Mutagenic Effect of 60Co γ-Irradiation on Rosa multiflora ‘Libellula’ and the Mechanism Underlying the Associated Leaf Changes
Abstract
1. Introduction
2. Results
2.1. Morphological Deformation of Rosa multiflora ‘Libellula’ Leaves Treated with 60Co γ-rays
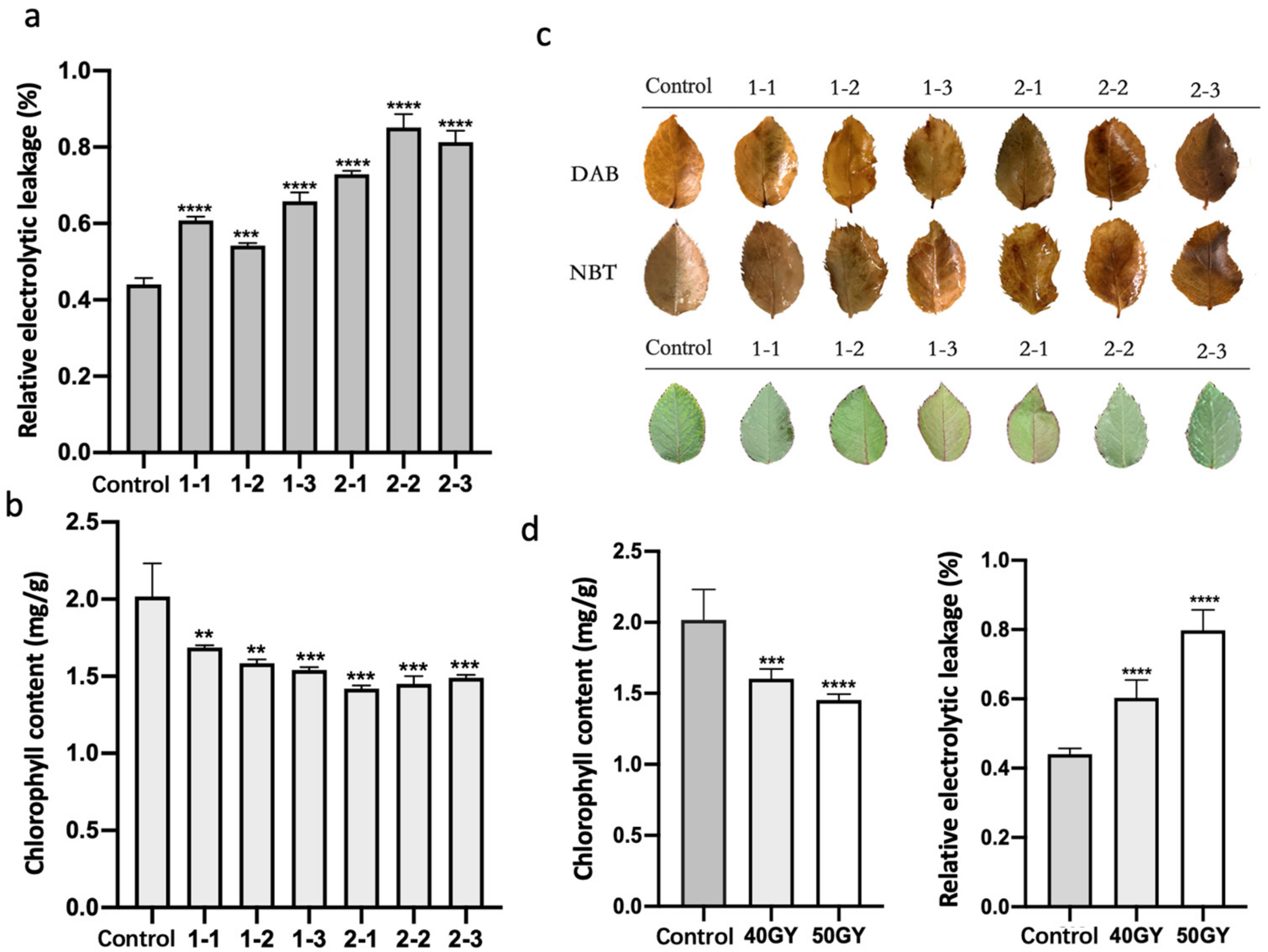
2.2. Irradiation of Plants with 60Co γ-rays Increased the Accumulation of Active Oxygen and Decreased the Chlorophyll Content
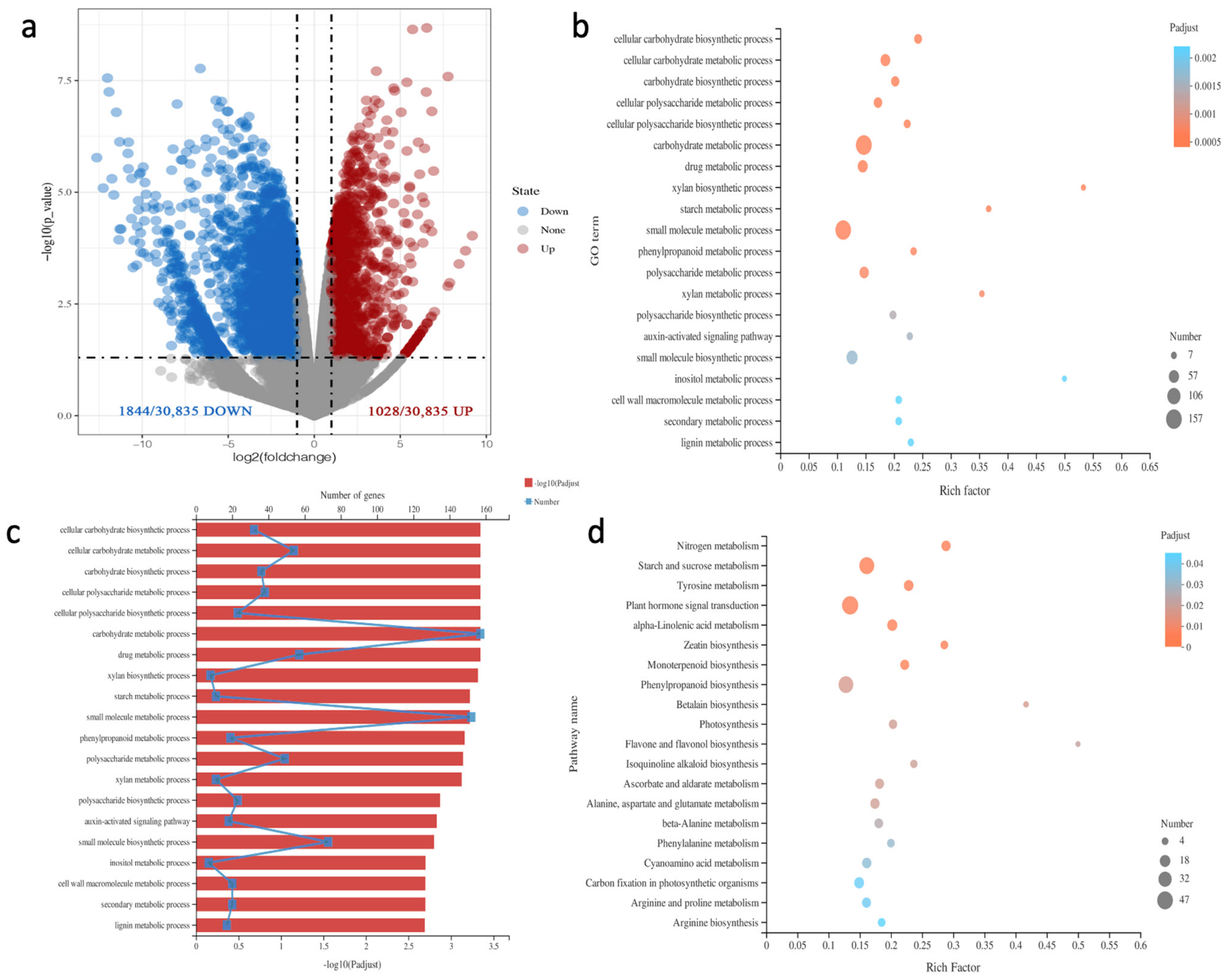
2.3. RNA-Seq Analysis of Differentially Expressed Genes in Irradiated Rose Leaves
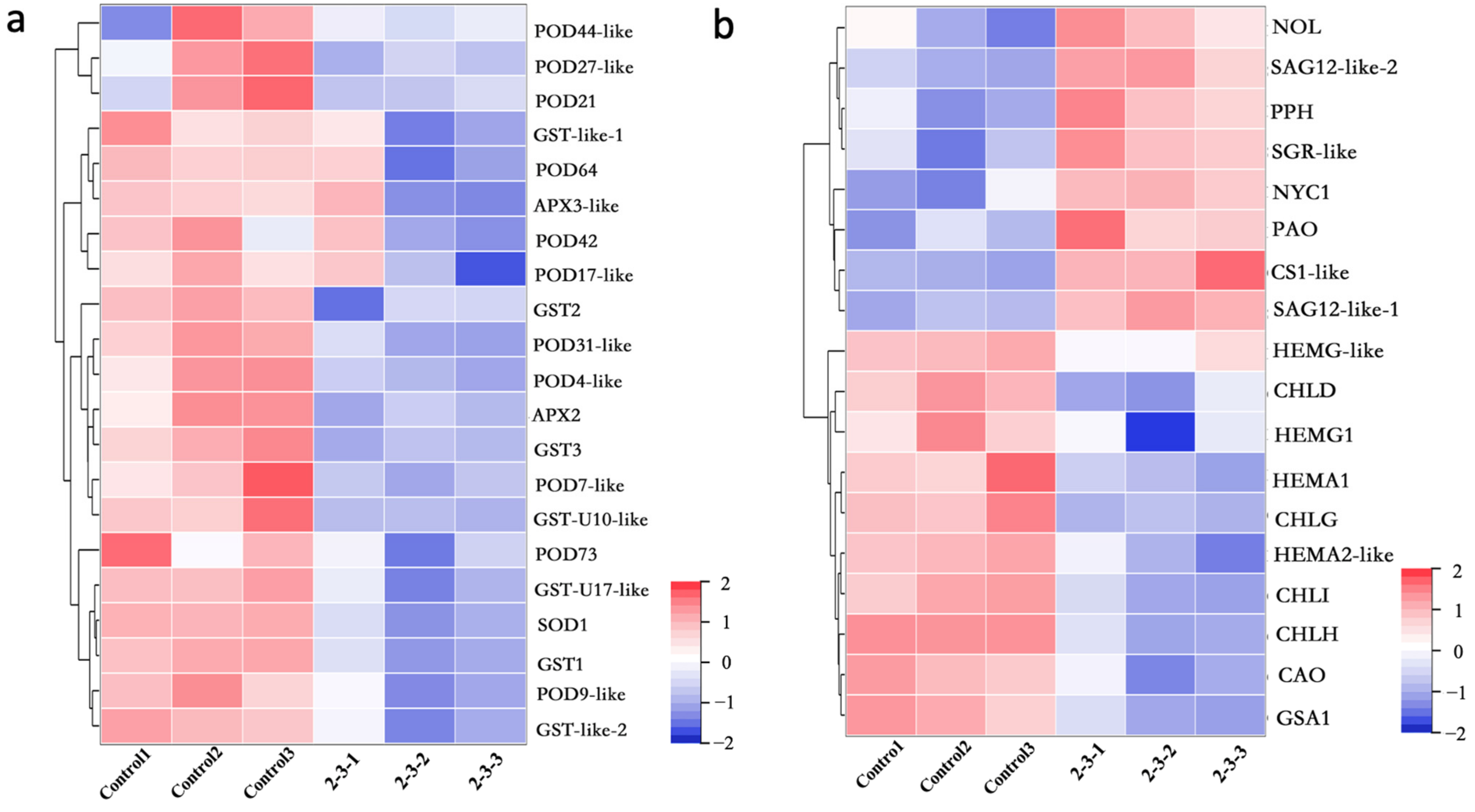
2.4. Antioxidant Functions in Irradiated Plants Were Weakened, and the Genes Related to Chlorophyll Synthesis and Metabolism Were Differentially Expressed
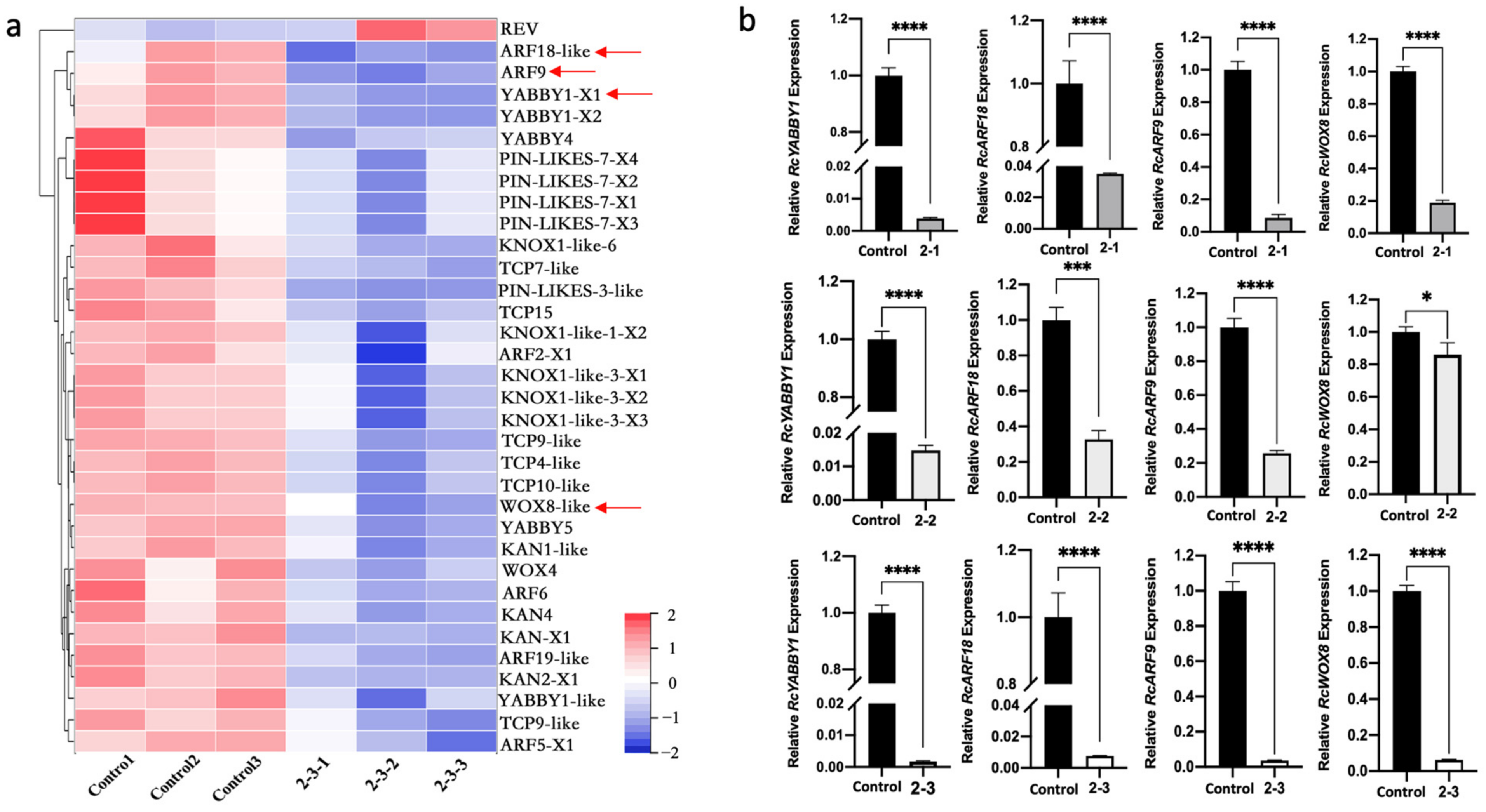
2.5. Differential Expression of Genes Related to Leaf Development and Morphogenesis in Irradiated Plants
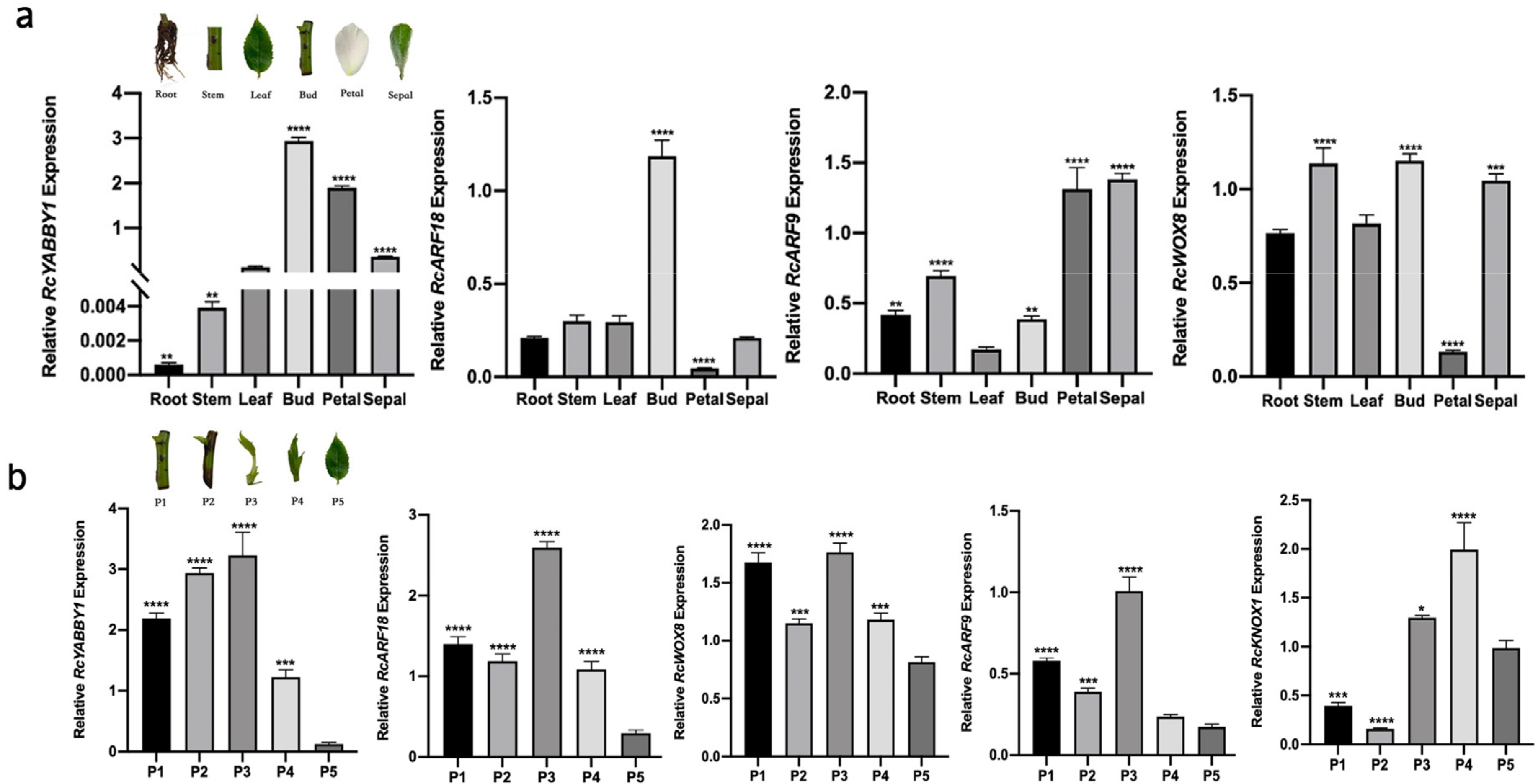
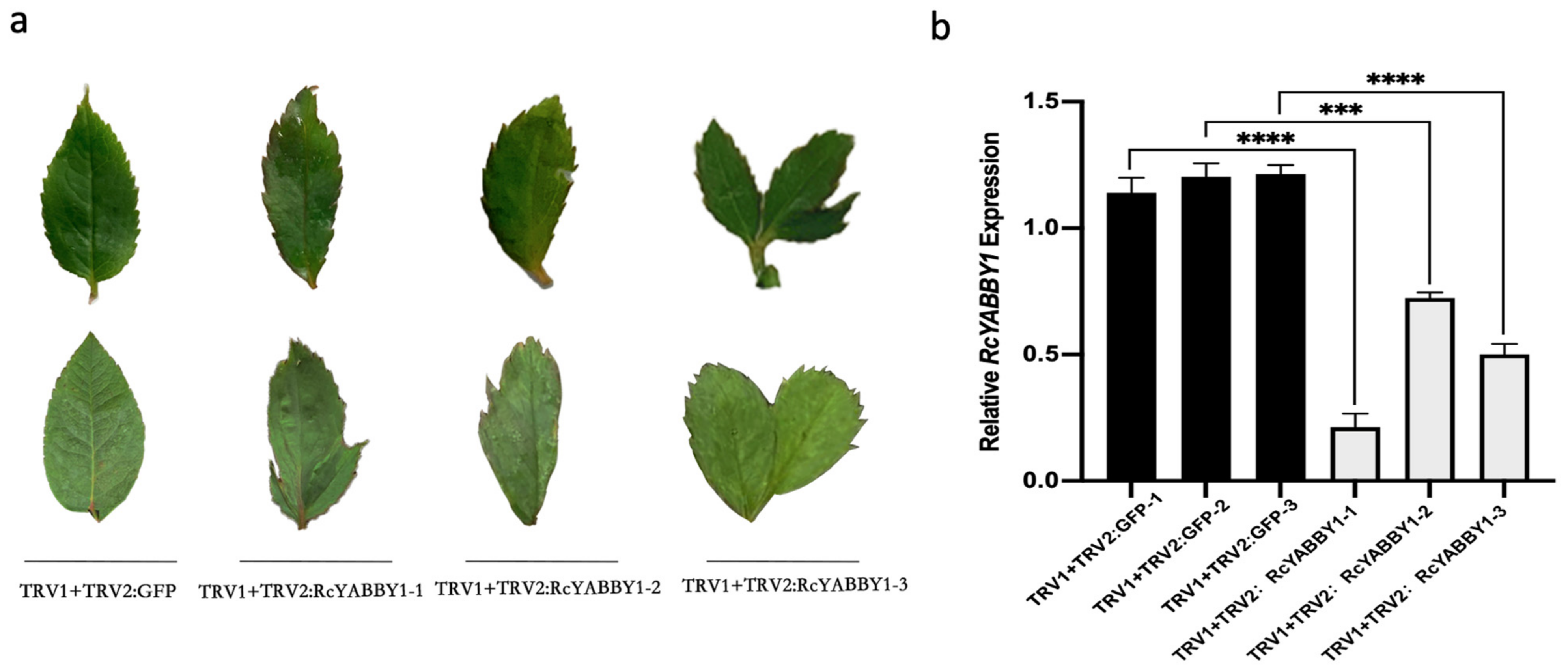
2.6. Identification and Functional Analysis of Candidate Genes for Leaf Development in Rosa multiflora ‘Libellula’
3. Discussion
4. Materials and Methods
4.1. Plant Material Culture Conditions and Irradiation Treatment Methods
4.2. Determination of Physiological Indicators
4.3. RNA Extraction and Quantitative Real-Time PCR Detection
4.4. Functional Verification of Virus-Induced Gene Silencing (VIGS)
4.5. RNA-Seq Data Processing, Reassembly, and Annotation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bendahmane, M.; Dubois, A.; Raymond, O.; Bris, M. Genetics and genomics of flower initiation and development in roses. J. Exp. Bot. 2013, 64, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, H. Mutation breeding of ornamental plants using ion beams. Breed. Sci. 2018, 68, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Shi, M.; Huang, J.; Xu, J.; Wang, Z.; Guo, D. Regulation of photosynthetic performance and antioxidant capacity by 60Coγ-irradiation in Zizania latifolia plants. J. Environ. Radioact. 2014, 129, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Esnault, M.; Legue, F.; Chenal, C. Ionizing radiation: Advances in plant response. Environ. Exp. Bot. 2010, 68, 231–237. [Google Scholar] [CrossRef]
- Calucci, L.; Pinzino, C.; Zandomeneghi, M.; Capocchi, A.; Ghiringhelli, S.; Saviozzi, F.; Tozzi, S.; Galleschi, L. Effects of γ-Irradiation on the Free Radical and Antioxidant Contents in Nine Aromatic Herbs and Spices. J. Agric. Food Chem. 2002, 51, 927–934. [Google Scholar] [CrossRef]
- Rodrigues, M.; Martins, A.; Desidério, J.; Bertoni, B.; Alves, M. Genetic characterization of fig tree mutants with molecular markers. Genet. Mol. Res. 2012, 11, 1990–1996. [Google Scholar] [CrossRef]
- Kim, J.; Baek, M.; Chung, B.; Wi, S.; Kim, J. Alterations in the photosynthetic pigments and antioxidant machineries of red pepper (Capsicum annuum L.) seedlings from gamma-irradiated seeds. J. Plant Biol. 2004, 47, 314–321. [Google Scholar] [CrossRef]
- Wi, S.; Chung, B.; Kim, J.; Baek, M.; Yang, D.; Lee, J.; Kim, J. Ultrastructural changes of cell organelles in Arabidopsis stems after gamma irradiation. J. Plant Biol. 2005, 48, 195–200. [Google Scholar] [CrossRef]
- Sakowska, K.; Alberti, G.; Genesio, L.; Peressotti, A.; Delle Vedove, G.; Gianelle, D.; Colombo, R.; Rodeghiero, M.; Panigada, C.; Juszczak, R.; et al. Leaf and canopy photosynthesis of a chlorophyll deficient soybean mutant. Plant Cell Environ. 2018, 41, 1427–1437. [Google Scholar] [CrossRef]
- Tanaka, A.; Shikazono, N.; Hase, Y. Studies on Biological Effects of Ion Beams on Lethality, Molecular Nature of Mutation, Mutation Rate, and Spectrum of Mutation Phenotype for Mutation Breeding in Higher Plants. J. Radiat. Res. 2010, 51, 223–233. [Google Scholar] [CrossRef]
- Tsukaya, H. Comparative leaf development in angiosperms. Curr. Opin. Plant Biol. 2014, 17, 103–109. [Google Scholar] [CrossRef]
- Wang, H.; Kong, F.; Zhou, C. From genes to networks: The genetic control of leaf development. J. Integr. Plant Biol. 2021, 63, 1181–1196. [Google Scholar] [CrossRef] [PubMed]
- Lodha, M.; Marco, C.; Timmermans, M. The ASYMMETRIC LEAVES complex maintains repression of KNOX homeobox genes via direct recruitment of Polycomb-repressive complex2. Genes Dev. 2013, 27, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Byrne, M.; Barley, R.; Curtis, M.; Arroyo, J.; Dunham, M.; Hudson, A.; Martienssen, R. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 2000, 408, 967–971. [Google Scholar] [CrossRef] [PubMed]
- Lin, W. The Arabidopsis LATERAL ORGAN BOUNDARIES-Domain Gene ASYMMETRIC LEAVES2 Functions in the Repression of KNOX Gene Expression and in Adaxial-Abaxial Patterning. Plant Cell 2003, 15, 2241–2252. [Google Scholar] [CrossRef]
- Sampathkumar, A.; Yan, A.; Krupinski, P.; Meyerowitz, E. Physical Forces Regulate Plant Development and Morphogenesis. Curr. Biol. 2014, 24, R475–R483. [Google Scholar] [CrossRef]
- Traas, J. Plant Development: From Dynamics to Mechanics. Curr. Biol. 2017, 27, R313–R315. [Google Scholar] [CrossRef][Green Version]
- Belles-Boix, E.; Hamant, O.; Witiak, S.; Morin, H.; Traas, J.; Pautot, V. KNAT6: An Arabidopsis Homeobox Gene Involved in Meristem Activity and Organ Separation. Plant Cell 2006, 18, 1900–1907. [Google Scholar] [CrossRef]
- McConnell, J.; Emery, J.; Eshed, Y.; Bao, N.; Bowman, J.; Barton, M. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 2001, 411, 709–713. [Google Scholar] [CrossRef]
- Emery, J.; Floyd, S.; Alvarez, J.; Eshed, Y.; Hawker, N.; Izhaki, A.; Baum, S.; Bowman, J. Radial Patterning of Arabidopsis Shoots by Class III HD-ZIP and KANADI Genes. Curr. Biol. 2003, 13, 1768–1774. [Google Scholar] [CrossRef]
- Kerstetter, R.; Bollman, K.; Taylor, R.; Bomblies, K.; Poethig, R. KANADI regulates organ polarity in Arabidopsis. Nature 2001, 411, 706–709. [Google Scholar] [CrossRef]
- Eshed, Y.; Baum, S.; Perea, J.; Bowman, J. Establishment of polarity in lateral organs of plants. Curr. Biol. 2001, 11, 1251–1260. [Google Scholar] [CrossRef]
- Bowman, J.; Floyd, S. Patterning and Polarity in Seed Plant Shoots. Annu. Rev. Plant Biol. 2008, 59, 67–88. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, C.; Li, D.; Liu, Y.; Yang, X. Roles of YABBY transcription factors in the modulation of morphogenesis, development, and phytohormone and stress responses in plants. J. Plant Res. 2020, 133, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Sieber, P.; Petrascheck, M.; Barberis, A.; Schneitz, K. Organ Polarity in Arabidopsis. NOZZLE Physically Interacts with Members of the YABBY Family. Plant Physiol. 2004, 135, 2172–2185. [Google Scholar] [CrossRef]
- Bresso, E.; Chorostecki, U.; Rodriguez, R.; Palatnik, J.; Schommer, C. Spatial Control of Gene Expression by miR319-Regulated TCP Transcription Factors in Leaf Development. Plant Physiol. 2017, 176, 1694–1708. [Google Scholar] [CrossRef] [PubMed]
- Efroni, I.; Blum, E.; Goldshmidt, A.; Eshed, Y. A Protracted and Dynamic Maturation Schedule Underlies Arabidopsis Leaf Development. Plant Cell 2008, 20, 2293–2306. [Google Scholar] [CrossRef]
- Wu, C.; Li, F.; Kramer, E. Large-scale phylogenomic analysis suggests three ancient superclades of the WUSCHEL-RELATED HOMEOBOX transcription factor family in plants. PLoS ONE. 2019, 14, e022352. [Google Scholar] [CrossRef]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis WUSCHEL Is a Bifunctional Transcription Factor That Acts as a Repressor in Stem Cell Regulation and as an Activator in Floral Patterning. Plant Cell 2009, 21, 3493–3505. [Google Scholar] [CrossRef]
- Austin, D.; Rice, H.; Lawson, A.; Daker, R. The English Roses; Conran: London, UK, 2011. [Google Scholar]
- Lee, M.; Moon, Y.; Chung, B.; Kim, J.; Lee, K.; Cho, J.; Kim, J. Practical use of chemical probes for reactive oxygen species produced in biological systems by γ-irradiation. Radiat. Phys. Chem. 2009, 78, 323–327. [Google Scholar] [CrossRef]
- Wi, S.; Chung, B.; Kim, J.; Kim, J.; Baek, M.; Lee, J. Localization of hydrogen peroxide in pumpkin (Cucurbita ficifolia bouché) seedlings exposed to high-dose gamma ray. J. Plant Biol. 2006, 49, 1. [Google Scholar] [CrossRef]
- Wada, H.; Koshiba, T.; Matsui, T.; Satô, M. Involvement of peroxidase in differential sensitivity to γ-radiation in seedlings of two Nicotiana species. Plant Sci. 1998, 132, 109–119. [Google Scholar] [CrossRef]
- Yue, Y.; Ruter, J. 60Co Irradiation Influences Germination and Phenotype of Three Pavonia Species. HortScience 2020, 55, 2037–2044. [Google Scholar] [CrossRef]
- Jia, C.; Li, A. Effect of gamma radiation on mutant induction of Fagopyrum dibotrys Hara. Photosynthetica 2008, 46, 363. [Google Scholar] [CrossRef]
- Okamoto, H. Effects of low-dose γ-irradiation on the cell cycle duration of barley roots. Environ. Exp. Bot. 1995, 35, 379–388. [Google Scholar] [CrossRef]
- Azigwe, C.; Zoryeku, P.; Asante, I.; Oppong-Adjei, F. Effect of Gamma Irradiation on Chlorophyll Content in the Cowpea (Vigna unguiculata (L.) Walp). Ghana J. Sci. 2021, 61, 113–117. [Google Scholar] [CrossRef]
- Ernawati; Oppong-Adjei, F.; Suryadi, H.; Mun’im, A. Effect of gamma irradiation on the caffeoylquinic acid derivatives content, antioxidant activity, and microbial contamination of Pluchea indica leaves. Heliyon 2021, 7, e07825. [Google Scholar] [CrossRef]
- Rhodes, D.; Rich, P.; Brunk, D.; Ju, G.; Rhodes, J.; Pauly, M.; Hansen, L. Development of Two Isogenic Sweet Corn Hybrids Differing for Glycinebetaine Content. Plant Physiol. 1989, 91, 1112. [Google Scholar] [CrossRef]
- Gill, S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Sato, Y.; Morita, R.; Katsuma, S.; Nishimura, M.; Tanaka, A.; Kusaba, M. Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll band light-harvesting complex II degradation during senescence in rice. Plant J. 2009, 57, 120–131. [Google Scholar] [CrossRef]
- Horie, Y.; Ito, H.; Kusaba, M.; Tanaka, R.; Tanaka, A. Participation of Chlorophyll b Reductase in the Initial Step of the Degradation of Light-harvesting Chlorophyll a/b-Protein Complexes in Arabidopsis. J. Biol. Chem. 2009, 284, 17449–17456. [Google Scholar] [CrossRef] [PubMed]
- Rodoni, S.; Schellenberg, M.; Matile, P. Chlorophyll breakdown in senescing barley leaves as correlated with phaeophorbidea oxygenase activity. J. Plant Physiol. 1998, 152, 139–144. [Google Scholar] [CrossRef]
- Balazadeh, S. Stay-Green Not Always Stays Green. Mol. Plant. 2014, 7, 1264–1266. [Google Scholar] [CrossRef]
- Beale, S. Green genes gleaned. Trends Plant Sci. 2005, 10, 309–312. [Google Scholar] [CrossRef]
- Burian, A.; Paszkiewicz, G.; Nguyen, K.; Meda, S.; Raczyska-Szajgin, M.; Timmermans, M. Specification of leaf dorsiventrality via a prepatterned binary readout of a uniform auxin input. Nat. Plants 2022, 8, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef]
- Wang, R.; Mao, C.; Jiang, C.; Zhang, L.; Peng, S.; Zhang, Y.; Feng, S.; Ming, F. One Heat Shock Transcription Factor Confers High Thermal Tolerance in Clematis Plants. Int. J. Mol. Sci. 2021, 22, 2900. [Google Scholar] [CrossRef]
- Livak, K.; Schmittgen, T. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2CT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
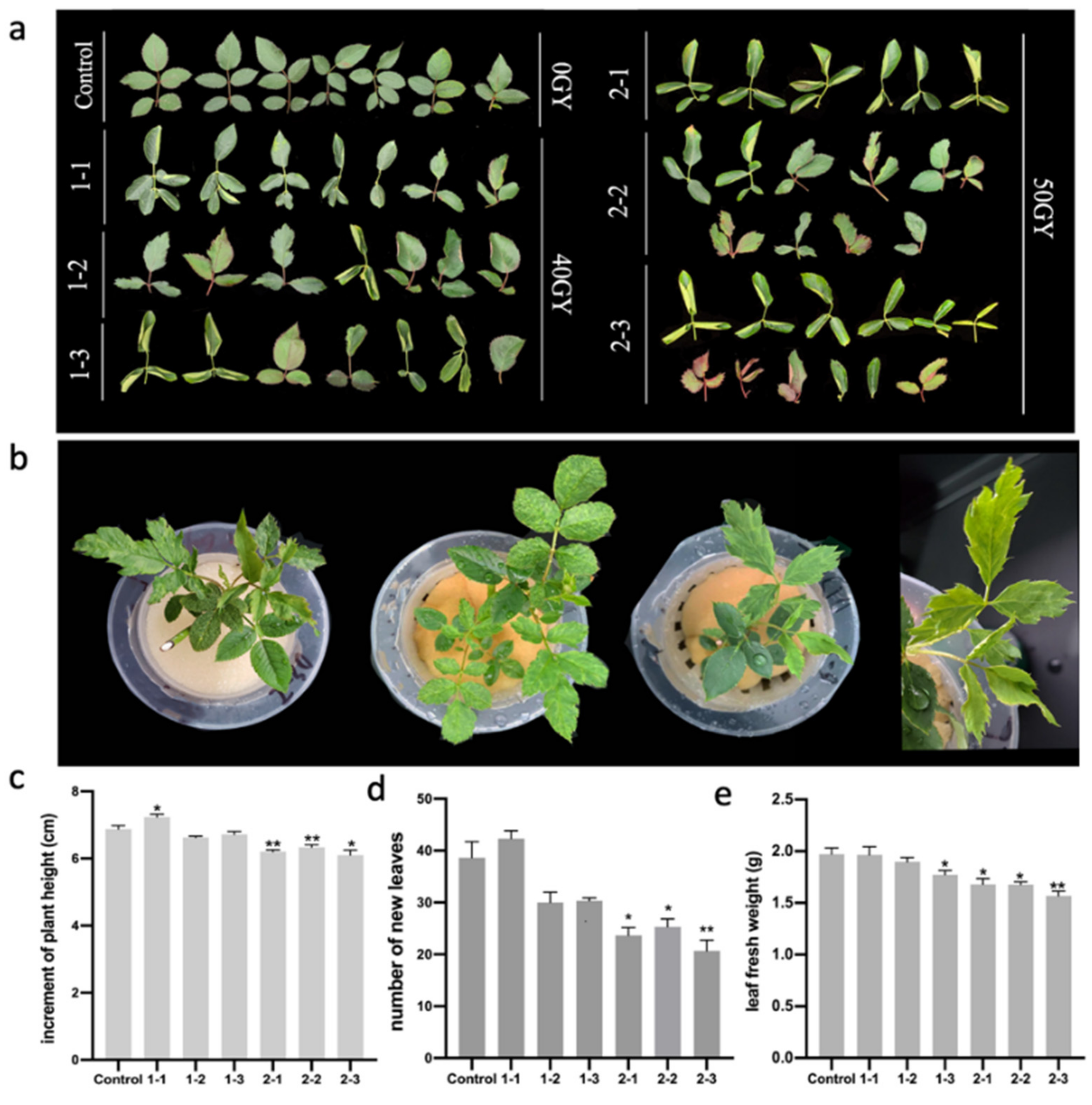
| Group Number | Variation Rate | Incised Leaves | Reduced Number of Compound Leaves | Rolling Leaf | Big Leaf | Biacuminate Leaf | Asymmetric Leaf |
|---|---|---|---|---|---|---|---|
| control | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1-1 | 84.89 ± 5.18 de | 17.55 ± 0.48 b | 18.57 ± 0.94 d | 22.92 ± 2.78 c | 9.96 ± 0.46 b | 5.89 ± 0.59 b | 10.00 ± 1.11 cd |
| 1-2 | 76.36 ± 1.18 e | 26.45 ± 0.47 a | 13.62 ± 0.98 e | 12.80 ± 2.08 d | 8.94 ± 1.11 bc | 3.89 ± 0.48 c | 10.66 ± 0.53 c |
| 1-3 | 94.26 ± 4.43 d | 3.02 ± 0.72 d | 20.71 ± 0.86 c | 48.70 ± 2.96 b | 14.75 ± 1.52 ab | 1.13 ± 0.21 de | 5.95 ± 0.40 d |
| 2-1 | 118.08 ± 4.46 b | 15.32 ± 0.57 cd | 32.49 ± 1.27 ab | 48.35 ± 4.06 bc | 5.81 ± 2.50 c | 3.79 ± 1.01 cd | 12.32 ± 0.77 b |
| 2-2 | 106.36 ± 1.63 c | 16.24 ± 0.39 c | 33.53 ± 1.48 a | 11.21 ± 2.55 de | 16.41 ± 1.58 a | 8.14 ± 1.01 a | 20.83 ± 1.43 a |
| 2-3 | 141.67 ± 2.71 a | 17.11 ± 1.16 bc | 27.28 ± 1.98 b | 81.06 ± 2.09 a | 4.53 ± 0.44 cd | 1.87 ± 0.50 d | 9.83 ± 1.30 cd |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, M.; Xu, Q.; Liu, Y.; Ming, F. Mutagenic Effect of 60Co γ-Irradiation on Rosa multiflora ‘Libellula’ and the Mechanism Underlying the Associated Leaf Changes. Plants 2022, 11, 1438. https://doi.org/10.3390/plants11111438
Xia M, Xu Q, Liu Y, Ming F. Mutagenic Effect of 60Co γ-Irradiation on Rosa multiflora ‘Libellula’ and the Mechanism Underlying the Associated Leaf Changes. Plants. 2022; 11(11):1438. https://doi.org/10.3390/plants11111438
Chicago/Turabian StyleXia, Meng, Qingyu Xu, Ying Liu, and Feng Ming. 2022. "Mutagenic Effect of 60Co γ-Irradiation on Rosa multiflora ‘Libellula’ and the Mechanism Underlying the Associated Leaf Changes" Plants 11, no. 11: 1438. https://doi.org/10.3390/plants11111438
APA StyleXia, M., Xu, Q., Liu, Y., & Ming, F. (2022). Mutagenic Effect of 60Co γ-Irradiation on Rosa multiflora ‘Libellula’ and the Mechanism Underlying the Associated Leaf Changes. Plants, 11(11), 1438. https://doi.org/10.3390/plants11111438







