Thermo-Sensitive Genic Male Sterile Lines of Neo-Tetraploid Rice Developed through Gene Editing Technology Revealed High Levels of Hybrid Vigor
Abstract
:1. Introduction
2. Results
2.1. Development of TGMS Lines by the Editing of Thermos-Sensitive Genic Male Sterile 5 (TMS5) Locus in Neo-Tetraploid Rice
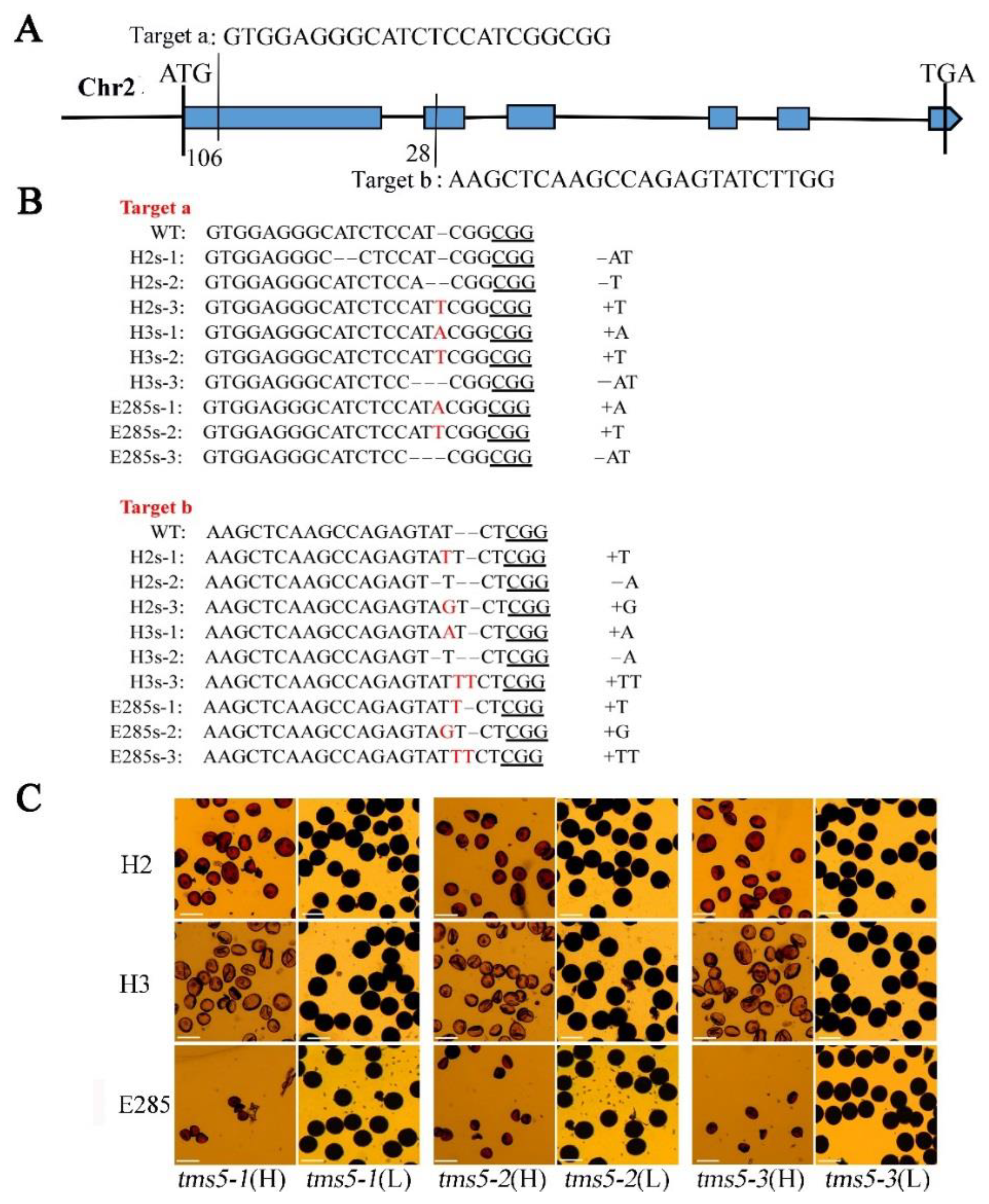
2.2. Cytological Observation of Pollen Development in H3s and E285s
2.3. Critical Sterility Temperature for the TGMS Lines of Neo-Tetraploid Rice
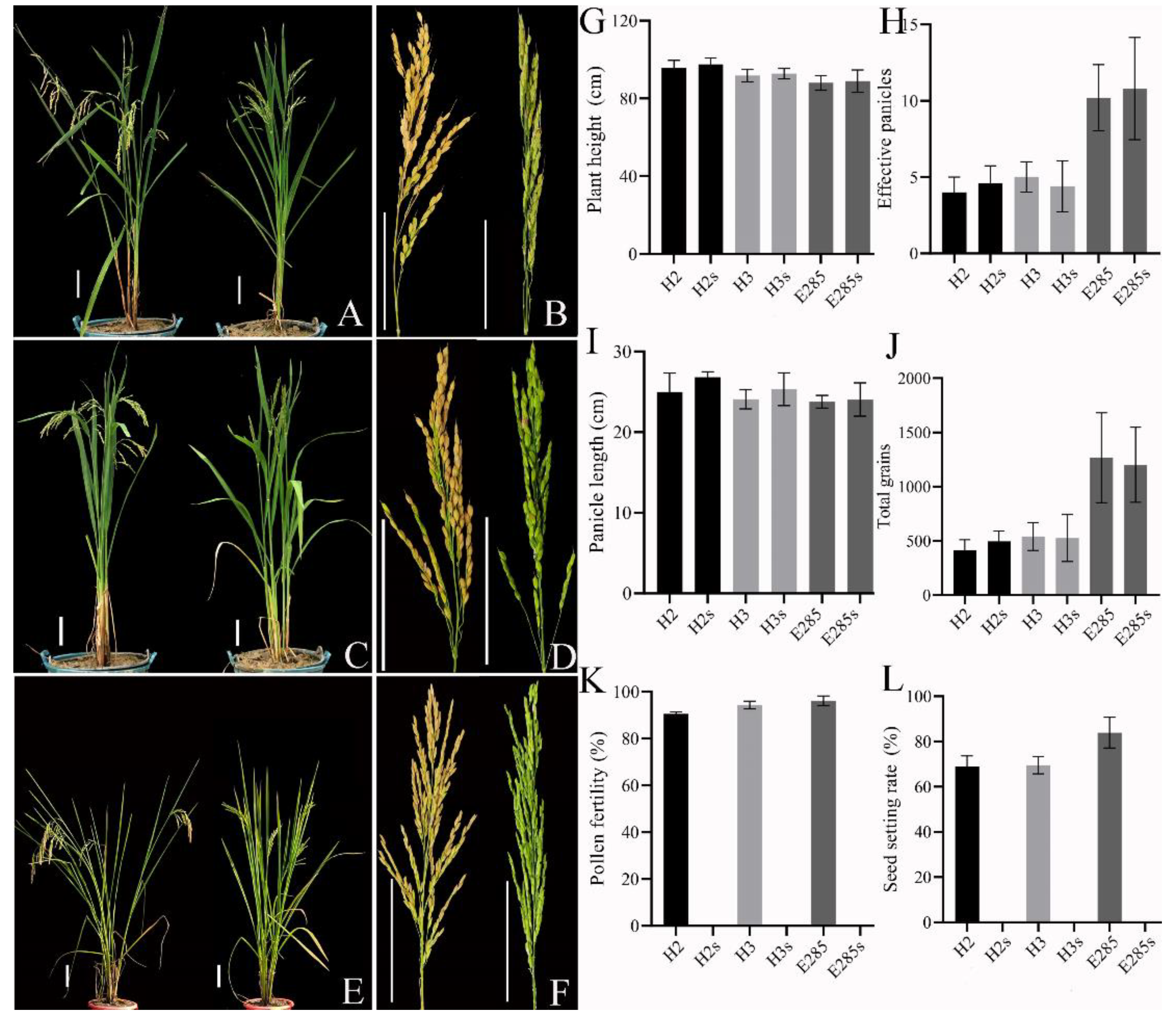
2.4. Heterosis Analysis of TGMS Lines of Neo-Tetraploid Rice Crossed with Autotetraploid Rice
3. Discussion
3.1. Gene Editing through CRISPR/Cas9 System Provides an Effective Way of Developing TGMS Lines of Tetraploid Rice
3.2. Pollen Sterility Is Different between the TGMS Lines of Neo-Tetraploid and Diploid Rice
3.3. Hybrids Generated by TGMS Lines of Neo-Tetraploid Rice Displayed Obvious Heterosis and Maintained for Several Generations
4. Materials and Methods
4.1. Plant Materials and Growing Conditions
4.2. Mutation-Sites Detection of Transgenic Plants Generated by CRISPR/Cas9 Gene Editing of TMS5
4.3. Cytological Observations of Pollen Fertility and Anther Development of tms5 Mutants and Wild Types
4.4. Analysis of Agronomic Traits in the Hybrids of TGMS Lines of Neo-Tetraploid Rice Crossed with Autotetraploid Rice
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shahid, M.Q.; Liu, G.; Li, J.; Naeem, M.; Liu, X.D. Heterosis and gene action study of agronomic traits in diploid and autotetraploid rice. Acta Agric. Scand. Sect. B Soil Plant Sci. 2011, 61, 23–32. [Google Scholar] [CrossRef]
- Wu, J.W.; Hu, C.Y.; Shahid, M.Q.; Guo, H.B.; Zeng, Y.X.; Liu, X.D.; Lu, Y.G. Analysis on genetic diversification and heterosis in autotetraploid rice. Springer Plus 2013, 2, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shahid, M.Q.; Sun, J.F.; Wei, C.M.; Zhang, P.; Liu, X.D. Studies on the abnormality of embryo sac and pollen fertility in autotetraploid rice during different growing seasons. Pak. J. Bot. 2010, 42, 7–19. [Google Scholar]
- Guo, H.B.; Mendrikahy, J.N.; Xie, L.; Deng, J.F.; Lu, Z.J.; Wu, J.W.; Li, X.; Shahid, M.Q.; Liu, X.D. Transcriptome analysis of neo-tetraploid rice reveals specific differential gene expressions associated with fertility and heterosis. Sci. Rep. 2017, 7, 40139. [Google Scholar] [CrossRef] [Green Version]
- Gan, L.; Huang, B.S.; Song, Z.J.; Zhang, Y.C.; Zhang, Y.J.; Chen, S.; Tong, L.Q.; Wei, Z.S.; Yu, L.X.; Luo, X.B.; et al. Unique glutelin expression patterns and seed endosperm structure facilitate glutelin accumulation in polyploid rice seed. Rice 2021, 14, 61. [Google Scholar] [CrossRef]
- Tu, S.B.; Kong, F.L.; Xu, Q.F.; He, T. Breakthrough in hybrid rice breeding with autotetraploid. Bull. Chin. Acad. Sci. 2003, 6, 426–428. [Google Scholar]
- Wang, X.; Liu, H.N.; Chen, Y.; Qiu, L.; Luan, L.; Long, W.B.; Tu, S.B.; Kong, F.L.; Sun, Y.S. Genetic improvement of the major agronomic traits in the parent materials of autotetraploid three-line rice. J. Southwest Univ. 2009, 31, 137–142. [Google Scholar]
- He, J.H.; Shahid, M.Q.; Li, Y.J.; Guo, H.B.; Cheng, X.A.; Liu, X.D.; Lu, Y.G. Allelic interaction of F1 pollen sterility loci and abnormal chromosome behavior caused pollen sterility in intersubspecific autotetraploid rice hybrids. J. Exp. Bot. 2011, 62, 4433–4445. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.W.; Shahid, M.Q.; Chen, L.; Chen, Z.X.; Wang, L.; Liu, X.D.; Lu, Y.G. Polyploidy enhances F1 pollen sterility loci interactions that increase meiosis abnormalities and pollen sterility in autotetraploid rice. Plant Physiol. 2015, 169, 2700–2717. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.W.; Chen, L.; Shahid, M.Q.; Chen, M.Y.; Dong, Q.L.; Li, J.R.; Xu, X.S.; Liu, X.D. Pervasive interactions of Sa and Sb loci cause high pollen sterility and abrupt changes in gene expression during meiosis that could be overcome by double neutral genes in autotetraploid rice. Rice 2017, 10, 49. [Google Scholar] [CrossRef]
- Shahid, M.Q.; Li, Y.J.; Saleem, M.F.; Naeem, M.; Wei, C.M.; Liu, X.D. Yield and yield components in autotetraploid and diploid rice genotypes (indica and japonica) sown in early and late seasons. Aust. J. Crop Sci. 2013, 7, 632–641. [Google Scholar]
- Lu, Z.J.; Guo, X.T.; Huang, Z.Y.; Xia, J.; Li, X.; Wu, J.W.; Yu, H.; Shahid, M.Q.; Liu, X.D. Transcriptome and gene editing analyses reveal MOF1a defect alters the expression of genes associated with tapetum development and chromosome behavior at meiosis stage resulting in low pollen fertility of tetraploid rice. Int. J. Mol. Sci. 2020, 21, 7489. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shahid, M.Q.; Xia, J.; Lu, Z.J.; Fang, N.; Wang, L.; Wu, J.W.; Chen, Z.X.; Liu, X.D. Analysis of small RNAs revealed differential expressions during pollen and embryo sac development in autotetraploid rice. BMC Genom. 2017, 18, 129. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.W.; Fan, H.; Hu, Y.F.; Guo, H.B.; Lin, H.; Jiao, Y.Z.; Lu, Z.J.; Du, S.S.; Liu, X.D.; Shahid, M.Q. Identification of stable pollen development related reference genes for accurate qRT-PCR analysis and morphological variations in autotetraploid and diploid rice. PLoS ONE 2021, 16, e0253244. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shahid, M.Q.; Wen, M.S.; Chen, S.L.; Yu, H.; Jiao, Y.M.; Lu, Z.J.; Li, Y.J.; Liu, X.D. Global identification and analysis revealed differentially expressed lncRNAs associated with meiosis and low fertility in autotetraploid rice. BMC Plant Biol. 2020, 20, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, D.T.; Yuan, L.P.; Lu, X.G. A new strategy of rice breeding in the 21st century: Searching a new pathway of rice breeding by utilization of double heterosis of wide cross and polyploidization. Acta Agron Sin. 2001, 1, 110–116. [Google Scholar]
- Cai, D.T.; Chen, J.G.; Chen, D.L.; Cheng, D.B.; Zhang, W.; Song, Z.J.; Yang, Z.F.; Du, C.Q.; Tang, Z.Q.; He, Y.C.; et al. The breeding of two polyploid rice lines with the characteristic of polyploid meiosis stability. Sci. China Ser. C Life Sci. 2007, 50, 356–366. [Google Scholar] [CrossRef]
- Luan, L.; Long, W.; Wang, X.; Chen, Y.; Liu, Y.; Tu, S.; Kong, F.; Li, W. Agronomic traits and cytogenetic behaviors of autotetraploid rice restorers with high seed set. Chin. J. Appl. Environ. Biol. 2008, 14, 11–17. [Google Scholar]
- Chen, L.; Guo, H.B.; Chen, S.L.; Yang, H.J.; Ghouri, F.; Shahid, M.Q. Comparative study on cytogenetics and transcriptome between diploid and autotetraploid rice hybrids harboring double neutral genes. PLoS ONE 2020, 15, e0239377. [Google Scholar] [CrossRef]
- Chen, L.; Yuan, Y.; Wu, J.W.; Chen, Z.X.; Wang, L.; Shahid, M.Q.; Liu, X.D. Carbohydrate metabolism and fertility related genes high expression levels promote heterosis in autotetraploid rice harboring double neutral genes. Rice 2019, 12, 34. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Shahid, M.Q.; Li, Q.H.; Li, Y.D.; Li, C.; Lu, Z.J.; Wu, J.W.; Zhang, Z.M.; Liu, X.D. Production assessment and genome comparison revealed high yield potential and novel specific alleles associated with fertility and yield in neo-tetraploid rice. Rice 2020, 13, 32. [Google Scholar] [CrossRef] [PubMed]
- Ghaleb, M.A.A.; Li, C.; Shahid, M.Q.; Yu, H.; Liang, J.H.; Chen, R.X.; Wu, J.W.; Liu, X.D. Heterosis analysis and underlying molecular regulatory mechanism in a wide-compatible neo-tetraploid rice line with long panicles. BMC Plant Biol. 2020, 20, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bei, X.J.; Shahid, M.Q.; Wu, J.W.; Chen, Z.X.; Wang, L.; Liu, X.D. Re-sequencing and transcriptome analysis reveal rich DNA variations and differential expressions of fertility-related genes in neo-tetraploid rice. PLoS ONE 2019, 14, e0214953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamara, N.; Jiao, Y.; Lu, Z.; Aloryi, K.D.; Wu, J.W.; Liu, X.D.; Shahid, M.Q. Cytological observations and bulked-segregant analysis coupled global genome sequencing reveal two genes associated with pollen fertility in tetraploid rice. Int. J. Mol. Sci. 2021, 22, 841. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Li, Q.H.; Li, Y.D.; Yang, H.J.; Lu, Z.J.; Wu, J.W.; Zhang, Z.M.; Shahid, M.Q.; Liu, X.D. Genomics analyses reveal unique classification, population structure and novel allele of neo-tetraploid rice. Rice 2021, 14, 16. [Google Scholar] [CrossRef]
- Huang, X.; Yang, S.; Gong, J.; Zhao, Q.; Feng, Q.; Zhan, Q.; Zhao, Y.; Li, W.; Cheng, B.; Xia, J. Genomic architecture of heterosis for yield traits in rice. Nature 2016, 537, 629–633. [Google Scholar] [CrossRef]
- Kato, S.; Kosaka, H.; Hara, S. On the affinity of rice varieties as shown by the fertility of hybrid plants. J. Dep. Agr. Kyushu Imp. Univ. 1928, 3, 132–147. [Google Scholar]
- Shahid, M.Q.; Chen, F.Y.; Li, H.Y.; Wang, S.Z.; Chen, P.F.; Lin, S.Q.; Liu, X.D.; Lu, Y.G. Double-neutral genes, San and Sbn, for pollen fertility in rice to overcome indica×japonica hybrid sterility. Crop Sci. 2013, 53, 164–176. [Google Scholar] [CrossRef]
- Huang, X.; Yang, S.; Gong, J.; Zhao, Y.; Feng, Q.; Gong, H.; Li, W.; Zhan, Q.; Cheng, B.; Xia, J. Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat Commun. 2015, 6, 6258. [Google Scholar] [CrossRef] [Green Version]
- Yuan, L.P. The strategy for hybrid rice development. Hybrid Rice 2018, 33, 1–2. [Google Scholar]
- Mou, T.M. The research progress and prospects of two-line hybrid rice in China. Chin. Sci. Bull. 2016, 61, 3761–3769. [Google Scholar] [CrossRef] [Green Version]
- Ce, H.Q.; Yong, Q.G.; Cheng, X.M.; Fang, L. Preliminary studies on male-sterile stability in autotetraploid rice with photo-thermal sensitive male-sterility. Acta Agron. Sin. 2004, 30, 183–185. [Google Scholar]
- Zhou, H.; Zhou, M.; Yang, Y.; Li, J.; Zhu, L.; Jiang, D.; Dong, J.; Liu, Q.; Gu, L.; Zhou, L. RNase Z S1 processes Ub L40 mRNAs and controls thermo-sensitive genic male sterility in rice. Nat Commun. 2014, 5, 4884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; He, M.; Li, J.; Chen, L.; Huang, Z.; Zheng, S.; Zhu, L.; Ni, E.; Jiang, D.; Zhao, B. Development of commercial thermo-sensitive genic male sterile rice accelerates hybrid rice breeding using the CRISPR/Cas9-mediated TMS5 editing system. Sci. Rep. 2016, 6, 37395. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.H.; Zuo, B.; Song, Z.J.; Wang, W.; He, Y.C.; Liu, Y.H.; Cai, D.T. Breeding and study of two new photoperiod-and thermo-sensitive genic male sterile lines of polyploid rice (Oryza sativa L.). Sci. Rep. 2017, 7, 14744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Z.; Zhou, Y.; Tang, X.; Zhao, X.; Zhou, Z.; Fu, X.; Wang, K.; Shi, J.; Li, Y.; Fu, C. Construction of tms5 mutants in rice based on CRISPR/Cas9 technology. Acta Agron. Sin. 2018, 44, 844–851. [Google Scholar] [CrossRef]
- Wu, M.; Lin, Y.; Liu, H.; Chen, J.; Fu, Y.; Yang, S.; Wang, F. Development of thermo-sensitive male sterile rice with CRISPR/Cas9 technology. Fujian J. Agric. Sci. 2018, 33, 1011–1015. [Google Scholar]
- Chen, R.R.; Zhou, Y.B.; Wang, D.J.; Zhao, X.H.; Tang, X.D.; Xu, S.C.; Tang, Q.Y.; Fu, X.X.; Wang, K.; Liu, X.M. CRISPR/Cas9-mediated editing of the thermo-sensitive genic male-sterile gene TMS5 in rice. Acta Agron. Sin. 2020, 46, 1157–1165. [Google Scholar]
- Barman, H.N.; Sheng, Z.; Fiaz, S.; Zhong, M.; Wu, Y.; Cai, Y.; Wang, W.; Jiao, G.; Tang, S.; Wei, X. Generation of a new thermo-sensitive genic male sterile rice line by targeted mutagenesis of TMS5 gene through CRISPR/Cas9 system. BMC Plant Biol. 2019, 19, 109. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.H.; Lu, Y.G.; Liu, X.D. Cytological mechanism of pollen abortion in photoperiod-temperature sensitive genic male sterile line Pei’ai 64S in rice (Oryza sativa L.). Chin. J. Rice Sci. 2000, 14, 7–14. [Google Scholar]
- Peng, G.Q.; He, Y.; Wang, M.M.; Ashraf, M.F.; Liu, Z.L.; Zhuang, C.X.; Zhou, H. The structural characteristics and the substrate recognition properties of RNase ZS1. Plant Physiol. Biochem. 2021, 158, 83–90. [Google Scholar]
- Jin, J.; Gui, S.T.; Li, Q.; Wang, Y.; Zhang, H.Y.; Zhu, Z.X.; Chen, H.; Sun, Y.Y.; Zou, Y.; Huang, X.G. The transcription factor GATA10 regulates fertility conversion of a two-line hybrid tms5 mutant rice via the modulation of UbL40 expression. J. Integr. Plant Biol. 2020, 62, 1034–1056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghouri, F.; Zhu, J.N.; Yu, H.; Wu, J.W.; Baloch, F.S.; Liu, L.; Shahid, M.Q. Deciphering global DNA variations and embryo sac fertility in autotetraploid rice line. Turk. J. Agric. For. 2019, 43, 554–568. [Google Scholar] [CrossRef] [Green Version]


| Material Name | PH (cm) | PL (cm) | EP | TGP | FGP | SS (%) | GYP (g) | KGW(g) |
|---|---|---|---|---|---|---|---|---|
| H2 | 94.67 ± 4.16 | 24.98 ± 2.33 | 4.40 ± 1.14 | 413.40 ± 97.23 | 285.20 ± 68.98 | 69.06 ± 4.58 | 10.68 ± 2.47 | 37.54 ± 2.52 |
| H2s × T437 | 101.33 ± 5.91 | 28.33 ± 1.65 | 13.33 ± 3.68 | 1546.00 ± 305.15 | 1180.00 ± 192.17 | 77.02 ± 4.36 | 39.60 ± 7.23 | 33.81 ± 4.30 |
| T437 | 94.60 ± 8.71 | 24.72 ± 0.73 | 5.00 ± 1.58 | 332.80 ± 108.98 | 65.00 ± 30.49 | 19.00 ± 5.84 | 2.83 ± 1.54 | 43.09 ± 6.82 |
| H2s × T445 | 97.80 ± 3.11 | 26.60 ± 0.98 | 6.60 ± 1.14 | 820.40 ± 264.19 | 536.00 ± 195.01 | 65.10 ± 7.67 | 17.10 ± 5.65 | 32.27 ± 1.93 |
| T445 | 90.40 ± 4.04 | 26.00 ± 1.80 | 5.20 ± 1.92 | 670.20 ± 229.73 | 297.20 ± 145.92 | 42.31 ± 7.99 | 10.21 ± 5.10 | 33.93 ± 4.47 |
| H2s × T473 | 104.33 ± 2.87 | 31.57 ± 0.42 | 9.67 ± 2.49 | 1042.67 ± 119.67 | 769.00 ± 113.39 | 73.46 ± 2.53 | 27.23 ± 4.67 | 35.32 ± 1.31 |
| T473 | 97.80 ± 3.19 | 25.52 ± 1.82 | 5.20 ± 1.64 | 566.60 ± 166.95 | 239.60 ± 49.94 | 44.27 ± 9.95 | 6.91 ± 1.22 | 29.04 ± 2.21 |
| H3 | 94.40 ± 1.14 | 24.08 ± 1.19 | 6.00 ± 1.22 | 541.20 ± 128.30 | 373.60 ± 78.64 | 69.49 ± 3.79 | 11.62 ± 2.53 | 31.19 ± 2.75 |
| H3s × T423 | 105.00 ± 2.35 | 23.92 ± 0.74 | 7.00 ± 2.00 | 1116.40 ± 421.80 | 840.40 ± 394.50 | 72.14 ± 17.07 | 31.11 ± 14.83 | 36.90 ± 0.55 |
| T423 | 86.00 ± 3.39 | 16.62 ± 0.33 | 5.60 ± 1.52 | 294.20 ± 51.22 | 51.80 ± 28.24 | 16.82 ± 0.92 | 1.42 ± 0.85 | 26.88 ± 4.50 |
| H3s × T424 | 97.00 ± 2.30 | 26.00 ± 2.72 | 9.00 ± 1.52 | 1429.00 ± 122.32 | 968.00 ± 59.72 | 67.74 ± 1.02 | 38.19 ± 3.54 | 39.45 ± 3.57 |
| T424 | 77.00 ± 7.87 | 23.14 ± 0.77 | 2.20 ± 0.45 | 176.40 ± 26.08 | 16.20 ± 6.76 | 9.08 ± 3.39 | 0.51 ± 0.23 | 31.85 ± 2.59 |
| H3s × T445 | 103.16 ± 3.86 | 28.22 ± 1.92 | 8.52 ± 2.11 | 1384.76 ± 797.14 | 1089.64 ± 691.34 | 77.45 ± 6.45 | 35.70 ± 19.83 | 33.58 ± 2.55 |
| T445 | 90.40 ± 4.04 | 26.00 ± 1.80 | 5.20 ± 1.92 | 670.20 ± 229.73 | 297.20 ± 145.92 | 42.31 ± 7.99 | 10.21 ± 5.10 | 33.93 ± 4.47 |
| H3s × T473 | 109.20 ± 4.87 | 30.76 ± 0.23 | 7.40 ± 1.52 | 1278.00 ± 421.72 | 893.60 ± 261.40 | 70.97 ± 7.92 | 32.09 ± 9.09 | 36.05 ± 1.70 |
| T473 | 97.80 ± 3.19 | 25.52 ± 1.82 | 5.20 ± 1.64 | 566.60 ± 166.95 | 239.60 ± 49.94 | 44.27 ± 9.95 | 6.91 ± 1.22 | 29.04 ± 2.21 |
| H3s × T485 | 107.76 ± 7.71 | 29.66 ± 0.92 | 9.20 ± 2.39 | 1241.24 ± 328.23 | 876.16 ± 242.68 | 69.95 ± 2.34 | 27.76 ± 6.86 | 32.01 ± 1.47 |
| T485 | 82.60 ± 1.52 | 27.94 ± 1.18 | 6.40 ± 1.14 | 498.40 ± 88.17 | 294.20 ± 66.86 | 58.64 ± 4.81 | 9.88 ± 2.37 | 33.47 ± 1.33 |
| Material Name | Biological Yield (Kg) | Economic Yield (Kg) | Harvest Index (%) |
|---|---|---|---|
| H2s × T437 (F2) | 6.93 ± 2.01 | 1.28 ± 0.12 | 19.12 ± 3.39 |
| H2s × T437 (F3) | 7.07 ± 1.79 | 1.29 ± 0.19 | 18.54 ± 2.03 |
| H2s × T445 (F2) | 5.90 ± 1.21 | 1.62 ± 0.25 | 27.79 ± 2.84 |
| H2s × T445 (F3) | 5.99 ± 1.22 | 1.53 ± 0.19 | 25.85 ± 1.97 |
| H2s × T473 (F2) | 7.20 ± 0.69 | 1.55 ± 0.20 | 21.55 ± 1.77 |
| H2s × T473 (F3) | 7.75 ± 1.01 | 1.45 ± 0.03 | 18.96 ± 2.75 |
| H3s × T423 (F2) | 7.42 ± 0.99 | 1.25 ± 0.23 | 16.90 ± 2.30 |
| H3s × T423 (F3) | 5.82 ± 0.38 | 0.96 ± 0.05 | 16.43 ± 0.74 |
| H3s × T424 (F2) | 7.58 ± 1.28 | 1.42 ± 0.43 | 18.73 ± 5.54 |
| H3s × T424 (F3) | 5.82 ± 0.38 | 0.96 ± 0.05 | 16.43 ± 0.74 |
| H3s × T445 (F2) | 7.35 ± 1.00 | 1.68 ± 0.16 | 23.02 ± 1.66 |
| H3s × T445 (F3) | 6.99 ± 1.38 | 1.55 ± 0.18 | 22.56 ± 3.97 |
| H3s × T473 (F2) | 8.62 ± 0.68 | 1.51 ± 0.12 | 17.59 ± 0.88 |
| H3s × T473 (F3) | 6.59 ± 1.05 * | 1.15 ± 0.15 * | 17.51 ± 0.60 |
| H3s × T485 (F2) | 6.82 ± 1.51 | 1.83 ± 0.30 | 27.14 ± 2.33 |
| H3s × T485 (F3) | 6.47 ± 1.02 | 2.34 ± 0.13 | 36.60 ± 4.08 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Shahid, M.Q.; Wu, J.; Deng, R.; Chen, Z.; Wang, L.; Liu, G.; Zhou, H.; Liu, X. Thermo-Sensitive Genic Male Sterile Lines of Neo-Tetraploid Rice Developed through Gene Editing Technology Revealed High Levels of Hybrid Vigor. Plants 2022, 11, 1390. https://doi.org/10.3390/plants11111390
Chen Y, Shahid MQ, Wu J, Deng R, Chen Z, Wang L, Liu G, Zhou H, Liu X. Thermo-Sensitive Genic Male Sterile Lines of Neo-Tetraploid Rice Developed through Gene Editing Technology Revealed High Levels of Hybrid Vigor. Plants. 2022; 11(11):1390. https://doi.org/10.3390/plants11111390
Chicago/Turabian StyleChen, Yang, Muhammad Qasim Shahid, Jinwen Wu, Ruilian Deng, Zhixiong Chen, Lan Wang, Guoqiang Liu, Hai Zhou, and Xiangdong Liu. 2022. "Thermo-Sensitive Genic Male Sterile Lines of Neo-Tetraploid Rice Developed through Gene Editing Technology Revealed High Levels of Hybrid Vigor" Plants 11, no. 11: 1390. https://doi.org/10.3390/plants11111390
APA StyleChen, Y., Shahid, M. Q., Wu, J., Deng, R., Chen, Z., Wang, L., Liu, G., Zhou, H., & Liu, X. (2022). Thermo-Sensitive Genic Male Sterile Lines of Neo-Tetraploid Rice Developed through Gene Editing Technology Revealed High Levels of Hybrid Vigor. Plants, 11(11), 1390. https://doi.org/10.3390/plants11111390








