Evaluation of Green Super Rice Lines for Agronomic and Physiological Traits under Salinity Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Experiments
2.2. Glasshouse Experiment
Seedling Evaluation at 200 mM NaCl
2.3. Field Experiment
2.3.1. Plant Growth and Field Conditions
2.3.2. Field Evaluation for Morphological Traits
2.3.3. Selection Criteria of Tolerant and Susceptible Lines
2.4. Molecular Characterization
2.4.1. RNA Sampling, Extraction, cDNA Synthesis
2.4.2. Selection of Differentially Expressed Genes (DEGs) and Expression Analysis
2.5. Statistical Analysis
3. Results
3.1. Glasshouse Experiment
3.1.1. Response of Seedlings Growth under Glasshouse Conditions
3.1.2. Mean Variability of Seedlings Parameters
3.1.3. Heritability (H2) of Seedlings Parameters
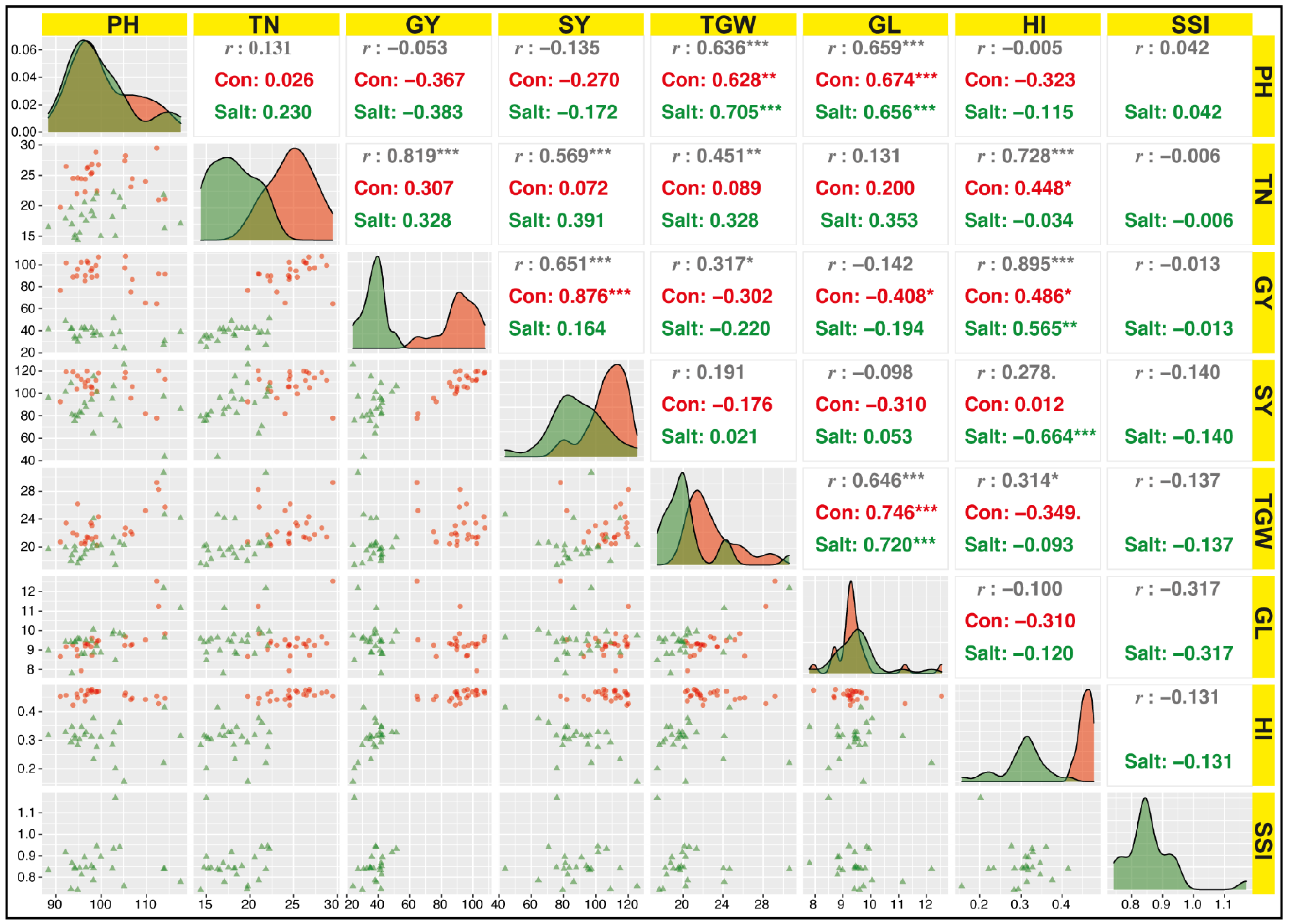
3.1.4. Correlation among Seedlings Parameters
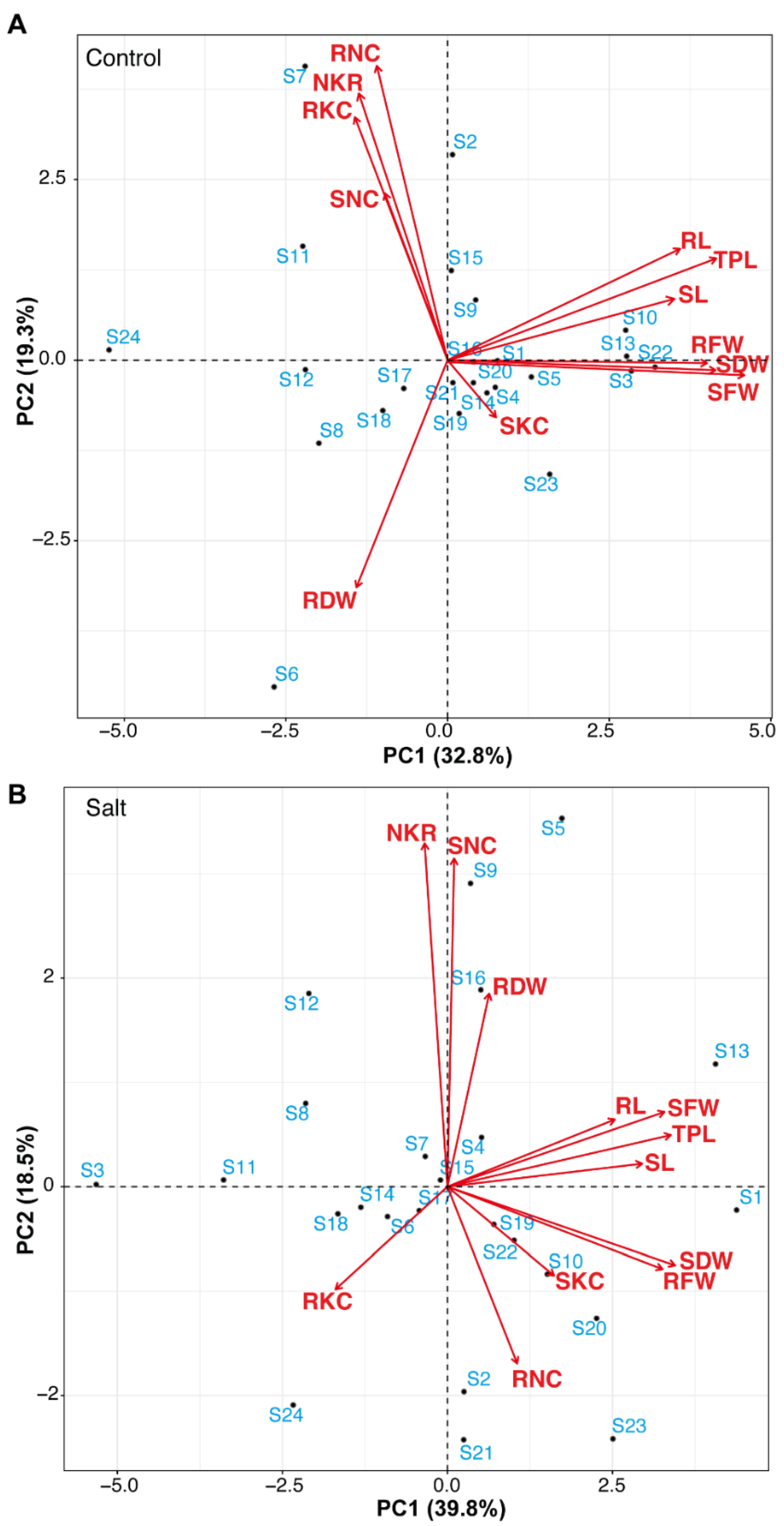
3.1.5. Principal Component Analysis of Seedlings Parameters
3.2. Field Experiment
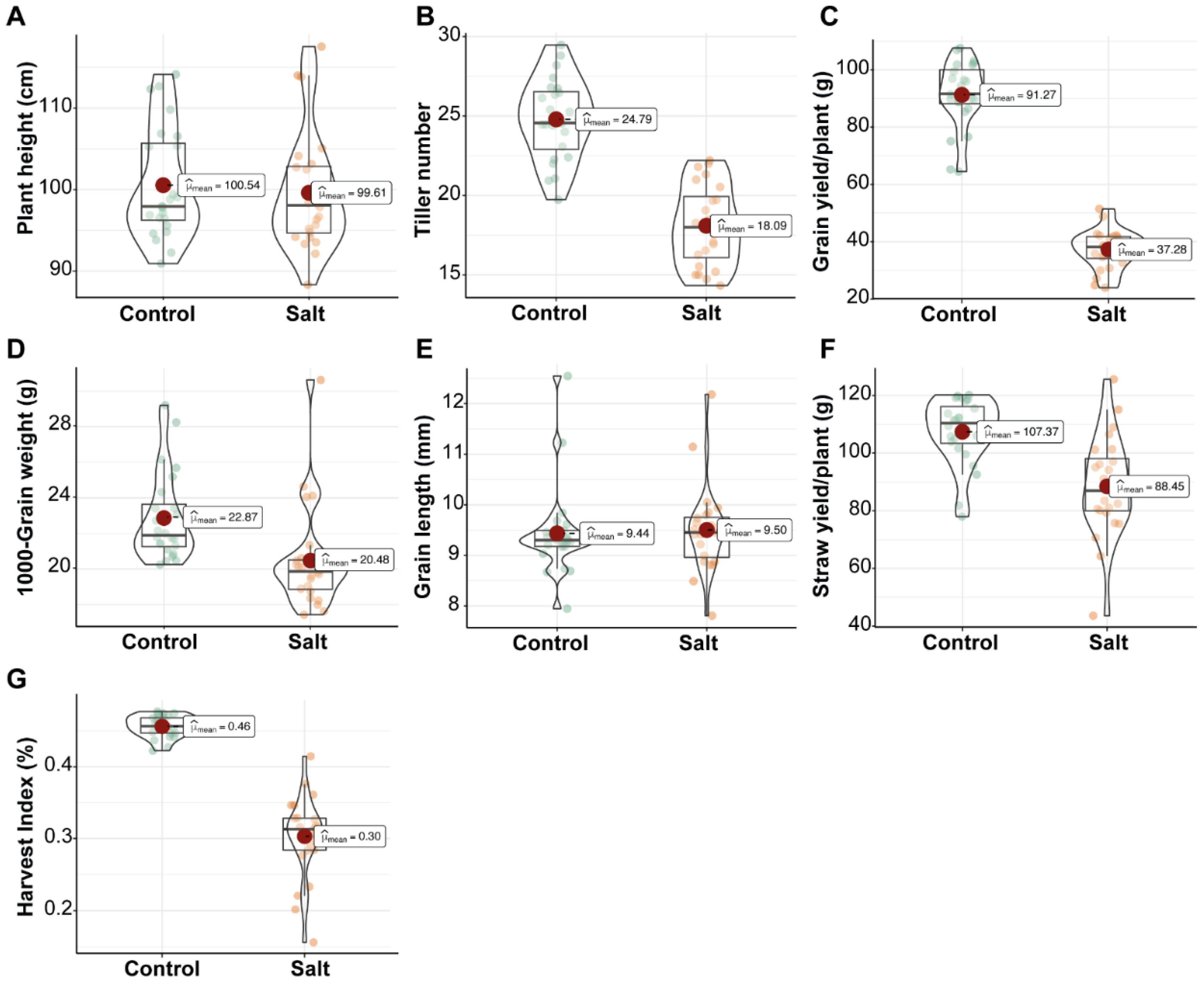
3.2.1. Effects of Salinity on Yield Components under Field Conditions
3.2.2. Association among Yield Parameters under Field Conditions
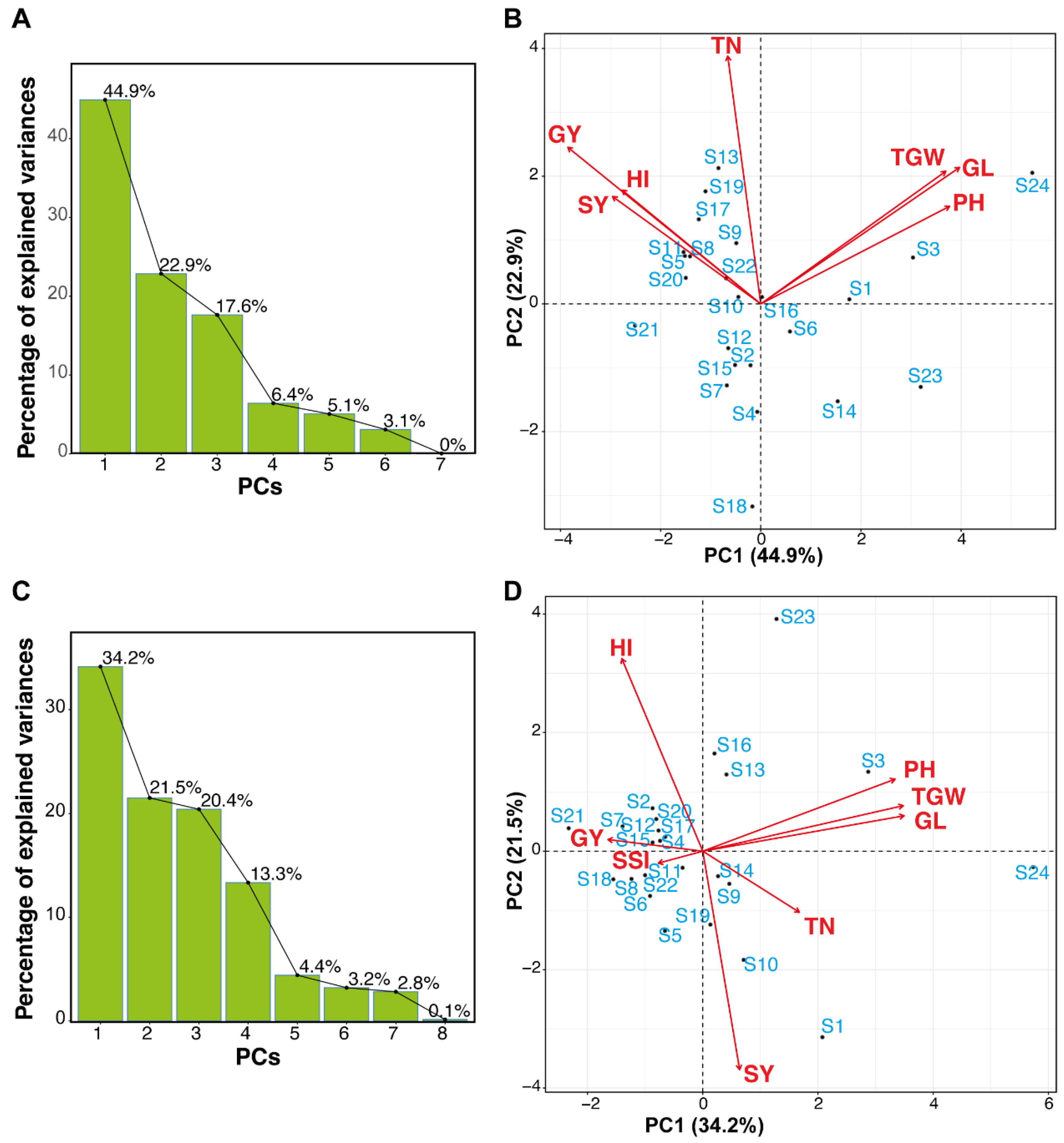
3.2.3. Assessment of Rice Genotypes Using Principal Component Analysis (PCA)
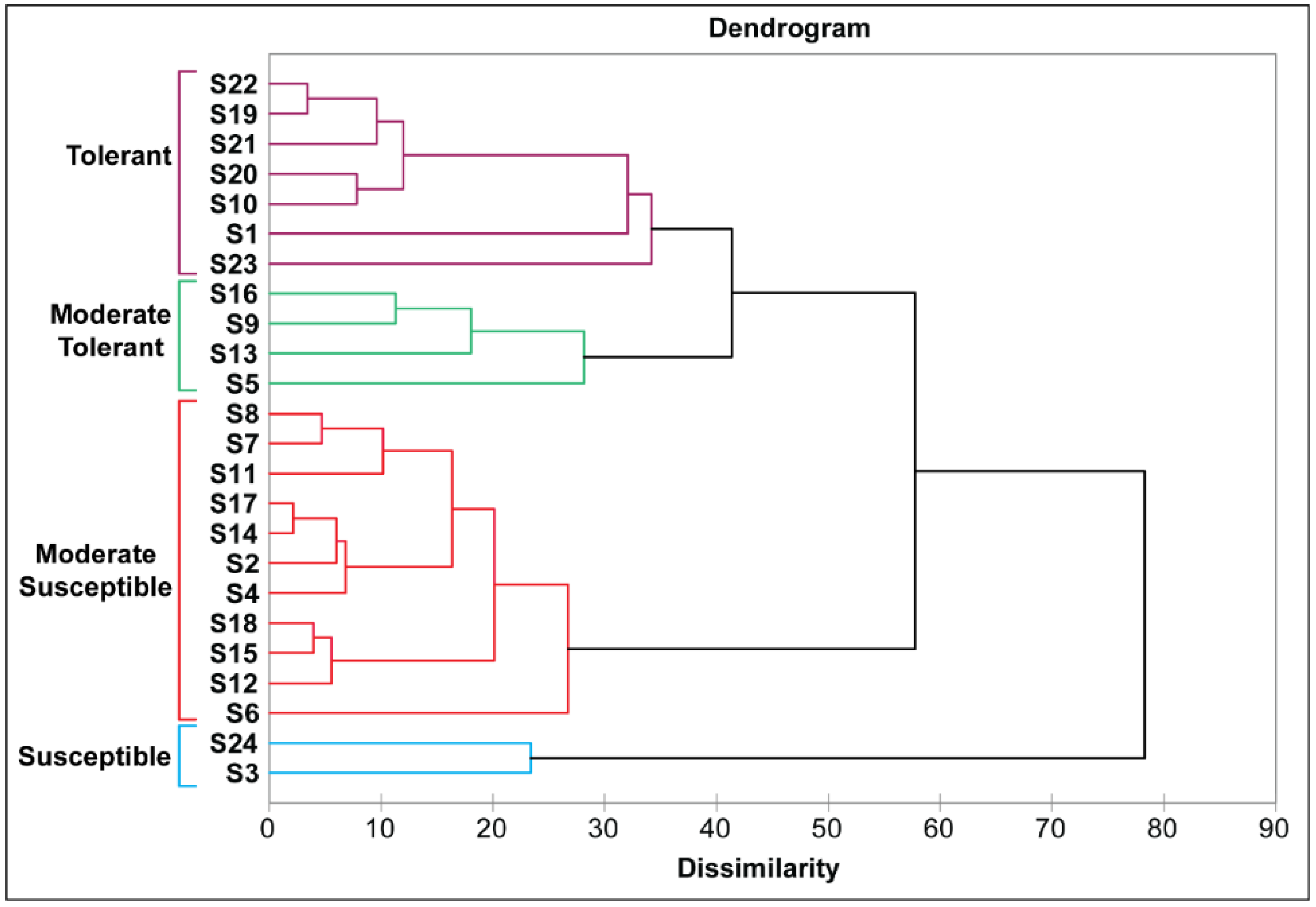
3.2.4. Cluster Analysis of Rice Genotypes
3.3. Molecular Characterization
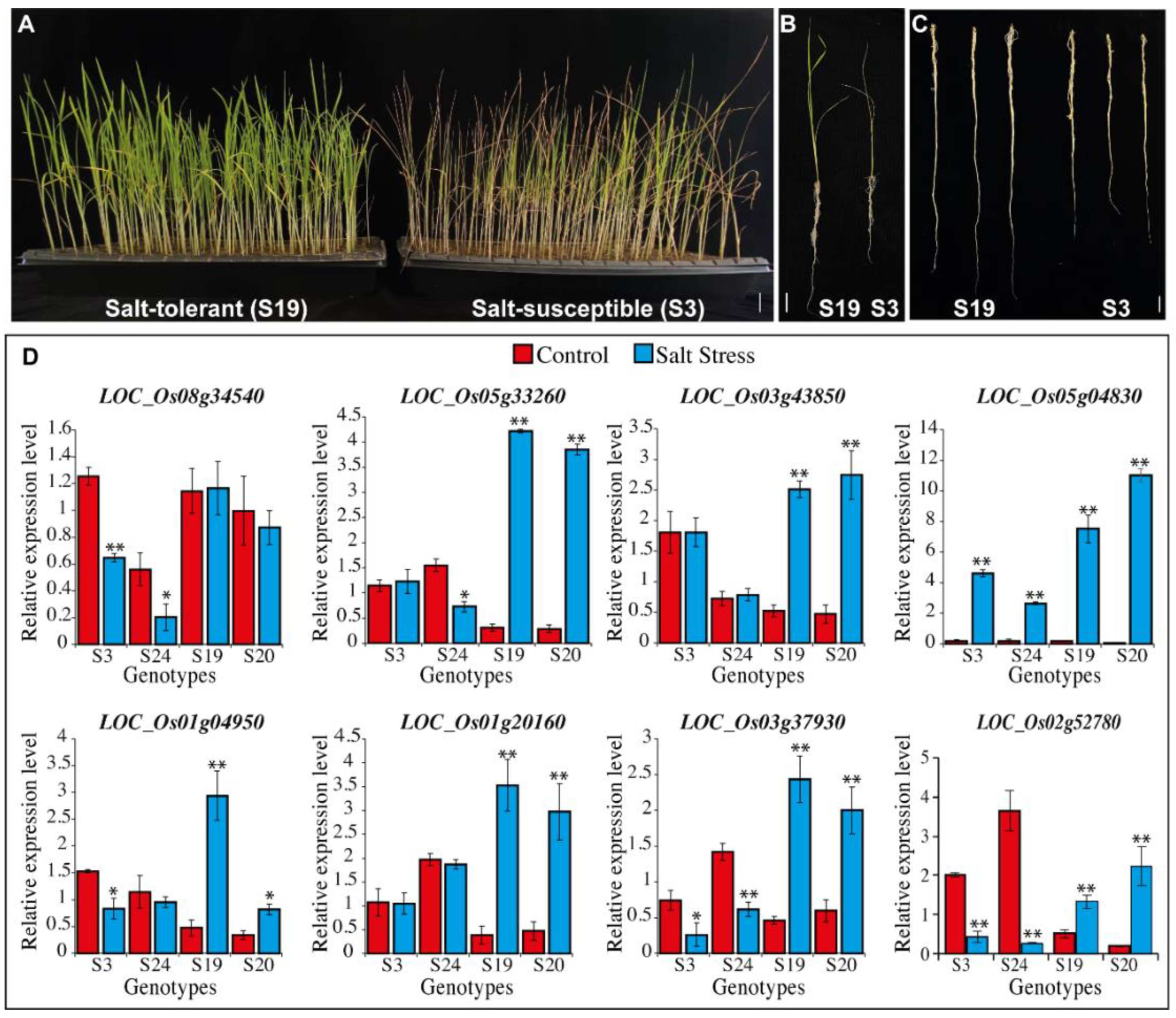
Expression Profiling Analysis in Salt-tolerant and Susceptible Genotypes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hakim, M.; Juraimi, A.; Hanafi, M.; Ismail, M.; Rafii, M.; Islam, M.; Selamat, A. The effect of salinity on growth, ion accumulation and yield of rice varieties. JAPS J. Anim. Plant Sci. 2014, 24, 874–885. [Google Scholar]
- Thomas, R.; Morini, S. Food and Agriculture Organization of the United Nations. 2005. Available online: https://www.fao.org/3/a0050e/a0050e.pdf (accessed on 1 January 2022).
- Yadav, S.; Irfan, M.; Ahmad, A.; Hayat, S. Causes of salinity and plant manifestations to salt stress: A review. J. Environ. Biol. 2011, 32, 667. [Google Scholar]
- Szabolcs, I. Soils and salinisation. In Handbook of Plant and Crop Stress; CRC Press: Boca Raton, FL, USA, 1994; pp. 3–11. [Google Scholar]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [Green Version]
- Zafar, S.A.; Shokat, S.; Ahmed, H.G.M.-D.; Khan, A.; Ali, M.Z.; Atif, R.M. Assessment of salinity tolerance in rice using seedling based morpho-physiological indices. Adv. Life Sci. 2015, 2, 142–149. [Google Scholar]
- Rehman, O.U.; Uzair, M.; Chao, H.; Fiaz, S.; Khan, M.R.; Chen, M. Role of the Type-B Authentic Response Regulator Gene Family in Fragrant Rice under Alkaline-salt Stress. Physiol. Plant. 2022, 174, e13696. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A.; Läuchli, A. Approaches to increasing the salt tolerance of wheat and other cereals. J. Exp. Bot. 2006, 57, 1025–1043. [Google Scholar] [CrossRef] [Green Version]
- Asch, F.; Dingkuhn, M.; Dörffling, K.; Miezan, K. Leaf K/Na ratio predicts salinity induced yield loss in irrigated rice. Euphytica 2000, 113, 109–118. [Google Scholar] [CrossRef]
- Uzair, M.; Ali, M.; Fiaz, S.; Attia, K.; Khan, N.; Al-Doss, A.A.; Khan, M.R.; Ali, Z. The characterization of wheat genotypes for salinity tolerance using morpho-physiological indices under hydroponic conditions. Saudi J. Biol. Sci. 2022, 29, 103299. [Google Scholar] [CrossRef]
- Flowers, T.; Flowers, S. Why does salinity pose such a difficult problem for plant breeders? Agric. Water Manag. 2005, 78, 15–24. [Google Scholar] [CrossRef]
- Thomson, M.J.; de Ocampo, M.; Egdane, J.; Rahman, M.A.; Sajise, A.G.; Adorada, D.L.; Tumimbang-Raiz, E.; Blumwald, E.; Seraj, Z.I.; Singh, R.K. Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice 2010, 3, 148–160. [Google Scholar] [CrossRef] [Green Version]
- Deng, P.; Shi, X.; Zhou, J.; Wang, F.; Dong, Y.; Jing, W.; Zhang, W. Identification and Fine Mapping of a Mutation Conferring Salt-Sensitivity in Rice (Oryza sativa L.). Crop Sci. 2015, 55, 219–228. [Google Scholar] [CrossRef]
- Ren, Z.-H.; Gao, J.-P.; Li, L.-G.; Cai, X.-L.; Huang, W.; Chao, D.-Y.; Zhu, M.-Z.; Wang, Z.-Y.; Luan, S.; Lin, H.-X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef]
- Demidchik, V.; Maathuis, F.J. Physiological roles of nonselective cation channels in plants: From salt stress to signalling and development. New Phytol. 2007, 175, 387–404. [Google Scholar] [CrossRef]
- Negrão, S.; Courtois, B.; Ahmadi, N.; Abreu, I.; Saibo, N.; Oliveira, M. Recent updates on salinity stress in rice: From physiological to molecular responses. Crit. Rev. Plant Sci. 2011, 30, 329–377. [Google Scholar] [CrossRef]
- Horie, T.; Motoda, J.; Kubo, M.; Yang, H.; Yoda, K.; Horie, R.; Chan, W.Y.; Leung, H.Y.; Hattori, K.; Konomi, M. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. Plant J. 2005, 44, 928–938. [Google Scholar]
- Lee, S.-K.; Kim, B.-G.; Kwon, T.-R.; Jeong, M.-J.; Park, S.-R.; Lee, J.-W.; Byun, M.-O.; Kwon, H.-B.; Matthews, B.F.; Hong, C.-B. Overexpression of the mitogen-activated protein kinase gene OsMAPK33 enhances sensitivity to salt stress in rice (Oryza sativa L.). J. Biosci. 2011, 36, 139–151. [Google Scholar] [CrossRef]
- Qi, W.; Sun, F.; Wang, Q.; Chen, M.; Huang, Y.; Feng, Y.-Q.; Luo, X.; Yang, J. Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol. 2011, 157, 216–228. [Google Scholar] [CrossRef] [Green Version]
- Kong, X.-Q.; Gao, X.-H.; Sun, W.; An, J.; Zhao, Y.-X.; Zhang, H. Cloning and functional characterization of a cation–chloride cotransporter gene OsCCC1. Plant Mol. Biol. 2011, 75, 567–578. [Google Scholar] [CrossRef]
- Xiang, Y.; Tang, N.; Du, H.; Ye, H.; Xiong, L. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 2008, 148, 1938–1952. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Yang, B.; He, Y.; Zhan, C.; Cheng, Y.; Zhang, J.; Zhang, H.; Cheng, J.; Wang, Z. A quantitative trait locus, qSE 3, promotes seed germination and seedling establishment under salinity stress in rice. Plant J. 2019, 97, 1089–1104. [Google Scholar] [CrossRef] [Green Version]
- Wing, R.A.; Purugganan, M.D.; Zhang, Q. The rice genome revolution: From an ancient grain to Green Super Rice. Nat. Rev. Genet. 2018, 19, 505–517. [Google Scholar] [CrossRef]
- Jewel, Z.A.; Ali, J.; Pang, Y.; Mahender, A.; Acero, B.; Hernandez, J.; Xu, J.; Li, Z. Developing green super rice varieties with high nutrient use efficiency by phenotypic selection under varied nutrient conditions. Crop J. 2019, 7, 368–377. [Google Scholar] [CrossRef]
- Ahmad, H.; Zafar, S.A.; Naeem, M.K.; Shokat, S.; Inam, S.; Naveed, A.S.; Xu, J.; Li, Z.; Ali, G.M.; Khan, M.R. Impact of pre-anthesis drought stress on physiology, yield-related traits and drought responsive genes in green super rice. Front. Genet. 2022. [Google Scholar] [CrossRef]
- Rahman, A.; Nahar, K.; Hasanuzzaman, M.; Fujita, M. Calcium supplementation improves Na+/K+ ratio, antioxidant defense and glyoxalase systems in salt-stressed rice seedlings. Front. Plant Sci. 2016, 7, 609. [Google Scholar] [CrossRef] [Green Version]
- Kushizaki, M. An extraction procedure of plant materials for the rapid determination of Mn, Cu, Zn and Mg by the atomic absorption analysis. J. Sci. Soil Manure 1968, 39, 489–490. [Google Scholar]
- Zhou, S.; Xu, Z.; Liu, F. Method for Determining the Optimal Number of Clusters Based on Agglomerative Hierarchical Clustering. IEEE Trans. Neural Netw. Learn. Syst. 2017, 28, 3007–3017. [Google Scholar] [CrossRef]
- Uzair, M.; Patil, S.B.; Zhang, H.; Kumar, A.; Mkumbwa, H.; Zafar, S.A.; Chun, Y.; Fang, J.; Zhao, J.; Khan, M.R. Screening Direct Seeding-Related Traits by Using an Improved Mesocotyl Elongation Assay and Association between Seedling and Maturity Traits in Rice. Agronomy 2022, 12, 975. [Google Scholar] [CrossRef]
- Sah, S.K.; Kaur, G.; Kaur, A. Rapid and reliable method of high-quality RNA extraction from diverse plants. Am. J. Plant Sci. 2014, 5, 3129. [Google Scholar] [CrossRef] [Green Version]
- Uzair, M.; Long, H.; Zafar, S.A.; Patil, S.B.; Chun, Y.; Li, L.; Fang, J.; Zhao, J.; Peng, L.; Yuan, S. Narrow Leaf21, encoding ribosomal protein RPS3A, controls leaf development in rice. Plant Physiol. 2021, 186, 497–518. [Google Scholar] [CrossRef]
- Garg, R.; Narayana Chevala, V.; Shankar, R.; Jain, M. Divergent DNA methylation patterns associated with gene expression in rice cultivars with contrasting drought and salinity stress response. Sci. Rep. 2015, 5, 14922. [Google Scholar] [CrossRef] [Green Version]
- Chandran, A.K.N.; Kim, J.-W.; Yoo, Y.-H.; Park, H.L.; Kim, Y.-J.; Cho, M.-H.; Jung, K.-H. Transcriptome analysis of rice-seedling roots under soil–salt stress using RNA-Seq method. Plant Biotechnol. Rep. 2019, 13, 567–578. [Google Scholar] [CrossRef]
- Lee, B.T.; Barber, G.P.; Benet-Pages, A.; Casper, J.; Clawson, H.; Diekhans, M.; Fischer, C.; Gonzalez, J.N.; Hinrichs, A.S.; Lee, C.M.; et al. The UCSC Genome Browser database: 2022 update. Nucleic Acids Res. 2022, 50, D1115–D1122. [Google Scholar] [CrossRef]
- Adeel Zafar, S.; Uzair, M.; Ramzan Khan, M.; Patil, S.B.; Fang, J.; Zhao, J.; Lata Singla-Pareek, S.; Pareek, A.; Li, X. DPS1 regulates cuticle development and leaf senescence in rice. Food Energy Secur. 2021, 10, e273. [Google Scholar] [CrossRef]
- Zaid, I.U.; Zahra, N.; Habib, M.; Naeem, M.K.; Asghar, U.; Uzair, M.; Latif, A.; Rehman, A.; Ali, G.M.; Khan, M.R. Estimation of Genetic Variances and Stability Components of Yield-Related Traits of Green Super Rice at Multi-Environmental Conditions in Pakistan. Agronomy 2022, 12, 1157. [Google Scholar] [CrossRef]
- Kruijer, W.; Flood, P.; Kooke, R. Heritability: Marker-Based Estimation of Heritability Using Individual Plant or Plot Data. 2016. Available online: https://cran.r-project.org/web/packages/heritability/heritability.pdf (accessed on 1 March 2022).
- Mohammadi, S.A.; Prasanna, B. Analysis of genetic diversity in crop plants—Salient statistical tools and considerations. Crop Sci. 2003, 43, 1235–1248. [Google Scholar] [CrossRef] [Green Version]
- Uzair, M.; Ali, Z.; Mahmood, T.; Karim, I.; Akram, U.; Mahmood, N.; Saeed, T.; Kalsoom, R. Genetic basis of some yield related traits in wheat (Triticum aestivum L.) under drought conditions. Imp. J. Interdiscip. Res. 2016, 2, 444–449. [Google Scholar]
- Zafar, S.A.; Patil, S.B.; Uzair, M.; Fang, J.; Zhao, J.; Guo, T.; Li, X. Degenerated panicle and partial sterility 1 (DPS 1) encodes a cystathionine β-synthase domain containing protein required for anther cuticle and panicle development in rice. New Phytol. 2020, 225, 356–375. [Google Scholar] [CrossRef] [Green Version]
- Zeng, L. Exploration of relationships between physiological parameters and growth performance of rice (Oryza sativa L.) seedlings under salinity stress using multivariate analysis. Plant Soil 2005, 268, 51–59. [Google Scholar] [CrossRef]
- Ali, M.; Yeasmin, L.; Gantait, S.; Goswami, R.; Chakraborty, S. Screening of rice landraces for salinity tolerance at seedling stage through morphological and molecular markers. Physiol. Mol. Biol. Plants 2014, 20, 411–423. [Google Scholar] [CrossRef] [Green Version]
- Bhowmik, U.; Kibria, M.G.; Rhaman, M.S.; Murata, Y.; Hoque, M.A. Screening of rice genotypes for salt tolerance by physiological and biochemical characters. Plant Sci. Today 2021, 8, 467–472. [Google Scholar] [CrossRef]
- Ali, Y.; Aslam, Z.; Ashraf, M.; Tahir, G. Effect of salinity on chlorophyll concentration, leaf area, yield and yield components of rice genotypes grown under saline environment. Int. J. Environ. Sci. Technol. 2004, 1, 221–225. [Google Scholar] [CrossRef] [Green Version]
- Bibha, R.; Sharma, V. Screening of rice genotypes for salt tolerance in relation to morphological traits and yield components under field condition. Int. J. Agric. Sci. 2017, 9, 4493–4497. [Google Scholar]
- Rad, H.E.; Aref, F.; Rezaei, M. Response of rice to different salinity levels during different growth stages. Res. J. Appl. Sci. Eng. Technol. 2012, 4, 3040–3047. [Google Scholar]
- Anshori, M.F.; Purwoko, B.S.; Dewi, I.S.; Ardie, S.W.; Suwarno, W.B. A new approach to select doubled haploid rice lines under salinity stress using indirect selection index. Rice Sci. 2021, 28, 368–378. [Google Scholar] [CrossRef]
- Farid, M.; Nasaruddin, M.Y.; Ridwan, I.; Anshori, M. Effective screening of tropical wheat mutant lines under hydroponically induced drought stress using multivariate analysis approach. Asian J. Plant Sci. 2021, 20, 172–182. [Google Scholar] [CrossRef]
- Arifuddin, M.; Musa, Y.; Farid, M.; Anshori, M.; Nasaruddin, N.; Nur, A.; Sakinah, A. Rice screening with hydroponic deep-flow technique under salinity stress. SABRAO J. Breed. Genet. 2021, 53, 435–446. [Google Scholar]
- Safitri, H.; Purwoko, B.S.; Dewi, I.S.; Ardie, S.W. Morpho-physiological response of rice genotypes grown under saline conditions. J. Int. Soc. Southeast Asian Agric. Sci. 2016, 22, 52–63. [Google Scholar]
- Ferdose, J.; Kawasaki, M.; Taniguchi, M.; Miyake, H. Differential sensitivity of rice cultivars to salinity and its relation to ion accumulation and root tip structure. Plant Prod. Sci. 2009, 12, 453–461. [Google Scholar] [CrossRef]
- Negrão, S.; Schmöckel, S.; Tester, M. Evaluating physiological responses of plants to salinity stress. Ann. Bot. 2017, 119, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Patil, S.B.; Barbier, F.F.; Zhao, J.; Zafar, S.A.; Uzair, M.; Sun, Y.; Fang, J.; Perez-Garcia, M.D.; Bertheloot, J.; Sakr, S.; et al. Sucrose promotes D53 accumulation and tillering in rice. New Phytol. 2022, 234, 122–136. [Google Scholar] [CrossRef]
- Gholizadeh, A.; Dehghania, H.; Dvorakb, J. Determination of the most effective traits on wheat yield under saline stress. Agric. Adv. 2014, 3, 103–110. [Google Scholar]
- Saade, S.; Maurer, A.; Shahid, M.; Oakey, H.; Schmöckel, S.M.; Negrão, S.; Pillen, K.; Tester, M. Yield-related salinity tolerance traits identified in a nested association mapping (NAM) population of wild barley. Sci. Rep. 2016, 6, 32586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barua, R.; De Ocampo, M.; Egdane, J.; Ismail, A.M.; Mondal, S. Phenotyping rice (Oryza sativa L.) genotypes for physiological traits associated with tolerance of salinity at seedling stage. Sci. Agric. 2015, 12, 156–162. [Google Scholar]
- Chen, T.; Shabala, S.; Niu, Y.; Chen, Z.-H.; Shabala, L.; Meinke, H.; Venkataraman, G.; Pareek, A.; Xu, J.; Zhou, M. Molecular mechanisms of salinity tolerance in rice. Crop J. 2021, 9, 506–520. [Google Scholar] [CrossRef]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, B.N.; Sadras, V.O.; Tester, M. A water-centred framework to assess the effects of salinity on the growth and yield of wheat and barley. Plant Soil 2010, 336, 377–389. [Google Scholar] [CrossRef]
- Katschnig, D.; Bliek, T.; Rozema, J.; Schat, H. Constitutive high-level SOS1 expression and absence of HKT1; 1 expression in the salt-accumulating halophyte Salicornia dolichostachya. Plant Sci. 2015, 234, 144–154. [Google Scholar] [CrossRef]
- Zhang, Y.; Fang, J.; Wu, X.; Dong, L. Na+/K+ balance and transport regulatory mechanisms in weedy and cultivated rice (Oryza sativa L.) under salt stress. BMC Plant Biol. 2018, 18, 375. [Google Scholar] [CrossRef]
- Assaha, D.V.; Ueda, A.; Saneoka, H.; Al-Yahyai, R.; Yaish, M.W. The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front. Physiol. 2017, 8, 509. [Google Scholar] [CrossRef]
- Fadhli, N.; Farid, M.; Effendi, R.; Azrai, M.; Anshori, M.F. Multivariate analysis to determine secondary characters in selecting adaptive hybrid corn lines under drought stress. Biodivers. J. Biol. Divers. 2020, 21. [Google Scholar] [CrossRef]
- Anshori, M.; Purwoko, B.; Dewi, I.; Ardie, S.; Suwarno, W. Selection index based on multivariate analysis for selecting doubled-haploid rice lines in lowland saline prone area. SABRAO J. Breed. Genet. 2019, 51, 161–174. [Google Scholar]
- Shuvo, I.I. A holistic decision-making approach for identifying influential parameters affecting sustainable production process of canola bast fibres and predicting end-use textile choice using principal component analysis (PCA). Heliyon 2021, 7, e06235. [Google Scholar] [CrossRef] [PubMed]
- Andrade, G.C.; Coelho, C.M.M.; Uarrota, V.G. Modelling the vigour of maize seeds submitted to artificial accelerated ageing based on ATR-FTIR data and chemometric tools (PCA, HCA and PLS-DA). Heliyon 2020, 6, e03477. [Google Scholar] [CrossRef] [PubMed]
- Azam, M.; Zhang, S.; Abdelghany, A.M.; Shaibu, A.S.; Feng, Y.; Li, Y.; Tian, Y.; Hong, H.; Li, B.; Sun, J. Seed isoflavone profiling of 1168 soybean accessions from major growing ecoregions in China. Food Res. Int. 2020, 130, 108957. [Google Scholar] [CrossRef] [PubMed]
- Das, B.; Manohara, K.; Mahajan, G.; Sahoo, R.N. Spectroscopy based novel spectral indices, PCA-and PLSR-coupled machine learning models for salinity stress phenotyping of rice. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 229, 117983. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Gao, L.; Wu, Z.; Zhang, X.; Wang, M.; Zhang, C.; Zhang, F.; Zhou, Y.; Li, Z. Genome-wide association study of salt tolerance at the seed germination stage in rice. BMC Plant Biol. 2017, 17, 92. [Google Scholar] [CrossRef] [Green Version]
- Pongprayoon, W.; Tisarum, R.; Theerawittaya, C.; Cha-Um, S. Evaluation and clustering on salt-tolerant ability in rice genotypes (Oryza sativa L. subsp. indica) using multivariate physiological indices. Physiol. Mol. Biol. Plants 2019, 25, 473–483. [Google Scholar] [CrossRef]
- Walia, H.; Wilson, C.; Zeng, L.; Ismail, A.M.; Condamine, P.; Close, T.J. Genome-wide transcriptional analysis of salinity stressed japonica and indica rice genotypes during panicle initiation stage. Plant Mol. Biol. 2007, 63, 609–623. [Google Scholar] [CrossRef] [Green Version]


| SOV | Genotype (G) | Treatment (T) | G × T | Error | Total | Heritability (H2) |
|---|---|---|---|---|---|---|
| DF | 23 | 1 | 23 | 92 | 143 | |
| SL | 15.15 *** | 80.27 *** | 4.72 * | 2.58 | 72.68 | |
| RL | 7.77 ns | 196.88 *** | 5.11 ns | 6.64 | 14.42 | |
| TPL | 27.22 ** | 528.6 *** | 14.1 ns | 13.07 | 50.09 | |
| SFW | 0.0022 *** | 0.2007 *** | 0.0005 ns | 0.008 | 65.60 | |
| RFW | 0.0012 ** | 0.2097 *** | 0.0003 ns | 0.006 | 52.69 | |
| SDW | 0.0002 *** | 0.0126 *** | 0.0001 *** | 0.000 | 72.39 | |
| RDW | 0.7205 ns | 0.0367 ns | 0.7828 ns | 0.7519 | 0.001 | |
| SNC | 4.05 *** | 665.64 *** | 6.6 *** | 0.0864 | 37.57 | |
| SKC | 3.65 *** | 108.16 *** | 1.44 *** | 0.1137 | 71.01 | |
| RNC | 0.211 *** | 15.34 *** | 0.224 *** | 0.0067 | 47.68 | |
| RKC | 0.424 *** | 5.46 *** | 0.455 *** | 0.0068 | 47.85 | |
| NKR | 0.23 *** | 38.74 *** | 0.29 *** | 0.0087 | 43.69 |
| Trait | Salt-Tolerant Genotypes Names and Mean Values | Salt-Susceptible Genotypes Names and Mean Values |
|---|---|---|
| SL | S13 (29.91), S1 (28.52), and S22 (27.14) | S3 (20.34), S11(22.92), and S8 (22.97) |
| RL | S5 (21.16), S3 (20.83), and S20 (20.41) | S8 (14.81), S24 (15.7), and S9 (16.51) |
| TPL | S4 (49.35), S5 (48.33), and S3 (47.6) | S24 (36.86), S9 (37.73), and S19 (39.83) |
| SFW | S4 (0.159), S5 (0.142), and S3 (0.141) | S24 (0.080), S9 (0.088), and S19 (0.094) |
| RFW | S12 (0.124), S24 (0.122), and S8 (0.113) | S17 (0.064), S7 (0.065), and S21 (0.066) |
| SDW | S5 (0.048), S2 (0.047), and S17 (0.045) | S9 (0.026), S19 (0.028), and S21 (0.029) |
| RDW | S18 (2.23), S10 (0.022), and S24 (0.020) | S23 (0.012), S3 (0.012), and S1 (0.013) |
| SNC | S21 (9.13), S5 (8.83), and S18 (8.63) | S15 (2.93), S22 (3.53), and S11 (3.6) |
| SKC | S24 (6.06), S15 (5.93), and S11 (5.46) | S20 (2.43), S7 (2.60), and S12 (2.63) |
| RNC | S17 (1.70), S22 (1.67), and S14 (1.62) | S24 (0.85), S12 (0.85), and S16 (0.92) |
| RKC | S10 (2.98), S1 (1.97), and S6 (1.91) | S17 (0.85), S23 (0.95), and S22 (1.15) |
| NKR | S15 (2.34), S10 (2.15), and S2 (2.12) | S3 (0.76), S16 (0.83), and S4 (1.01) |
| SOV | Genotype (G) | Treatment (T) | G × T | Error | Total | Heritability |
|---|---|---|---|---|---|---|
| DF | 23 | 1 | 23 | 92 | 143 | |
| PH | 282.9 *** | 31.0 ns | 14.0 ns | 13.63 | 93.21 | |
| TN | 28.3 ** | 1614.7 *** | 10.19 ns | 12.10 | 57.23 | |
| GY | 391.56 *** | 104959.36 *** | 151.45 ns | 151.40 | 61.32 | |
| SY | 861.98 ** | 12886 *** | 439 ns | 339.18 | 54.35 | |
| TGW | 39.21 *** | 205.76 *** | 3.73 ns | 2.01 | 90.89 | |
| GL | 4.30 *** | 0.166 ns | 0.155 ns | 0.10 | 96.43 | |
| HI | 0.0056 *** | 0.8445 *** | 0.0045 *** | 0.001 | 48.10 |
| Trait | Salt-Tolerant Genotypes Names and Mean Values | Salt-Susceptible Genotypes Names and Mean Values |
|---|---|---|
| PH | S3 (117.53), S23 (114), and S24 (113.8) | S18 (88.33), S8 (92.13), and S5 (93.33) |
| TN | S9 (22.2), S19 (22), and S24 (21.8) | S4 (14.33), S2 (14.73), and S6 (15) |
| GY | S9 (51.46), S8 (48.8), and S22 (42.66) | S1 (23.93), S6 (24.83), and S24 (27.2) |
| SY | S1 (125.6), S20 (115.06), and S23 (108.93) | S8 (43.6), S17 (64.26), and S19 (70.66) |
| TGW | S1 (30.62), S20 (24.63), and S19 (24.12) | S4 (17.43), S6 (17.63), and S12 (17.99) |
| GL | S1 (12.18), S12 (11.14), and S15 (10.05) | S2 (7.81), S10 (8.49), and S23 (8.81) |
| HI | S12 (0.41), S14 (0.37), and S9 (0.36) | S2 (0.15), S5 (0.20), and S1 (0.22) |
| SSI | S19 (1.16), S18 (0.94), and S23 (0.94) | S16 (0.743), S17 (0.746), and S2 (0.759) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amanat, M.A.; Naeem, M.K.; Algwaiz, H.I.M.; Uzair, M.; Attia, K.A.; AlKathani, M.D.F.; Zaid, I.U.; Zafar, S.A.; Inam, S.; Fiaz, S.; et al. Evaluation of Green Super Rice Lines for Agronomic and Physiological Traits under Salinity Stress. Plants 2022, 11, 1461. https://doi.org/10.3390/plants11111461
Amanat MA, Naeem MK, Algwaiz HIM, Uzair M, Attia KA, AlKathani MDF, Zaid IU, Zafar SA, Inam S, Fiaz S, et al. Evaluation of Green Super Rice Lines for Agronomic and Physiological Traits under Salinity Stress. Plants. 2022; 11(11):1461. https://doi.org/10.3390/plants11111461
Chicago/Turabian StyleAmanat, Muhammad Ammar, Muhammad Kashif Naeem, Hussah I. M. Algwaiz, Muhammad Uzair, Kotb A. Attia, Muneera D. F. AlKathani, Imdad Ulah Zaid, Syed Adeel Zafar, Safeena Inam, Sajid Fiaz, and et al. 2022. "Evaluation of Green Super Rice Lines for Agronomic and Physiological Traits under Salinity Stress" Plants 11, no. 11: 1461. https://doi.org/10.3390/plants11111461
APA StyleAmanat, M. A., Naeem, M. K., Algwaiz, H. I. M., Uzair, M., Attia, K. A., AlKathani, M. D. F., Zaid, I. U., Zafar, S. A., Inam, S., Fiaz, S., Arif, M. H., Ahmad, D., Zahra, N., Saleem, B., & Khan, M. R. (2022). Evaluation of Green Super Rice Lines for Agronomic and Physiological Traits under Salinity Stress. Plants, 11(11), 1461. https://doi.org/10.3390/plants11111461








