Genetic Mapping to Detect Stringent QTLs Using 1k-RiCA SNP Genotyping Platform from the New Landrace Associated with Salt Tolerance at the Seedling Stage in Rice
Abstract
:1. Introduction
2. Results
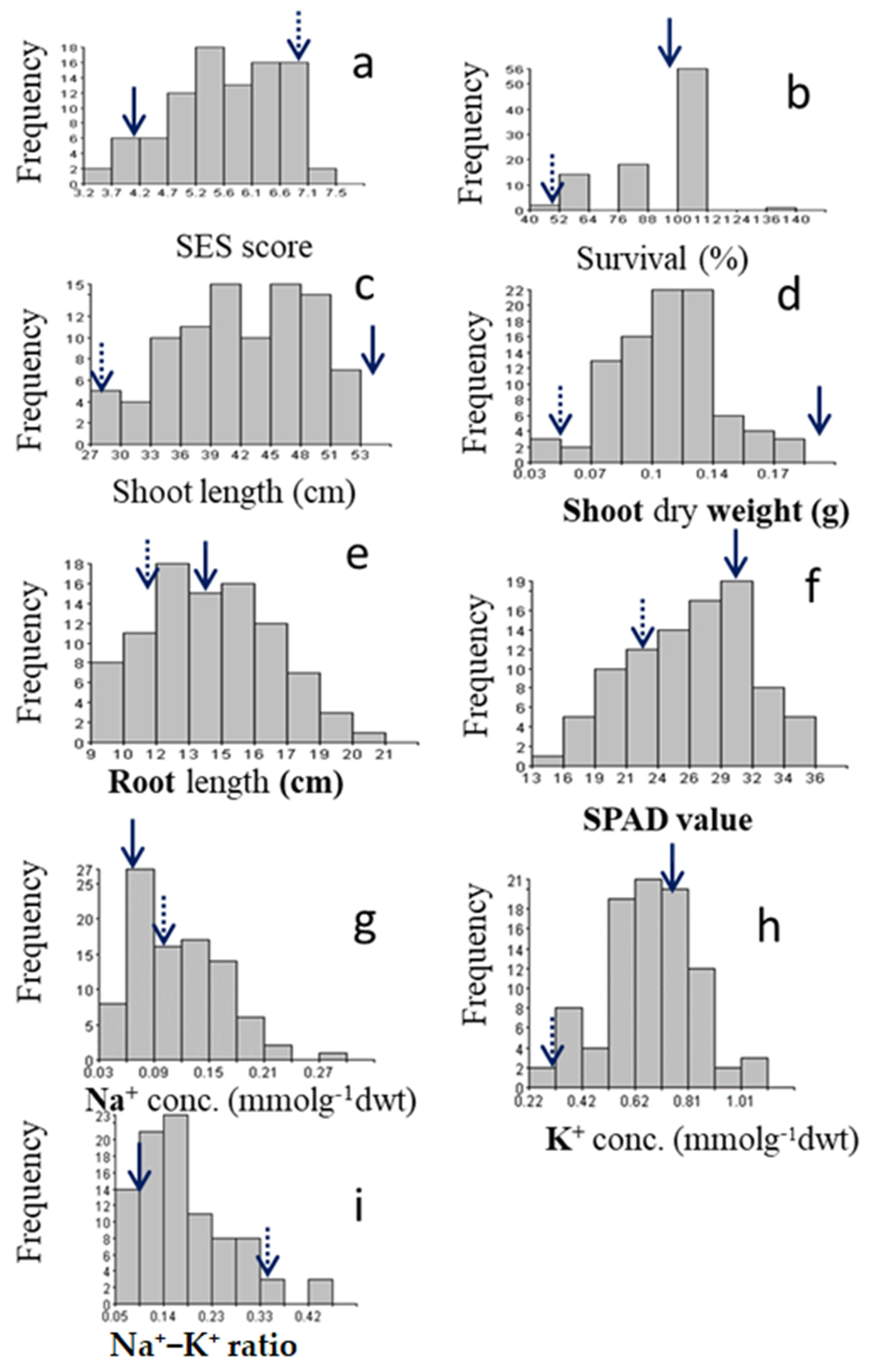
2.1. Investigating the Salt Stress Responses
2.1.1. Characterizing Agronomic Traits under Salt Stress
2.1.2. Physiological Traits under Salt Stress
2.2. Correlation Analysis between Traits
2.3. Path Analysis to Assess the Contribution of Traits (Independent Variables) to Salt Tolerance (Dependent Variable) through Partitioning
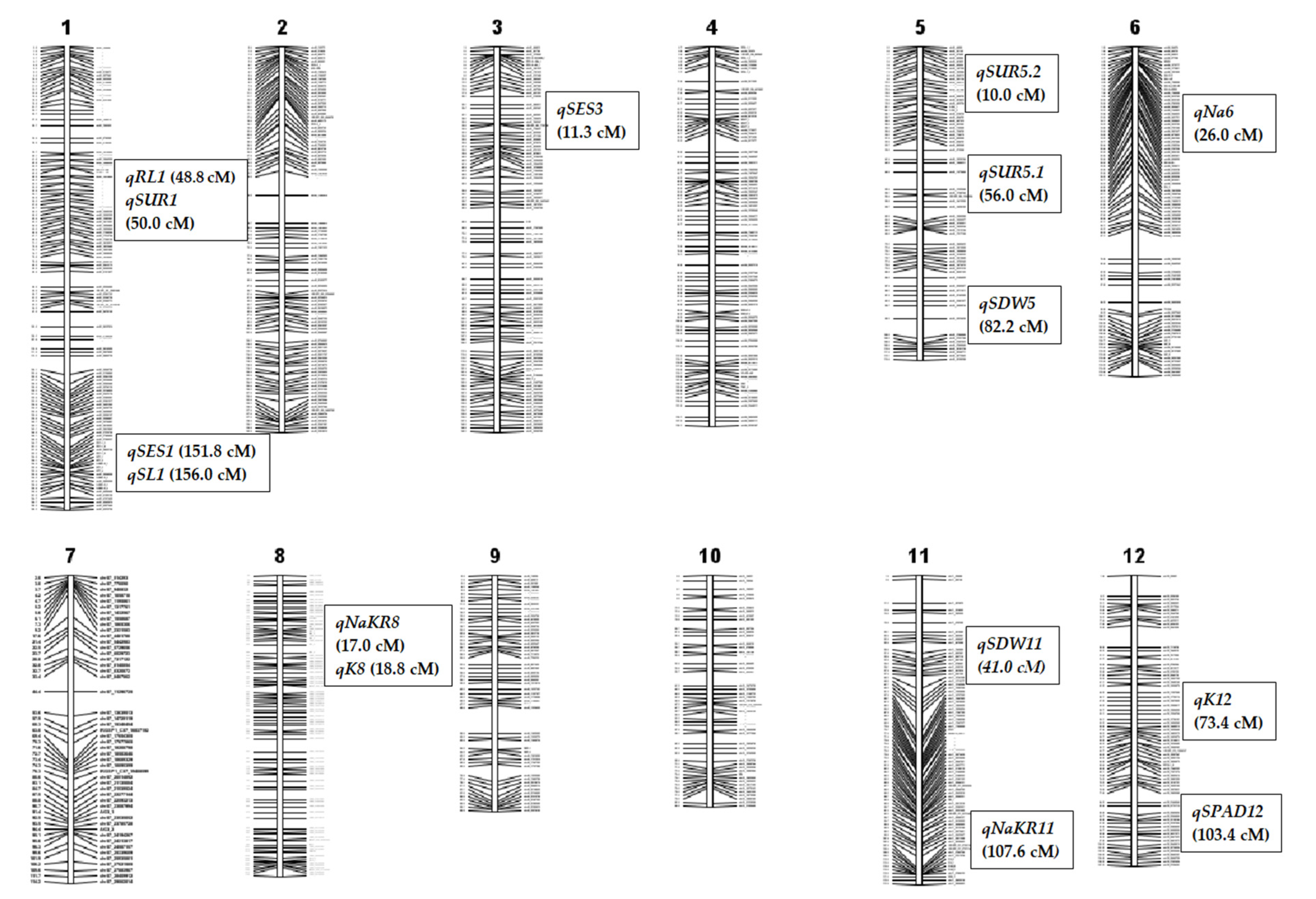
2.4. Marker Segregation and Genetic Linkage Mapping
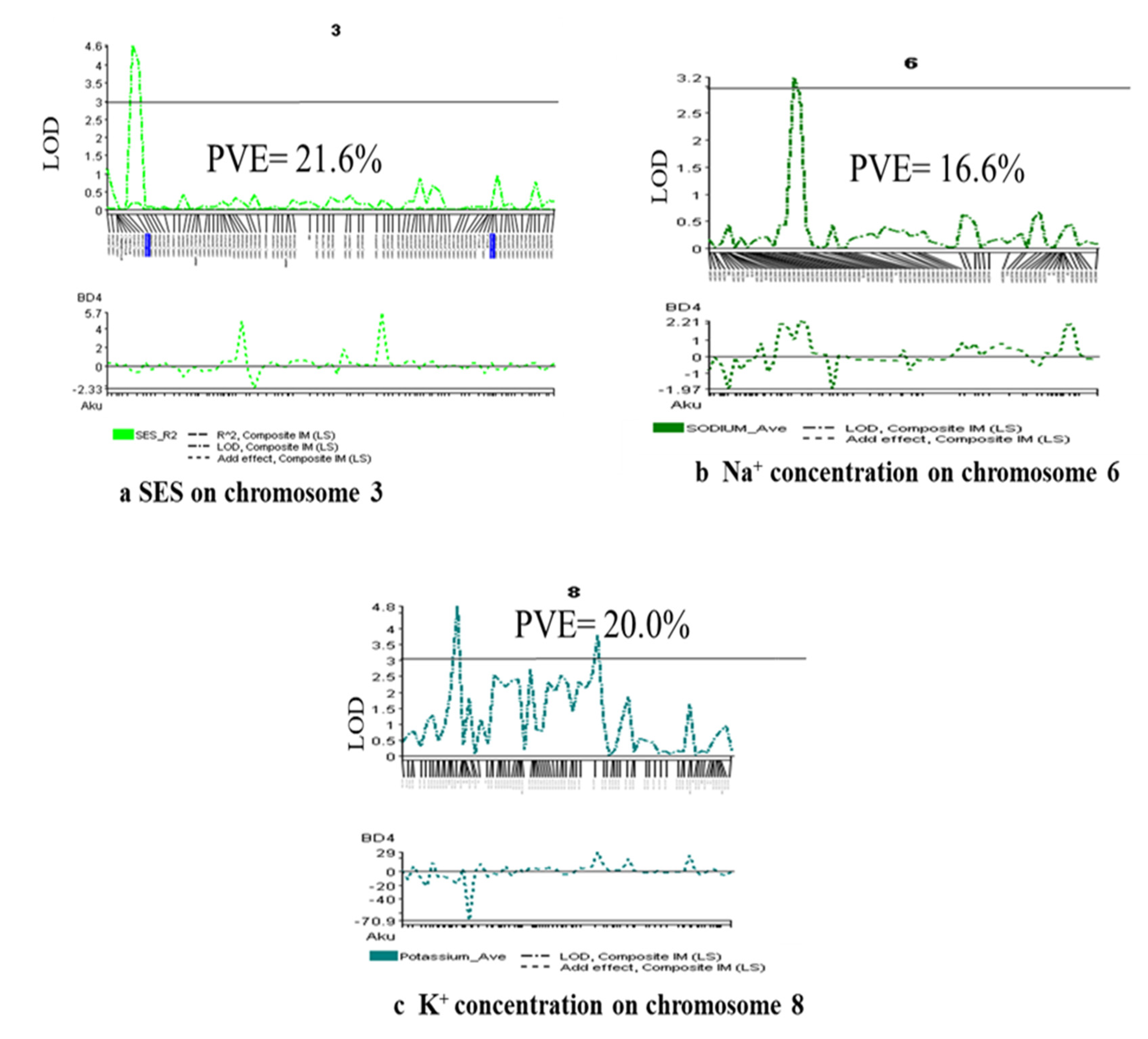
2.5. Salinity Tolerance QTL for Component Agronomic Traits
2.6. QTL for Physiological Traits
2.7. Identification of Functional Genes in the QTL Region
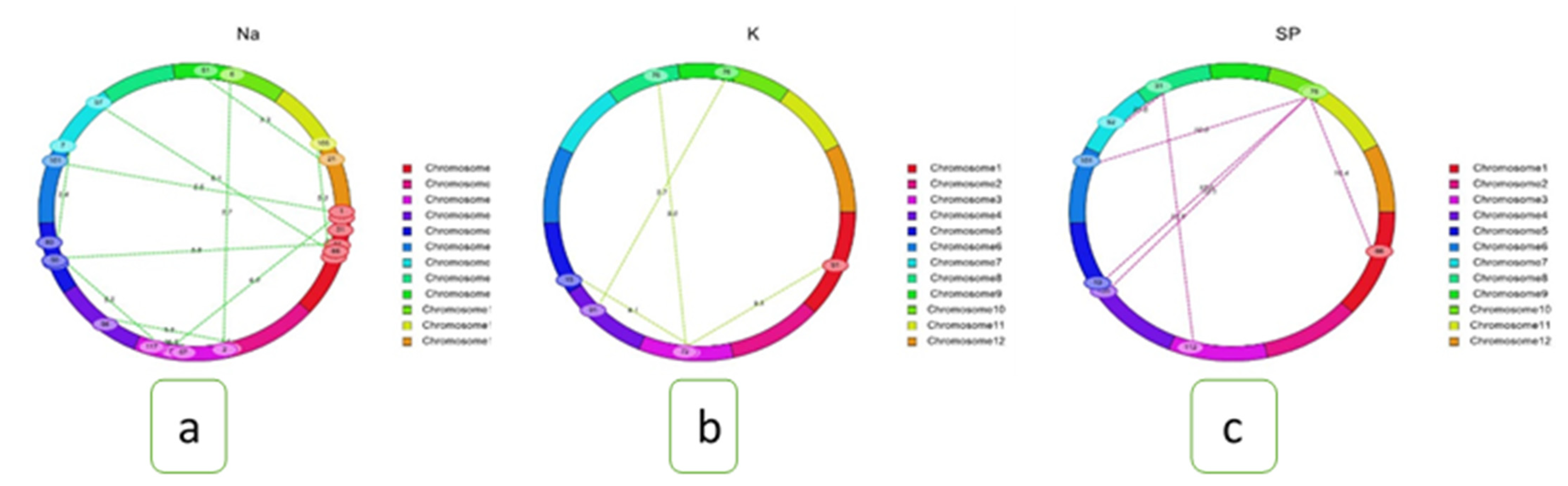
2.8. Epistasis Interaction
3. Discussion
3.1. Salt Stress Responses of the Parental Lines and Selected F2:3 Progenies and Path Coefficient Analysis
3.2. Genetic Map Construction and QTL Detection
3.3. Genomic Regions for Salinity Tolerance
3.4. Comparison between New QTL from the Current and Previously Mapped QTLs
4. Materials and Methods
4.1. Parent Selection
4.2. Details of the Cross, Confirmation and Management of the F1s and the Segregating Population
4.3. Growing Conditions
4.4. Characterization of Agronomic Traits
4.5. Physiological Characterization
4.6. SPAD Reading
4.7. Correlation Analysis for Trait Associations
4.8. Path Coefficient Analysis
4.9. Residual Effect
4.10. Genotyping and Construction of a Genetic Linkage Map
4.11. QTL Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Birla, D.S.; Malik, K.; Sainger, M.; Chaudhary, D.; Jaiwal, R.; Jaiwal, P.K. Progress and challenges in improving the nutritional quality of rice (Oryza sativa L.). Crit. Rev. Food Sci. Nutr. 2017, 57, 2455–2481. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.S.; Ali, M.A.; Khan, M.A.I.; Maniruzzaman, S. Prevalence and Transmission of Fusarium moniliforme: A Seed Borne Pathogen of Rice. Bangladesh Rice J. 2020, 24, 11–19. [Google Scholar] [CrossRef]
- Barker, R.; Herdt, R.W.; Rose, B. The Rice Economy of Asia; Routledge: London, UK, 2014. [Google Scholar]
- Mondal, S.; Borromeo, T.H. Screening of salinity tolerance of rice at early seedling stage. J. Biosci. Agric. Res. 2016, 10, 843–847. [Google Scholar] [CrossRef] [Green Version]
- Rahman, M.A.; Khatun, H.; Sarker, M.R.A.; Hossain, H.; Quddus, M.R.; Iftekharuddaula, K.M.; Kabir, M.S. Enhancing abiotic stress tolerance to develop climate-smart rice using holistic breeding approach. Cereal Grains 2021, 2, 91. [Google Scholar] [CrossRef]
- Munns, R. Plant adaptations to salt and water stress: Differences and commonalities. Adv. Bot. Res. 2021, 57, 1–32. [Google Scholar]
- Islam, F.; Wang, J.; Farooq, M.A.; Yang, C.; Jan, M.; Mwamba, T.M.; Hannan, F.; Xu, L.; Zhou, W. Rice responses and tolerance to salt stress: Deciphering the physiological and molecular mechanisms of salinity adaptation. In Advances in Rice Research for Abiotic Stress Tolerance; Woodhead Publishing: Cambridge, UK, 2019; pp. 791–819. [Google Scholar]
- Rahman, M.A.; Thomson, M.J.; De Ocampo, M.; Egdane, J.A.; Salam, M.A.; Ismail, A.M. Assessing trait contribution and mapping novel QTL for salinity tolerance using the Bangladeshi rice landrace Capsule. Rice 2019, 12, 63. [Google Scholar] [CrossRef] [Green Version]
- Rasel, M.; Tahjib-Ul-Arif, M.; Hossain, M.A.; Hassan, L.; Farzana, S.; Brestic, M. Screening of salt-tolerant rice landraces by seedling stage phenotyping and dissecting biochemical determinants of tolerance mechanism. J. Plant Growth Regul. 2021, 40, 1853–1868. [Google Scholar] [CrossRef]
- Al Tamimi, N.; Brien, C.; Oakey, H.; Berger, B.; Saade, S.; Ho, Y.S.; Schmöckel, S.M.; Tester, M.; Negrão, S. Data from: Salinity Tolerance Loci Revealed in Rice Using High-Throughput Non-Invasive Phenotyping. Available online: https://datadryad.org/stash/dataset/doi:10.5061/dryad.3118j (accessed on 22 April 2022).
- Zeng, L.; Shannon, M.C.; Lesch, S.M. Timing of salinity stress affects rice growth and yield components. Agric. Water Manag. 2001, 48, 191–206. [Google Scholar] [CrossRef]
- Hossain, H.; Rahman, M.A.; Alam, M.S.; Singh, R.K. Mapping of quantitative trait loci associated with reproductive-stage salt tolerance in rice. J. Agron. Crop Sci. 2015, 201, 17–31. [Google Scholar] [CrossRef]
- Ahmadizadeh, M.; Vispo, N.A.; Calapit-Palao, C.D.O.; Pangaan, I.D.; Viña, C.D.; Singh, R.K. Reproductive stage salinity tolerance in rice: A complex trait to phenotype. Indian J. Plant Physiol. 2016, 21, 528–536. [Google Scholar] [CrossRef]
- Rahman, M.A.; Thomson, M.; Shah-E-Alam, M.; De Ocampo, M.; Egdane, J.; Ismail, A.M. Exploring novel genetic sources of salinity tolerance in rice through molecular and physiological characterization. Ann. Bot. 2016, 117, 1083–1097. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.R.; Hassan, L.; Salam, M.A.; Collard, B.C.Y.; Singh, R.K.; Gregorio, G.B. QTL mapping for salinity tolerance at seedling stage in rice. Emir. J. Food Agric. 2021, 23, 137–146. [Google Scholar] [CrossRef]
- Wang, Z.; Cheng, J.; Chen, Z.; Huang, J.; Bao, Y.; Wang, J.; Zhang, H. Identification of QTLs with main, epistatic and QTL× environment interaction effects for salt tolerance in rice seedlings under different salinity conditions. Theor. Appl. Genet. 2012, 125, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, R.; Mendioro, M.S.; Diaz, G.Q.; Gregorio, G.B.; Singh, R.K. Mapping quantitative trait loci associated with yield and yield components under reproductive stage salinity stress in rice (Oryza sativa L.). J. Genet. 2013, 92, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Nayyeripasand, L.; Garoosi, G.A.; Ahmadikhah, A. Genome-Wide Association Study (GWAS) to Identify Salt-Tolerance QTLs Carrying Novel Candidate Genes in Rice During Early Vegetative Stage. Rice 2021, 14, 9. [Google Scholar] [CrossRef]
- Thomson, M.J.; de Ocampo, M.; Egdane, J.; Rahman, M.A.; Sajise, A.G.; Adorada, D.L.; Tumimbang, E.; Blumwald, E.; Seraj, Z.; ZebaIsmail, A.M. Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice 2010, 3, 148–160. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.; Shen, Q.; Kuang, L.; Yu, J.; Wu, D.; Zhang, G. Metabolite profiling and gene expression of Na/K transporter analyses reveal mechanisms of the difference in salt tolerance between barley and rice. Plant Physiol. Biochem. 2018, 130, 248–257. [Google Scholar] [CrossRef]
- De Souza Freitas, W.E.; de Oliveira, A.B.; Mesquita, R.O.; de Carvalho, H.H.; Prisco, J.T.; Gomes-Filho, E. Sulfur-induced salinity tolerance in lettuce is due to a better P and K uptake, lower Na/K ratio and an efficient antioxidative defense system. Sci. Hortic. 2019, 257, 108764. [Google Scholar] [CrossRef]
- Mackay, T.F.; Huang, W. Charting the genotype–phenotype map: Lessons from the Drosophila melanogaster Genetic Reference Panel. Wiley Interdiscip. Rev. Dev. Biol. 2018, 7, e289. [Google Scholar] [CrossRef]
- Harushima, Y.; Yano, M.; Shomura, A.; Sato, M.; Shimano, T.; Kuboki, Y.; Yamamoto, T.; Lin, S.Y.; Antonio, B.A.; Parco, A.; et al. A highdensity rice.genetic linkage map with 2275 markers using a single F2 population. Genetics 1998, 148, 479–494. [Google Scholar] [CrossRef] [PubMed]
- Kosambi, D.D. The estimation of map distance from recombination values. Ann. Eugen. 1944, 12, 172–175. [Google Scholar] [CrossRef]
- Ke, Y.; Han, G.; He, H.; Li, J. Differential regulation of proteins and phosphoproteins in rice under drought stress. Biochem. Biophys. Res. Commun. 2009, 379, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Sarhadi, E.; Bazargani, M.M.; Sajise, A.G.; Abdolahi, S.; Vispo, N.A.; Arceta, M.; Mohammadi-Nejad, G.; Singh, R.K.; Salekdeh, G.H. Proteomic analysis of rice anthers under salt stress. Plant Physiol. Biochem. 2012, 58, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhao, X.; Pan, Y.; Zhu, L.; Fu, B.; Li, Z. DNA methylation changes detected by methylation-sensitive amplified polymorphism in two contrasting rice genotypes under salt stress. J. Genet. Genom. 2011, 38, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Cortés, T.; Pomar, F.; Novo-Uzal, E. Evolutionary Implications of a Peroxidase with High Affinity for Cinnamyl Alcohols from Physcomitrium patens, a Non-Vascular Plant. Plants 2021, 10, 1476. [Google Scholar] [CrossRef]
- Lelandais-Brière, C.; Jovanovic, M.; Torres, G.A.M.; Perrin, Y.; Lemoine, R.; Corre-Menguy, F.; Hartmann, C. Disruption of AtOCT1, an organic cation transporter gene, affects root development and carnitine-related responses in Arabidopsis. Plant J. 2007, 51, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Strohm, A.K.; Vaughn, L.M.; Masson, P.H. Natural variation in the expression of ORGANIC CATION TRANSPORTER 1 affects root length responses to cadaverine in Arabidopsis. J. Exp. Bot. 2015, 66, 853–862. [Google Scholar] [CrossRef] [Green Version]
- Radwan, A.; Ali, R.M.I.A.; Nada, A.; Hashem, W.; Assem, S.; Husein, E. Isolation and characterization of some drought-related ESTs from barley. Afr. J. Biotechnol. 2015, 14, 794–810. [Google Scholar]
- Grandbastien, M.-A. Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 1998, 3, 181–187. [Google Scholar] [CrossRef]
- Schapire, A.L.; Voigt, B.; Jasik, J.; Rosado, A.; Lopez-Cobollo, R.; Menzel, D.; Salinas, J.; Mancuso, S.; Valpuesta, V.; Baluska, F. Arabidopsis synaptotagmin 1 is required for the maintenance of plasma membrane integrity and cell viability. Plant Cell 2008, 20, 3374–3388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, M.; Pandey, G.K. Expansion and function of repeat domain proteins during stress and development in plants. Front. Plant Sci. 2016, 6, 1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Zhao, X.; Yu, H.; Ouyang, Y.; Wang, L.; Zhang, Q. The ankyrin repeat gene family in rice: Genome-wide identification, classification and expression profiling. Plant Mol. Biol. 2009, 71, 207–226. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Ortiz, C.; Peña-Garcia, Y.; Natarajan, P.; Bhandari, M.; Abburi, V.; Dutta, S.K.; Yadav, L.; Stommel, J.; Nimmakayala, P.; Reddy, U.K. The ankyrin repeat gene family in Capsicum spp: Genome-wide survey, characterization and gene expression profile. Sci. Rep. 2020, 10, 4044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wicker, T.; Guyot, R.; Yahiaoui, N.; Keller, B. CACTA transposons in Triticeae. A diverse family of high-copy repetitive elements. Plant Physiol. 2003, 132, 52–63. [Google Scholar] [CrossRef] [Green Version]
- Rossi, M.; Araujo, P.G.; Van Sluys, M.-A. Survey of transposable elements in sugarcane expressed sequence tags (ESTs). Genet. Mol. Biol. 2001, 24, 147–154. [Google Scholar] [CrossRef]
- Zhu, J.K.; Shi, J.; Bressan, R.A.; Hasegawa, P.M. Expression of an Atriplex nummularia gene encoding a protein homologous to the bacterial molecular chaperone DnaJ. Plant Cell 1993, 5, 341–349. [Google Scholar]
- Zhong, X.; Yang, J.; Shi, Y.; Wang, X.; Wang, G.L. The DnaJ protein OsDjA6 negatively regulates rice innate immunity to the blast fungus Magnaporthe oryzae. Mol. Plant Pathol. 2018, 19, 607–614. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Guo, Y.; Wu, C.; Yang, G.; Li, Y.; Zheng, C. Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genom. 2008, 9, 44. [Google Scholar] [CrossRef] [Green Version]
- De Leon, T.B.; Linscombe, S.; Subudhi, P.K. Identification and validation of QTLs for seedling salinity tolerance in introgression lines of a salt tolerant rice landrace ‘Pokkali’. PLoS ONE 2017, 12, e0175361. [Google Scholar] [CrossRef]
- Lv, W.; Wu, J.; Xu, Z.; Dai, H.; Ma, Z.; Wang, Z. The putative histone-like transcription factor FgHltf1 is required for vegetative growth, sexual reproduction, and virulence in Fusarium graminearum. Curr. Genet. 2019, 65, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Sakrikar, S.; Schmid, A.K. An archaeal histone-like protein regulates gene expression in response to salt stress. Nucleic Acids Res. 2021, 49, 12732–12743. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Lu, C.; Guo, J.; Qiao, Z.; Sui, N.; Qiu, N.; Wang, B. C2H2 zinc finger proteins: Master regulators of abiotic stress responses in plants. Front. Plant Sci. 2020, 11, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Zhang, H.; Allen, R.D. Overexpression of an Arabidopsis peroxisomal ascorbate peroxidase gene in tobacco increases protection against oxidative stress. Plant Cell Physiol. 1999, 40, 725–732. [Google Scholar] [CrossRef] [Green Version]
- Ahmadi, N.; Negrão, S.; Katsantonis, D.; Frouin, J.; Ploux, J.; Letourmy, P.; Droc, G.; Babo, P.; Trindade, H.; Bruschi, G.; et al. Targeted association analysis identified japonica rice varieties achieving Na+/K+ homeostasis without the allelic make-up of the salt tolerant indica variety Nona Bokra. Theor. Appl. Genet. 2011, 123, 881–895. [Google Scholar] [CrossRef]
- Abdelaziz, M.N.; Xuan, T.D.; Mekawy, A.M.M.; Wang, H.; Khanh, T.D. Relationship of Salinity Tolerance to Na+ Exclusion, Proline Accumulation, and Antioxidant Enzyme Activity in Rice Seedlings. Agriculture 2018, 8, 166. [Google Scholar] [CrossRef] [Green Version]
- Ishikawa, T.; Shabala, S. Control of xylem Na+ loading and transport to the shoot in rice and barley as a determinant of differential salinity stress tolerance. Physiol. Plant. 2019, 165, 619–631. [Google Scholar] [CrossRef] [Green Version]
- Rahman, S.; Bimpong, I.K.; Bizimana, J.B.; Pascual, E.D.; Arceta, M.; Swamy, B.P.M.; Diaw, F.; Singh, R.K. Mapping QTLs using a novel source of salinity tolerance from Hasawi and their interaction with environments in rice. Rice 2017, 10, 47. [Google Scholar] [CrossRef] [Green Version]
- Nakhla, W.R.; Sun, W.; Fan, K.; Yang, K.; Zhang, C.; Yu, S. Identification of QTLs for salt tolerance at the germination and seedling stages in rice. Plants 2021, 10, 428. [Google Scholar] [CrossRef]
- Puvanitha, S.; Mahendran, S. Effect of salinity on plant height, shoot and root dry weight of selected rice cultivars. Sch. J. Agric. Vet. Sci. 2017, 4, 126–131. [Google Scholar]
- Pires, I.S.; Negrão, S.; Oliveira, M.M.; Purugganan, M.D. Comprehensive phenotypic analysis of rice (Oryza sativa) response to salinity stress. Physiol. Plant. 2015, 155, 43–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, Y.-C.; Chen, K.-C.; Cheng, T.-S.; Lee, C.; Lin, S.-H.; Tung, C.-W. Chlorophyll fluorescence analysis in diverse rice varieties reveals the positive correlation between the seedlings salt tolerance and photosynthetic efficiency. BMC Plant Biol. 2019, 19, 403. [Google Scholar] [CrossRef] [Green Version]
- Eswaran, R.; Anandan, A. Investigation of correlation between traits and path analysis of rice (Oryza sativa L.) grain yield under coastal salinity. Electron. J. Plant Breed. 2011, 2, 538–542. [Google Scholar]
- Gopikannan, M.; Ganesh, S.K. Inter-relationship and path analysis in rice (Oryza sativa L.) under sodicity. Indian J. Sci. Technol. 2013, 6, 5223–5227. [Google Scholar] [CrossRef]
- Manohara, K.K.; Singh, N.P. Genetic variability, correlation and path analysis in rice (Oryza sativa L.) under coastal salinity conditions of Goa. J. Indian Soc. Coast. Agric. Res. 2015, 33, 34–39. [Google Scholar]
- Rasel; Hassan, L.; Hoque, I.U.; Saha, S.R. Estimation of genetic variability, correlation and path coefficient analysis in local landraces of rice (Oryza sativa L.) for the improvement of salinity tolerance. J. Bangladesh Agric. Univ. 2018, 16, 41–46. [Google Scholar] [CrossRef] [Green Version]
- Hossain, M.; Ivy, N.; Raihan, M.; Kayesh, E.; Maniruzzaman, S. Genetic Variability, Correlation and Path Analysis of Floral, Yield and its Component Traits of Maintainer Lines of Rice (Oryza sativa L.). Bangladesh Rice J. 2021, 24, 1–9. [Google Scholar] [CrossRef]
- Anshori, M.F.; Purwoko, B.S.; Dewi, I.S.; Ardie, S.W.; Suwarno, W.B.; Safitri, H. Determination of selection criteria for screening of rice genotypes for salinity tolerance. SABRAO J. Breed. Genet. 2018, 50, 279–294. [Google Scholar]
- Kiruthikadevi, U.; Banumathy, S.; Arunachalam, P.; Renuka, R.; Thirumurugan, T. Correlation, path analysis and stress indices studies of Saltol introgressed lines of rice for salinity tolerance. Electron. J. Plant Breed. 2020, 11, 230–237. [Google Scholar]
- Rajasekar, R.; Jeyaprakash, P.; Manonmani, K.; Nithila, S.; Thirumurugan, T. Trait relationship and path analysis under sodicity in Nagina 22 rice mutants. Electron. J. Plant Breed. 2021, 12, 963–968. [Google Scholar]
- Ismail, A.M.; Horie, T. Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu. Rev. Plant Biol. 2017, 68, 405–434. [Google Scholar] [CrossRef] [Green Version]
- Tan, C.; Han, Z.; Yu, H.; Zhan, W.; Xie, W.; Chen, X.; Zhao, H.; Zhou, F.; Xing, Y. QTL scanning for rice yield using a whole genome SNP array. J. Genet. Genom. 2013, 40, 629–638. [Google Scholar] [CrossRef] [PubMed]
- Temnykh, S.; Park, W.D.; Ayres, N.; Cartinhour, S.; Hauck, N.; Lipovich, L.; Cho, Y.G.; Ishii, T.; McCouch, S.R. Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Appl. Genet. 2000, 100, 697–712. [Google Scholar] [CrossRef]
- Xing, Y.; Tan, Y.; Hua, J.; Sun, X.; Xu, C.; Zhang, Q. Characterization of the main effects epistatic effects and their environmental interactions of QTLs on the genetic basis of yield traits in rice. Appl. Genet. 2002, 105, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Waziri, A.; Kumar, P.; Purty, R.S. Saltol QTL and their role in salinity tolerance in rice. Austin J. Biotechnol. Bioeng. 2016, 3, 1067. [Google Scholar]
- Krishnamurthy, S.L.; Pundir, P.; Warraich, A.S.; Rathor, S.; Lokeshkumar, B.M.; Singh, N.K.; Sharma, P.C. Introgressed Saltol QTL lines improves the salinity tolerance in rice at seedling stage. Front. Plant Sci. 2020, 11, 833. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, E.; Negm, M.; Abdelrahman, M. Molecular Analysis For Salt Tolerance QTLs Emphasizing Saltol QTL in Some Egyptian and International Rice Genotypes (Oryza sativa L.). Egypt. Acad. J. Biol. Sci. C Physiol. Mol. Biol. 2020, 12, 57–69. [Google Scholar] [CrossRef]
- Manohara, K.K.; Morajkar, S.; Shanbhag, Y.; Phadte, P.; Singh, N.K. Haplotype analysis of Saltol QTL region in diverse landraces, wild rice and introgression lines of rice (Oryza sativa L.). Plant Genet. Resour. 2021, 19, 289–298. [Google Scholar] [CrossRef]
- Emon, R.M.; Islam, M.M.; Halder, J.; Fan, Y. Genetic diversity and association mapping for salinity tolerance in Bangladeshi rice landraces. Crop J. 2015, 3, 440–444. [Google Scholar] [CrossRef] [Green Version]
- Thuy, N.T.T.; Tokuyasu, M.; Mai, N.S.; Hirai, Y. Identification and characterization of chromosome regions associated with salinity tolerance in rice. J. Agric. Sci. 2018, 10, 57–68. [Google Scholar] [CrossRef] [Green Version]
- Xie, L.; Zheng, C.; Li, W.; Pu, M.; Zhou, G.; Sun, W.; Wu, X.; Zhao, X.; Xie, X. Mapping and Identification a Salt-Tolerant QTL in a Salt-Resistant Rice Landrace, Haidao86. J. Plant Growth Regul. 2021, 1–12. [Google Scholar] [CrossRef]
- Mondal, S.; Borromeo, T.H.; Diaz, M.G.Q.; Amas, J.; Rahman, M.A.; Thomson, M.; Gregorio, G.B. Dissecting QTLs for Reproductive Stage Salinity Tolerance in Rice from BRRI dhan47. Plant Breed. Biotechnol. 2019, 7, 302–312. [Google Scholar] [CrossRef]
- Jahan, N.; Zhang, Y.; Lv, Y.; Song, M.; Zhao, C.; Hu, H.; Cui, Y.; Wang, Z.; Yang, S.; Zhang, A.; et al. QTL analysis for rice salinity tolerance and fine mapping of a candidate locus qSL7 for shoot length under salt stress. Plant Growth Regul. 2019, 90, 307–319. [Google Scholar] [CrossRef] [Green Version]
- O’Connor, K.; Hayes, B.; Hardner, C.; Nock, C.; Baten, A.; Alam, M.; Henry, R.; Topp, B. Genome-wide association studies for yield component traits in a macadamia breeding population. BMC Genom. 2020, 21, 199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- BRRI. Adhunik Dhaner Chash, 23rd ed.; Bangladesh Rice Research Institute: Gazipur, Bangladesh, 2020. [Google Scholar]
- Yoshida, S.; Forno, D.A.; Cock, J.H. Laboratory Manual for Physiological Studies of Rice; IRRI: Laguna, Philippines, 1976. [Google Scholar]
- IRRI. Standard Evaluation System for Rice (SES), 5th ed.; International Rice Research Institute: Los Banos, Philippines, 2014; p. 57. [Google Scholar]
- Flowers, T.J.; Yeo, A.R. Variability in the resistance of sodium chloride salinity within rice (Oryza sativa l.) varieties. New Phytol. 1981, 88, 363–373. [Google Scholar] [CrossRef]
- Moradi, F.; Ismail, A.M. Responses of Photosynthesis, Chlorophyll Fluorescence and ROS-Scavenging Systems to Salt Stress During Seedling and Reproductive Stages in Rice. Ann. Bot. 2007, 99, 1161–1173. [Google Scholar] [CrossRef] [Green Version]
- Wright, S. Coefficients of Inbreeding and Relationship. Am. Nat. 1922, 56, 330–338. [Google Scholar] [CrossRef] [Green Version]
- Dewey, D.R.; Lu, K. A correlation and path-coefficient analysis of components of crested wheatgrass seed production 1. Agron. J. 1959, 51, 515–518. [Google Scholar] [CrossRef]
- Arbelaez, J.D.; Dwiyanti, M.S.; Tandayu, E.; Llantada, K.; Jarana, A.; Ignacio, J.C.; Platten, J.D.; Cobb, J.; Rutkoski, J.E.; Thomson, M.J.; et al. 1k-RiCA (1K-Rice Custom Amplicon) a novel genotyping amplicon-based SNP assay for genetics and breeding applications in rice. Rice 2019, 12, 55. [Google Scholar] [CrossRef] [Green Version]
- Joehanes, R.; Nelson, J.C. QGene 4.0, an extensible Java QTL-analysis platform. Bioinformatics 2008, 24, 2788–2789. [Google Scholar] [CrossRef] [Green Version]
- Churchill, G.A.; Doerge, R.W. Empirical threshold values for quantitative trait mapping. Genetics 1994, 138, 963–971. [Google Scholar] [CrossRef] [PubMed]

| Variable | Correlation | Survival Rate | Shoot Length | Shoot Dry Weight | Root Length | SPAD Value | Na+ Conc. | K+ Conc. | Na+/K+ Ratio | Total Effect |
|---|---|---|---|---|---|---|---|---|---|---|
| Survival rate | −0.257 | −0.301 | 0.148 | 0.009 | −0.014 | −0.011 | −0.003 | −0.088 | 0.002 | −0.257 |
| Shoot length | 0.226 | −0.074 | 0.605 | 0.024 | −0.134 | 0.010 | −0.004 | −0.190 | −0.011 | 0.226 |
| Shoot dry weight | 0.141 | −0.059 | 0.304 | 0.047 | −0.096 | −0.008 | −0.010 | −0.083 | 0.046 | 0.141 |
| Root length | −0.306 | −0.011 | 0.208 | 0.012 | −0.390 | −0.008 | −0.001 | −0.103 | −0.012 | −0.306 |
| SPAD value | −0.310 | −0.046 | −0.089 | 0.006 | −0.046 | −0.068 | −0.002 | −0.058 | −0.005 | −0.310 |
| Na+ conc. | 0.114 | −0.060 | 0.151 | 0.028 | −0.026 | −0.007 | −0.016 | −0.044 | 0.088 | 0.114 |
| K+ conc. | −0.183 | −0.090 | 0.388 | 0.013 | −0.136 | −0.013 | −0.002 | −0.296 | −0.045 | −0.183 |
| Na+/K+ ratio | 0.218 | −0.007 | −0.060 | 0.019 | 0.042 | 0.003 | −0.013 | 0.120 | 0.112 | 0.218 |
| Trait | QTL | Chr. | Peak Marker | QTL Position (cM) | Add Effect | LOD | PVE (%) | Method of QTL Detection | Contributor of Favorable Allele | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| QGene | ICIM | QGene | ICIM | QGene | ICIM | |||||||
| SES | qSES1 | 1 | Chr01_38632196 | 151.8 | - | −0.61 | 3.4 | - | 15.6 | 3.80 | SMR | Akundi |
| qSES3 | 3 | Chr03_2885423 | 11.3 | - | −0.70 | 4.8 | - | 21.6 | 4.50 | SMR, CIM | Akundi | |
| Survival rate (%) | qSUR1 | 1 | chr01_12941687 | 50 | 54.8 | 31.91 | 4.3 | 5.0 | 19.9 | 0.98 | IM, CIM | BR49 |
| qSUR5.1 | 5 | chr05_14412355 | 56 | - | 20.25 | 7.5 | - | 31.6 | - | IM, CIM | BR49 | |
| qSUR5.2 | 5 | chr05_2291156 | 10 | 11.2 | −10.36 | 3.1 | 5.0 | 14.9 | 0.94 | IM, CIM | Akundi | |
| Shoot length | qSL1 | 1 | QSES1-2_2 | 156 | 151.8 | 4.86 | 7.2 | 4.0 | 30.7 | 16.28 | IM, CIM | BR49 |
| Shoot dry weight | qSDW 5 | 5 | chr05_20551103 | 82.2 | - | 0.35 | 3.5 | - | 16.3 | - | IM, CIM | BR49 |
| qSDW11 | 11 | Chr11_10741559 | 41.0 | - | 0.27 | 3.0 | - | 14 | - | IM, CIM | BR49 | |
| Root length | qRL1 | 1 | Chr01_12335190 | 48.8 | - | −1.59 | 3.3 | - | 15.8 | - | IM | Akundi |
| SPAD | qSPAD12 | 12 | Chr12_26259494 | 103.4 | - | 3.05 | 3.5 | - | 16.3 | - | IM, CIM | BR49 |
| Na+ conc. | qNa6 | 6 | SSIIA-3B | 26 | 27 | −1.98 | 3.6 | 4.0 | 16.8 | 8.18 | CIM, IM | Akundi |
| K+ conc. | qK8 | 8 | chr08_4853081 | 18.8 | - | −0.34 | 4.7 | - | 21.2 | - | IM, CIM | Akundi |
| qK12 | 12 | Chr12_18881059 | 73.4 | - | 11.4 | 3.5 | - | 16.3 | - | IM, CIM, | BR49 | |
| Na+/K+ ratio | qNaKR8 | 8 | DTH8-IR24 | 17 | - | 0.08 | 3.0 | - | 0.1 | - | SMR | BR49 |
| qNaKR11 | 11 | IRGSP1_C11_27391141 | 107.6 | 108 | −0.04 | 3.0 | 3.0 | 12.9 | 16.80 | SMR` | Akundi | |
| Traits | QTL | Chr. | Position (bp) | Total no. of Locus | Candidate Genes | Putative Function | References |
|---|---|---|---|---|---|---|---|
| SES | qSES1 | 1 | 38,723,347–38,724,165 | 16 | LOC_Os01g66670 | Expressed protein (drought-induced proteins, anther and pollen wall remodeling/metabolism proteins contribute to the tolerance of rice to salt stress). | [25,26] |
| qSES3 | 3 | 2,878,828–2,880,890 | 20 | LOC_Os03g05770 | Peroxidase precursor, putative, expressed (increases protection against oxidative stress and is highly tolerant to different stresses, allowing survival when water supply is a limiting factor). | [27,28] | |
| Survival rate (%) | qSUR1 | 1 | 12,756,935–12,757,588 | 17 | LOC_Os01g22700 | Organic cation transporter-related, putative, expressed (affects root development, carnitine-related responses under stress, and numerous biological processes, including transcription, translation, cell signaling, and ion channel activity). | [29,30] |
| qSUR5.1 | 5 | 14,280,156–14,292,389 | 10 | LOC_Os05g24680 | Retrotransposon protein, putative, Ty3-gypsy subclass, expressed (enable plants to cope with drought stress conditions; impact on biological process under stress response). | [31,32] | |
| qSUR5.2 | 5 | 2,548,592–2,559,171 | 17 | LOC_Os05g05230 | Expressed protein (drought-induced proteins, anther and pollen wall remodeling/metabolism proteins contribute to the tolerance of rice to salt stress). | [25,26] | |
| Shoot length | qSL1 | 1 | 39,794,226–39,799,341 | 18 | LOC_Os01g68490 | Tetratricopeptide-like helical, putative, expressed (abscisic acid responses and osmotic stress tolerance enable plants to cope with adverse environmental conditions). | [33,34] |
| Shoot dry weight | qSDW 5 | 5 | 20,966,622–20,969,373 | 17 | LOC_Os05g35310 | Ankyrin repeat family protein, putative, expressed (implicated in plant growth, development and signal transduction; response to biotic and abiotic stresses). | [35,36] |
| qSDW11 | 11 | 10,456,526–10,459,838 | 18 | LOC_Os11g18550 | Transposon protein, putative, CACTA, En/Spm sub-class, expressed (contributes significantly to genome size, produces a large number of cDNA sequences in plant tissues different conditions of stress). | [37,38] | |
| Root length | qRL1 | 1 | 12,444,630–12,446,150 | 12 | LOC_Os01g22150 | Transposon protein, putative, CACTA, En/Spm sub-class (contributes significantly to genome size, produces a large number of cDNA sequences in plant tissues under different conditions of stress). | [37,38] |
| SPAD | qSPAD12 | 12 | 26,377,868–26,379,849 | 17 | LOC_Os12g42440 | Chaperone protein dnaJ, putative, expressed (response to NaCl stress, involved in basal resistance to M. oryzae in rice). | [39,40] |
| Na+ Conc. | qNa6 | 6 | 6,643,235–6,643,552 | 11 | LOC_Os06g12300 | Expressed protein (drought-induced proteins, anther and pollen wall remodeling/metabolism proteins contribute to the tolerance of rice to salt stress). | [25,26] |
| K+ Conc. | qK8 | 8 | 4,794,164–4,799,199 | 12 | LOC_Os08g08350 | Retrotransposon protein, putative, Ty1-copia subclass, expressed (tuning gene expression during plant development salinity; plays a major role in shaping genome structure and size during salinity). | [41,42] |
| qK12 | 12 | 18,717,286–18,718,979 | 10 | LOC_Os12g31120 | Transposon protein, putative, CACTA, En/Spm sub-class, expressed (contributes significantly to genome size, produces a large number of cDNA sequences in plant tissues different conditions of stress). | [37,38] | |
| NaK ratio | qNaK-R8 | 8 | 4,333,717–4,335,434 | 13 | LOC_Os08g07740 | Histone-like transcription factor and archaeal histone, putative, expressed (regulating vegetative growth, sexual reproduction, virulence and hyperosmotic stresses, response to salt stress). | [43,44] |
| qNaK-R11 | 11 | 27,449,823–27,452,792 | 12 | LOC_Os11g45380 | Zinc finger family protein, putative, expressed (plant growth, development, and stress signal transduction, effective role in stress tolerance). | [45,46] |
| Traits | Chr1 | Position1 (cM) | FM1 | Chr2 | Position2 (cM) | FM2 | LOD | PVE (%) | Add1 | Add2 | Add by Add | Type of interaction |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SES | 4 | 10.7 | C308–C309 | 7 | 27 | C548–C549 | 42 | 2.628 | −0.003 | −0.953 | −0.951 | Between complementary loci |
| 2 | 120.4 | C190–C191 | 12 | 26.4 | C865–C866 | 48 | 2.637 | −0.706 | 1.198 | −0.206 | Between complementary loci | |
| 7 | 67 | C557–C558 | 12 | 26.4 | C865–C866 | 45 | 2.638 | −0.322 | 0.322 | −0.674 | Between complementary loci | |
| Survival | 3 | 111.9 | C276–C277 | 8 | 30.8 | C608–C609 | 21 | 0.056 | 3.260 | −0.444 | −2.893 | Between complementary loci |
| 4 | 130.7 | C380–C381 | 10 | 70.6 | C763–C764 | 10 | 1.826 | 12.950 | 4.923 | −10.412 | Between complementary loci | |
| 5 | 10.2 | C394–C395 | 10 | 70.6 | C763–C764 | 10 | 1.947 | 4.950 | −6.390 | 1.673 | Between complementary loci | |
| 6 | 101 | C516–C517 | 10 | 70.6 | C763–C764 | 10 | 1.584 | 3.374 | 2.100 | −14.695 | Between complementary loci | |
| 1 | 65.8 | C47–C48 | 10 | 75.6 | C770–C771 | 10 | 2.222 | 1.557 | 7.981 | 6.373 | Between QTLs and background | |
| Shoot Length | 2 | 15.4 | C132–C134 | 2 | 75.4 | C160–C161 | 6 | 9.206 | 4.177 | 0.113 | −4.192 | Between complementary loci |
| 2 | 75.4 | C160–C161 | 8 | 70.8 | C640–C641 | 6 | 6.744 | 1.225 | −5.466 | 3.291 | Between complementary loci | |
| 8 | 15.8 | C595–C596 | 8 | 70.8 | C640–C641 | 5 | 9.325 | −1.393 | −1.560 | 6.540 | Between complementary loci | |
| 6 | 71 | C507–C508 | 11 | 56 | C799–C800 | 5 | 7.587 | −2.065 | −1.355 | 5.903 | Between complementary loci | |
| 7 | 7 | C542–C543 | 11 | 96 | C836–C839 | 5 | 7.724 | 0.024 | 0.512 | 3.333 | Between complementary loci | |
| Shoot dry weight | 9 | 30.6 | C694–C695 | 10 | 75.6 | C770–C771 | 6 | 9.656 | −0.001 | 0.000 | −0.017 | Between complementary loci |
| 2 | 75.4 | C160–C161 | 11 | 21 | C782–C783 | 6 | 9.723 | −0.004 | 0.000 | 0.003 | Between complementary loci | |
| 12 | 11.4 | C857–C858 | 12 | 56.4 | C884–C885 | 8 | 8.903 | −0.013 | 0.017 | 0.012 | Between complementary loci | |
| 6 | 86 | C512–C513 | 12 | 76.4 | C901–C902 | 19 | 1.480 | 0.003 | 0.004 | 0.007 | Between complementary loci | |
| 1 | 155.8 | C104–C105 | 12 | 86.4 | C906–C907 | 6 | 9.417 | −0.003 | 0.011 | −0.011 | Between complementary loci | |
| Root length | 4 | 135.7 | C381–C382 | 9 | 0.6 | C671–C672 | 10 | 1.423 | 0.074 | 0.209 | 0.074 | Between complementary loci |
| 8 | 75.8 | C644–C645 | 12 | 76.4 | C901–C902 | 89 | 7.637 | −0.409 | −0.427 | −0.427 | Between complementary loci | |
| SPAD | 1 | 115.8 | C73–C74 | 3 | 136.9 | C296–C297 | 6 | 8.485 | −0.478 | 2.305 | 3.562 | Between complementary loci |
| 1 | 115.8 | C73–C74 | 4 | 55.7 | C335–C336 | 5 | 5.614 | 2.343 | 4.534 | 1.190 | Between complementary loci | |
| 3 | 56.9 | C246–C247 | 4 | 55.7 | C335–C336 | 6 | 3.118 | −2.093 | 0.707 | 4.291 | Between complementary loci | |
| 3 | 11.9 | C217–C218 | 5 | 50.2 | C417–C418 | 5 | 7.909 | 2.494 | 0.180 | −1.053 | Between complementary loci | |
| 1 | 115.8 | C73–C74 | 6 | 101 | C516–C517 | 5 | 8.103 | 1.306 | 2.094 | 3.784 | Between complementary loci | |
| 4 | 25.7 | C314–C315 | 7 | 27 | C548–C549 | 6 | 6.784 | −2.153 | −0.795 | 1.959 | Between complementary loci | |
| 11 | 21 | C782–C783 | 11 | 96 | C836–C839 | 6 | 6.641 | 2.780 | 1.330 | 0.550 | Between complementary loci | |
| 9 | 45.6 | C702–C703 | 12 | 81.4 | C904–C905 | 6 | 1.772 | −0.103 | −0.503 | 0.583 | Between complementary loci | |
| Na+ Conc. | 2 | 135.4 | C199–C200 | 4 | 55.7 | C335–C336 | 6 | 4.625 | 0.012 | −0.028 | −0.013 | Between complementary loci |
| 3 | 116.9 | C279–C280 | 5 | 45.2 | C416–C417 | 5 | 0.533 | 0.006 | 0.006 | −0.016 | Between complementary loci | |
| 1 | 55.8 | C38–C39 | 5 | 50.2 | C417–C418 | 6 | 10.06 | −0.020 | −0.022 | 0.019 | Between complementary loci | |
| 1 | 0.8 | C1–C2 | 6 | 101 | C516–C517 | 6 | 5.709 | −0.015 | 0.012 | −0.018 | Between complementary loci | |
| 1 | 65.8 | C47–C48 | 7 | 97 | C577–C578 | 6 | 8.641 | −0.013 | −0.046 | 0.037 | Between complementary loci | |
| 3 | 1.9 | C208–C209 | 10 | 5.6 | C728–C729 | 6 | 6.317 | 0.015 | 0.024 | 0.012 | Between complementary loci | |
| 1 | 65.8 | C47–C48 | 11 | 106 | C842–C843 | 5 | 9.005 | 0.019 | −0.018 | −0.049 | Between complementary loci | |
| 9 | 50.6 | C706–C707 | 12 | 21.4 | C865–C866 | 5 | 8.375 | 0.010 | −0.011 | −0.002 | Between complementary loci | |
| K+ conc. | 1 | 90.8 | C62–C63 | 3 | 66.9 | C251–C252 | 9 | 12.65 | −0.025 | −0.014 | 0.014 | Between complementary loci |
| 3 | 66.9 | C251–C252 | 5 | 15.2 | C397–C398 | 9 | 12.69 | −0.014 | −0.025 | 0.014 | Between complementary loci | |
| 3 | 71.9 | C254–C255 | 8 | 75.8 | C644–C645 | 9 | 1.718 | 0.002 | −0.011 | 0.002 | Between complementary loci | |
| 4 | 90.7 | C354–C355 | 9 | 75.6 | C715–C716 | 6 | 8.798 | 0.026 | 0.000 | 0.000 | Between complementary loci | |
| Na+/K+ ratio | 8 | 20.8 | C599–C600 | 8 | 60.8 | C637–C638 | 7 | 1.569 | −0.016 | −0.002 | 0.002 | Between QTLs and background |
| 3 | 71.9 | C254–C255 | 8 | 75.8 | C644–C645 | 9 | 5.615 | 0.004 | −0.017 | 0.004 | Between complementary loci | |
| 4 | 90.7 | C354–C355 | 9 | 75.6 | C715–C716 | 6 | 27.54 | 0.041 | 0.002 | 0.002 | Between complementary loci |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maniruzzaman, S.; Rahman, M.A.; Hasan, M.; Rasul, M.G.; Molla, A.H.; Khatun, H.; Akter, S. Genetic Mapping to Detect Stringent QTLs Using 1k-RiCA SNP Genotyping Platform from the New Landrace Associated with Salt Tolerance at the Seedling Stage in Rice. Plants 2022, 11, 1409. https://doi.org/10.3390/plants11111409
Maniruzzaman S, Rahman MA, Hasan M, Rasul MG, Molla AH, Khatun H, Akter S. Genetic Mapping to Detect Stringent QTLs Using 1k-RiCA SNP Genotyping Platform from the New Landrace Associated with Salt Tolerance at the Seedling Stage in Rice. Plants. 2022; 11(11):1409. https://doi.org/10.3390/plants11111409
Chicago/Turabian StyleManiruzzaman, Sheikh, Mohammad Akhlasur Rahman, Mehfuz Hasan, Mohammad Golam Rasul, Abul Hossain Molla, Hasina Khatun, and Salma Akter. 2022. "Genetic Mapping to Detect Stringent QTLs Using 1k-RiCA SNP Genotyping Platform from the New Landrace Associated with Salt Tolerance at the Seedling Stage in Rice" Plants 11, no. 11: 1409. https://doi.org/10.3390/plants11111409






