rhizobium-induced infection and nodule organogenesis in pea based on ENOD12A and ENOD5 expression analysis. Plant Biol. 2011, 13, 285–296. [Google Scholar] [CrossRef]
- Xiao, A.; Yu, H.; Fan, Y.; Kang, H.; Ren, Y.; Huang, X.; Gao, X.; Wang, C.; Zhang, Z.; Zhu, H.; et al. Transcriptional regulation of NIN expression by IPN2 is required for root nodule symbiosis in Lotus japonicus. New Phytol. 2020, 227, 513–528. [Google Scholar] [CrossRef]
- Miwa, H.; Sun, J.; Oldroyd, G.E.D.; Downie, J.A. Analysis of Nod-factor-induced calcium signaling in root hairs of symbiotically defective mutants of Lotus japonicus. Mol. Plant-Microbe Interact. 2006, 19, 914–923. [Google Scholar] [CrossRef] [PubMed]
- Middleton, P.H.; Jakab, J.; Penmetsa, R.V.; Starker, C.G.; Doll, J.; Kaló, P.; Prabhu, R.; Marsh, J.F.; Mitra, R.M.; Kereszt, A.; et al. An ERF transcription factor in Medicago truncatula that is essential for nod factor signal transduction. Plant Cell 2007, 19, 1221–1234. [Google Scholar] [CrossRef]
- Cerri, M.R.; Frances, L.; Laloum, T.; Auriac, M.C.; Niebel, A.; Oldroyd, G.E.D.; Barker, D.G.; Fournier, J.; de Carvalho-Niebel, F. Medicago truncatula ERN transcription factors: Regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiol. 2012, 160, 2155–2172. [Google Scholar] [CrossRef] [PubMed]
- Cerri, M.R.; Frances, L.; Kelner, A.; Fournier, J.; Middleton, P.H.; Auriac, M.C.; Mysore, K.S.; Wen, J.; Erard, M.; Barker, D.G.; et al. The symbiosis-related ERN transcription factors act in concert to coordinate rhizobial host root infection. Plant Physiol. 2016, 171, 1037–1054. [Google Scholar] [CrossRef] [PubMed]
- Fonouni-Farde, C.; Tan, S.; Baudin, M.; Brault, M.; Wen, J.; Mysore, K.S.; Niebel, A.; Frugier, F.; Diet, A. DELLA-mediated gibberellin signalling regulates Nod factor signalling and rhizobial infection. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Liu, H.; Luo, D.; Yu, N.; Dong, W.; Wang, C.; Zhang, X.; Dai, H.; Yang, J.; Wang, E. DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat. Commun. 2016, 7, 12433. [Google Scholar] [CrossRef]
- Dolgikh, A.V.; Kirienko, A.N.; Tikhonovich, I.A.; Foo, E.; Dolgikh, E.A. The DELLA Proteins Influence the Expression of Cytokinin Biosynthesis and Response Genes During Nodulation. Front. Plant Sci. 2019, 10, 432. [Google Scholar] [CrossRef] [PubMed]
- Soyano, T.; Kouchi, H.; Hirota, A.; Hayashi, M. Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet. 2013, 9, e1003352. [Google Scholar] [CrossRef]
- Laloum, T.; Baudin, M.; Frances, L.; Lepage, A.; Billault-Penneteau, B.; Cerri, M.R.; Ariel, F.; Jardinaud, M.F.; Gamas, P.; De Carvalho-Niebel, F.; et al. Two CCAAT-box-binding transcription factors redundantly regulate early steps of the legume-rhizobia endosymbiosis. Plant J. 2014, 79, 757–768. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, M.E.; Blanco, F.A.; Beker, M.P.; Battaglia, M.; Aguilar, O.M. A C Subunit of the Plant Nuclear Factor NF-Y Required for Rhizobial Infection and Nodule Development Affects Partner Selection in the Common Bean–Rhizobium etli Symbiosis. Plant Cell 2010, 22, 4142–4157. [Google Scholar] [CrossRef] [PubMed]
- Suzaki, T.; Ito, M.; Yoro, E.; Sato, S.; Hirakawa, H.; Takeda, N.; Kawaguchi, M. Endoreduplication-mediated initiation of symbiotic organ development in Lotus japonicus. Development 2014, 141, 2441–2445. [Google Scholar] [CrossRef]
- Azarakhsh, M.; Kirienko, A.N.; Zhukov, V.A.; Lebedeva, M.A.; Dolgikh, E.A.; Lutova, L.A. KNOTTED1-LIKE HOMEOBOX 3: A new regulator of symbiotic nodule development. J. Exp. Bot. 2015, 66, 7181–7195. [Google Scholar] [CrossRef] [PubMed]
- Vernié, T.; Kim, J.; Frances, L.; Ding, Y.; Sun, J.; Guan, D.; Niebel, A.; Gifford, M.L.; de Carvalho-Niebel, F.; Oldroyd, G.E.D. The NIN transcription factor coordinates diverse nodulation programs in different tissues of the Medicago truncatula root. Plant Cell 2015, 27, 3410–3424. [Google Scholar] [CrossRef] [PubMed]
- Plet, J.; Wasson, A.; Ariel, F.; Le Signor, C.; Baker, D.; Mathesius, U.; Crespi, M.; Frugier, F. MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J. 2011, 65, 622–633. [Google Scholar] [CrossRef] [PubMed]
- Dolgikh, E.A.; Kusakin, P.G.; Kitaeva, A.B.; Tsyganova, A.V.; Kirienko, A.N.; Leppyanen, I.V.; Dolgikh, A.V.; Ilina, E.L.; Demchenko, K.N.; Tikhonovich, I.A.; et al. Mutational analysis indicates that abnormalities in rhizobial infection and subsequent plant cell and bacteroid differentiation in pea (pisum sativum) nodules coincide with abnormal cytokinin responses and localization. Ann. Bot. 2020, 125, 905–923. [Google Scholar] [CrossRef] [PubMed]
- Tsyganov, V.E.; Morzhina, E.V.; Stefanov, S.Y.; Borisov, A.Y.; Lebsky, V.K.; Tikhonovich, I.A. The pea (Pisum sativum L.) genes sym33 and sym40 control infection thread formation and root nodule function. Mol. Gen. Genet. 1998, 259, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Tsyganov, V.E.; Seliverstova, E.V.; Voroshilova, V.A.; Tsyganova, A.V.; Pavlova, Z.B.; Lebskii, V.K.; Borisov, A.Y.; Brewin, N.J.; Tikhonovich, I.A. Double mutant analysis of sequential functioning of pea (Pisum sativum L.) genes Sym13, Sym33, and Sym40 during symbiotic nodule development. Russ. J. Genet. Appl. Res. 2011, 1, 343–348. [Google Scholar] [CrossRef]
- Voroshilova, V.A.; Boesten, B.; Tsyganov, V.E.; Borisov, A.Y.; Tikhonovich, I.A.; Priefer, U.B. Effect of mutations in Pisum sativum L. genes blocking different stages of nodule development on the expression of late symbiotic genes in Rhizobium leguminosarum bv. viciae. Mol. Plant Microbe Interact. 2001, 14, 471–476. [Google Scholar] [CrossRef]
- Voroshilova, V.A.; Demchenko, K.N.; Brewin, N.J.; Borisov, A.Y.; Tikhonovich, I.A. Initiation of a legume nodule with an indeterminate meristem involves proliferating host cells that harbour infection threads. New Phytol. 2009, 181, 913–923. [Google Scholar] [CrossRef]
- Bauer, P.; Ratet, P.; Crespi, M.D.; Schultze, M.; Kondorosi, A. Nod factors and cytokinins induce similar cortical cell division, amyloplast deposition and MsEnod12A expression patterns in alfalfa roots. Plant J. 1996, 10, 91–105. [Google Scholar] [CrossRef]
- Lorteau, M.A.; Ferguson, B.J.; Guinel, F.C. Effects of cytokinin on ethylene production and nodulation in pea (Pisum sativum) cv. Sparkle. Physiol. Plant. 2001, 112, 421–428. [Google Scholar] [CrossRef]
- Tsyganov, V.E.; Belimov, A.A.; Borisov, A.Y.; Safronova, V.I.; Georgi, M.; Dietz, K.-J.; Tikhonovich, I.A. A Chemically Induced New Pea (Pisum sativum) Mutant SGECd t with Increased Tolerance to, and Accumulation of, Cadmium. Ann. Bot. 2007, 99, 227–237. [Google Scholar] [CrossRef]
- Singh, S.P.; Thomason, P.A.; Lilla, S.; Schaks, M.; Tang, Q.; Goode, B.L.; Machesky, L.M.; Rottner, K.; Insall, R.H. Cell–substrate adhesion drives Scar/WAVE activation and phosphorylation by a Ste20-family kinase, which controls pseudopod lifetime. PLoS Biol. 2020, 18, e3000774. [Google Scholar] [CrossRef]
- Schmidt, R.R.; Weits, D.A.; Feulner, C.F.J.; Van Dongen, J.T. Oxygen sensing and integrative stress signaling in plants. Plant Physiol. 2018, 176, 1131–1142. [Google Scholar] [CrossRef]
- van Velzen, R.; Holmer, R.; Bu, F.; Rutten, L.; van Zeijl, A.; Liu, W.; Santuari, L.; Cao, Q.; Sharma, T.; Shen, D.; et al. Comparative genomics of the nonlegume Parasponia reveals insights into evolution of nitrogen-fixing rhizobium symbioses. Proc. Natl. Acad. Sci. USA 2018, 115, E4700–E4709. [Google Scholar] [CrossRef]
- Ovchinnikova, E.; Journet, E.P.; Chabaud, M.; Cosson, V.; Ratet, P.; Duc, G.; Fedorova, E.; Liu, W.; Op Den Camp, R.; Zhukov, V.; et al. IPD3 controls the formation of nitrogen-fixing symbiosomes in pea and Medicago spp. Mol. Plant-Microbe Interact. 2011, 24, 1333–1344. [Google Scholar] [CrossRef]
- Liu, J.; Rasing, M.; Zeng, T.; Klein, J.; Kulikova, O.; Bisseling, T. NIN is essential for development of symbiosomes, suppression of defence and premature senescence in Medicago truncatula nodules. New Phytol. 2021, 230, 290–303. [Google Scholar] [CrossRef] [PubMed]
- Berrabah, F.; Ratet, P.; Gourion, B. Multiple steps control immunity during the intracellular accommodation of rhizobia. J. Exp. Bot. 2015, 66, 1977–1985. [Google Scholar] [CrossRef] [PubMed]
- Sinharoy, S.; Torres-Jerez, I.; Bandyopadhyay, K.; Kereszt, A.; Pislariu, C.I.; Nakashima, J.; Benedito, V.A.; Kondorosi, E.; Udvardi, M.K. The C2H2 transcription factor REGULATOR OF SYMBIOSOME DIFFERENTIATION represses transcription of the secretory pathway gene VAMP721a and promotes symbiosome development in Medicago truncatula. Plant Cell 2013, 25, 3584–3601. [Google Scholar] [CrossRef]
- Bao, F.; Azhakanandam, S.; Franks, R.G. SEUSS and SEUSS-LIKE transcriptional adaptors regulate floral and embryonic development in arabidopsis. Plant Physiol. 2010, 152, 821–836. [Google Scholar] [CrossRef]
- Gregis, V.; Sessa, A.; Colombo, L.; Kater, M.M. AGL24, SHORT VEGETATIVE PHASE, and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis. Plant Cell 2006, 18, 1373–1382. [Google Scholar] [CrossRef]
- Sridhar, V.V.; Surendrarao, A.; Liu, Z. Erratum: APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 2006, 133, 3496. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, H.; Lu, Z.; Wen, L.; Gu, Z.; Zhang, X.; Yu, G.; Wang, H.; Zhou, C.; Han, L. Developmental Analysis of the GATA Factor HANABA TARANU Mutants in Medicago truncatula Reveals Their Roles in Nodule Formation. Front. Plant Sci. 2021, 12, 597. [Google Scholar] [CrossRef]
- Pan, H.; Wang, D. Nodule cysteine-rich peptides maintain a working balance during nitrogen-fixing symbiosis. Nat. Plants 2017, 3, 17048. [Google Scholar] [CrossRef]
- Guefrachi, I.; Nagymihaly, M.; Pislariu, C.I.; Van de Velde, W.; Ratet, P.; Mars, M.; Udvardi, M.K.; Kondorosi, E.; Mergaert, P.; Alunni, B. Extreme specificity of NCR gene expression in Medicago truncatula. BMC Genom. 2014, 15, 712. [Google Scholar] [CrossRef]
- Mergaert, P. Differentiation of symbiotic nodule cells and their rhizobium endosymbionts. Adv. Bot. Res. 2020, 94, 149–180. [Google Scholar]
- Combier, J.P.; Frugier, F.; De Billy, F.; Boualem, A.; El-Yahyaoui, F.; Moreau, S.; Vernié, T.; Ott, T.; Gamas, P.; Crespi, M.; et al. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 2006, 20, 3084–3088. [Google Scholar] [CrossRef]
- Combier, J.P.; De Billy, F.; Gamas, P.; Niebel, A.; Rivas, S. Trans-regulation of the expression of the transcription factor MtHAP2-1 by a uORF controls root nodule development. Genes Dev. 2008, 22, 1549–1559. [Google Scholar] [CrossRef]
- Laloum, T.; De Mita, S.; Gamas, P.; Baudin, M.; Niebel, A. CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 2013, 18, 157–166. [Google Scholar] [CrossRef]
- Laporte, P.; Lepage, A.; Fournier, J.; Catrice, O.; Moreau, S.; Jardinaud, M.F.; Mun, J.H.; Larrainzar, E.; Cook, D.R.; Gamas, P.; et al. The CCAAT box-binding transcription factor NF-YA1 controls rhizobial infection. J. Exp. Bot. 2014, 65, 481–494. [Google Scholar] [CrossRef]
- Baudin, M.; Laloum, T.; Lepage, A.; Rípodas, C.; Ariel, F.; Frances, L.; Crespi, M.; Gamas, P.; Blanco, F.A.; Zanetti, M.E.; et al. A phylogenetically conserved group of nuclear factor-Y transcription factors interact to control nodulation in legumes. Plant Physiol. 2015, 169, 2761–2773. [Google Scholar] [CrossRef]
- Rípodas, C.; Castaingts, M.; Clúa, J.; Villafañe, J.; Blanco, F.A.; Zanetti, M.E. The PvNF-YA1 and PvNF-YB7 subunits of the heterotrimeric nf-y transcription factor influence strain preference in the Phaseolus vulgaris–rhizobium etli symbiosis. Front. Plant Sci. 2019, 10, 221. [Google Scholar] [CrossRef]
- Iantcheva, A.; Boycheva, I.; Vassileva, V.; Revalska, M.; Zechirov, G. Cyclin-like F-box protein plays a role in growth and development of the three model species Medicago truncatula, Lotus japonicus, and Arabidopsis thaliana. Res. Rep. Biol. 2015, 6, 117–130. [Google Scholar] [CrossRef][Green Version]
- Zühl, L.; Volkert, C.; Ibberson, D.; Schmidt, A. Differential activity of F-box genes and E3 ligases distinguishes sexual versus apomictic germline specification in Boechera. J. Exp. Bot. 2019, 70, 5643–5657. [Google Scholar] [CrossRef]
- Bonhomme, M.; André, O.; Badis, Y.; Ronfort, J.; Burgarella, C.; Chantret, N.; Prosperi, J.M.; Briskine, R.; Mudge, J.; Debéllé, F.; et al. High-density genome-wide association mapping implicates an F-box encoding gene in Medicago truncatula resistance to Aphanomyces euteiches. New Phytol. 2014, 201, 1328–1342. [Google Scholar] [CrossRef]
- Vivek, A.T. In silico identification and characterization of microRNAs based on EST and GSS in orphan legume crop, Lens culinaris medik. (Lentil). Agri Gene 2018, 8, 45–56. [Google Scholar] [CrossRef]
- Shu, Y.; Liu, Y.; Zhang, J.; Song, L.; Guo, C. Genome-wide analysis of the AP2/ERF superfamily genes and their responses to abiotic stress in Medicago truncatula. Front. Plant Sci. 2016, 6, 1247. [Google Scholar] [CrossRef]
- Andriankaja, A.; Boisson-Dernier, A.; Frances, L.; Sauviac, L.; Jauneau, A.; Barker, D.G.; De Carvalho-Niebel, F. AP2-ERF transcription factors mediate nod factor-dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 2007, 19, 2866–2885. [Google Scholar] [CrossRef]
- Íñiguez, L.P.; Nova-Franco, B.; Hernández, G. Novel players in the AP2-miR172 regulatory network for common bean nodulation. Plant Signal. Behav. 2015, 10, e1062957. [Google Scholar] [CrossRef]
- Alvarez-Buylla, E.R.; García-Ponce, B.; de la Paz Sánchez, M.; Espinosa-Soto, C.; García-Gómez, M.L.; Piñeyro-Nelson, A.; Garay-Arroyo, A. MADS-box genes underground becoming mainstream: Plant root developmental mechanisms. New Phytol. 2019, 223, 1143–1158. [Google Scholar] [CrossRef]
- Ayra, L.; del Rocio Reyero-Saavedra, M.; Isidra-Arellano, M.C.; Lozano, L.; Ramírez, M.; Leija, A.; Fuentes, S.I.; Girard, L.; Valdés-López, O.; Hernández, G. Control of the Rhizobia Nitrogen-Fixing Symbiosis by Common Bean MADS-Domain/AGL Transcription Factors. Front. Plant Sci. 2021, 12, 679463. [Google Scholar] [CrossRef]
- Reid, D.E.; Nadzieja, M.; Novak, O.; Heckmann, A.B.; Sandal, N.; Stougaard, J. Cytokinin biosynthesis promotes cortical cell responses during nodule development. Plant Physiol. 2017, 175, 361–375. [Google Scholar] [CrossRef]
- Eklund, D.M.; Thelander, M.; Landberg, K.; Ståldal, V.; Nilsson, A.; Johansson, M.; Valsecchi, I.; Pederson, E.R.A.; Kowalczyk, M.; Ljung, K.; et al. Homologues of the Arabidopsis thaliana SHI/STY/LRP1 genes control auxin biosynthesis and affect growth and development in the moss Physcomitrella patens. Development 2010, 137, 1275–1284. [Google Scholar] [CrossRef]
- Gomariz-Fernández, A.; Sánchez-Gerschon, V.; Fourquin, C.; Ferrándiz, C. The role of SHI/STY/SRS genes in organ growth and carpel development is conserved in the distant eudicot species Arabidopsis thaliana and Nicotiana benthamiana. Front. Plant Sci. 2017, 8, 814. [Google Scholar] [CrossRef]
- Estornell, L.H.; Landberg, K.; Cierlik, I.; Sundberg, E. SHI/STY genes affect pre- and post-meiotic anther processes in auxin sensing domains in arabidopsis. Front. Plant Sci. 2018, 9, 150. [Google Scholar] [CrossRef]
- Shrestha, A.; Zhong, S.; Therrien, J.; Huebert, T.; Sato, S.; Mun, T.; Andersen, S.U.; Stougaard, J.; Lepage, A.; Niebel, A.; et al. Lotus japonicus Nuclear Factor YA1, a nodule emergence stage-specific regulator of auxin signalling. New Phytol. 2020, 229, 1535–1552. [Google Scholar] [CrossRef]
- Hossain, M.S.; Shrestha, A.; Zhong, S.; Miri, M.; Austin, R.S.; Sato, S.; Ross, L.; Huebert, T.; Tromas, A.; Torres-Jerez, I.; et al. Lotus japonicus NF-YA1 plays an essential role during nodule differentiation and targets members of the SHI/STY gene family. Mol. Plant-Microbe Interact. 2016, 29, 950–964. [Google Scholar] [CrossRef]
- Kim, M.H.; Cho, J.S.; Park, E.J.; Lee, H.; Choi, Y.I.; Bae, E.K.; Han, K.H.; Ko, J.H. Overexpression of a poplar Ring-H2 zinc finger, Ptxerico, confers enhanced drought tolerance via reduced water loss and ion leakage in Populus. Int. J. Mol. Sci. 2020, 21, 9454. [Google Scholar] [CrossRef]
- Orosz, L.; Sváb, Z.; Kondorosi, Á.; Sik, T. Genetic studies on Rhizobiophage 16-3. Mol. Gen. Genet. MGG 1973, 125, 341–350. [Google Scholar] [CrossRef]
- van Brussel, A.A.; Tak, T.; Wetselaar, A.; Pees, E.; Wijffelman, C. Small leguminosae as test plants for nodulation of Rhizobium leguminosarum and other rhizobia and agrobacteria harbouring a leguminosarum sym-plasmid. Plant Sci. Lett. 1982, 27, 317–325. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Kreplak, J.; Madoui, M.A.; Cápal, P.; Novák, P.; Labadie, K.; Aubert, G.; Bayer, P.E.; Gali, K.K.; Syme, R.A.; Main, D.; et al. A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 2019, 51, 1411–1422. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Boccacci, P.; Beltramo, C.; Sandoval Prando, M.A.; Lembo, A.; Sartor, C.; Mehlenbacher, S.A.; Botta, R.; Torello Marinoni, D. Gene ontology: Tool for the unification of biology. Mol. Breed. 2015, 25, 25–29. [Google Scholar] [CrossRef]
- Morgan, A.M.; Falcon, S.; Gentleman, R. GSEABase: Gene Set Enrichment Data Structures and Methods, R Package Version 1.56.0; 2021. Available online: http://www.bioconductor.org (accessed on 25 October 2021).


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rudaya, E.S.; Kozyulina, P.Y.; Pavlova, O.A.; Dolgikh, A.V.; Ivanova, A.N.; Dolgikh, E.A. Regulation of the Later Stages of Nodulation Stimulated by IPD3/CYCLOPS Transcription Factor and Cytokinin in Pea Pisum sativum L. Plants 2022, 11, 56. https://doi.org/10.3390/plants11010056
Rudaya ES, Kozyulina PY, Pavlova OA, Dolgikh AV, Ivanova AN, Dolgikh EA. Regulation of the Later Stages of Nodulation Stimulated by IPD3/CYCLOPS Transcription Factor and Cytokinin in Pea Pisum sativum L. Plants. 2022; 11(1):56. https://doi.org/10.3390/plants11010056
Chicago/Turabian StyleRudaya, Elizaveta S., Polina Yu. Kozyulina, Olga A. Pavlova, Alexandra V. Dolgikh, Alexandra N. Ivanova, and Elena A. Dolgikh. 2022. "Regulation of the Later Stages of Nodulation Stimulated by IPD3/CYCLOPS Transcription Factor and Cytokinin in Pea Pisum sativum L." Plants 11, no. 1: 56. https://doi.org/10.3390/plants11010056
APA StyleRudaya, E. S., Kozyulina, P. Y., Pavlova, O. A., Dolgikh, A. V., Ivanova, A. N., & Dolgikh, E. A. (2022). Regulation of the Later Stages of Nodulation Stimulated by IPD3/CYCLOPS Transcription Factor and Cytokinin in Pea Pisum sativum L. Plants, 11(1), 56. https://doi.org/10.3390/plants11010056






