Genetic Structure of Native Blue Honeysuckle Populations in the Western and Eastern Eurasian Ranges
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
| Sample Number | Site | Code | Habitat | Coordinates (Latitude N, Longitude E) | Altitude, m | Climate Zone 1 |
|---|---|---|---|---|---|---|
| JAPAN | ||||||
| 1 | Taiki, Hokkaido | JP | Alder forest, Quercus crispula and Q. dentata forest | 42.31–42.33, 143.28–143.30 | 0–70 | Dfb |
| 2–3 | Yokotsu-dake, Hokkaido | JP | Rocky area around mountaintop | 41.56, 140.46 | 1120–1150 | Dfb |
| 4 | Taisetsu, Hokkaido | JP | Around mountainous mire | 43.36–43.89, 142.53 | 1700–1800 | Dfb |
| 5 | Betsukai, Hokkaido | JP | Area drained by agricultural development in Fuhren mire | 43.16, 145.11 | 20 | Dfb |
| 6 | Bekanbe, Hokkaido | JP | Alder forest | 43.10, 144.51 | 0–10 | Dfb |
| 7–14 | Kiritappu, Hokkaido | JP | Intermediate moor | 43.02–43.08, 145.01–145.06 | 0–10 | Dfb |
| 15–18 | Nikko, Tochigi, Honshu | JP | Area surrounding Senjogahara mire | 36.46, 139.26 | 1390–1400 | Dfa |
| 19 | Tomakomai, Hokkaido | JP | Area drained by industrial development | No data | No data | Dfb |
| 20 | Yufutsu, Hokkaido | JP | Area drained by industrial development | 42.40,141.45 | No data | Dfb |
| 21–23 | Apoi-dake, Hokkaido | JP | Mountain area | 42.06, 143.01 | 600 | Dfb |
| LATVIA | ||||||
| 24–35 | Kemeri | LV1 | Fen with birch and willow | 56.97, 23.54 | 0–79 | Dfb |
| 36–47 | Kandava | LV2 | Abava river valley, open place, shrubs with Pentaphylloides fruticosa | 57.02, 22.78 | 38–41 | Dfb |
| 48–58 | Mikeltornis | LV3 | Baltic Sea coastal pinewood | 57.60, 21.97 | 8 | Dfb |
| 59–69 | Venspils | LV4 | Baltic Sea coastal pinewood, dunes | 57.45, 21.60 | 8 | Dfb |
| ESTONIA | ||||||
| 70–83 | Kalli | EE | Fen with alder and birch | 58.50, 24.09 | 22 | Dfb |
| RUSSIA | ||||||
| 84–104 | Elizovo | RU1 | Swamp, peat soils, bumps, puddles Mossy marsh with thickets of alder and willow | 53.24 158.40 | 21 | Dfc |
| 105–121 | Petropavlovsk-Kamchatsky 1 | RU2 | Krasnaya Sopka, uphill, southeastern slope. Dry soils, peat, volcanic slag. Forest with birch, alder, willow, wild rose | 52.99, 158.67 | 200 | Dfc |
| 122–140 | Petropavlovsk-Kamchatsky 2 | RU3 | Zerkalnaya Sopka, on the slope of the hill. Forest with birch, wild rose, alder, rowan | 53.04, 158.67 | 147 | Dfc |
2.2. Genomic DNA Extraction and ISSR-PCR Analysis
2.3. Chloroplast DNA Analysis
2.4. Data Analysis
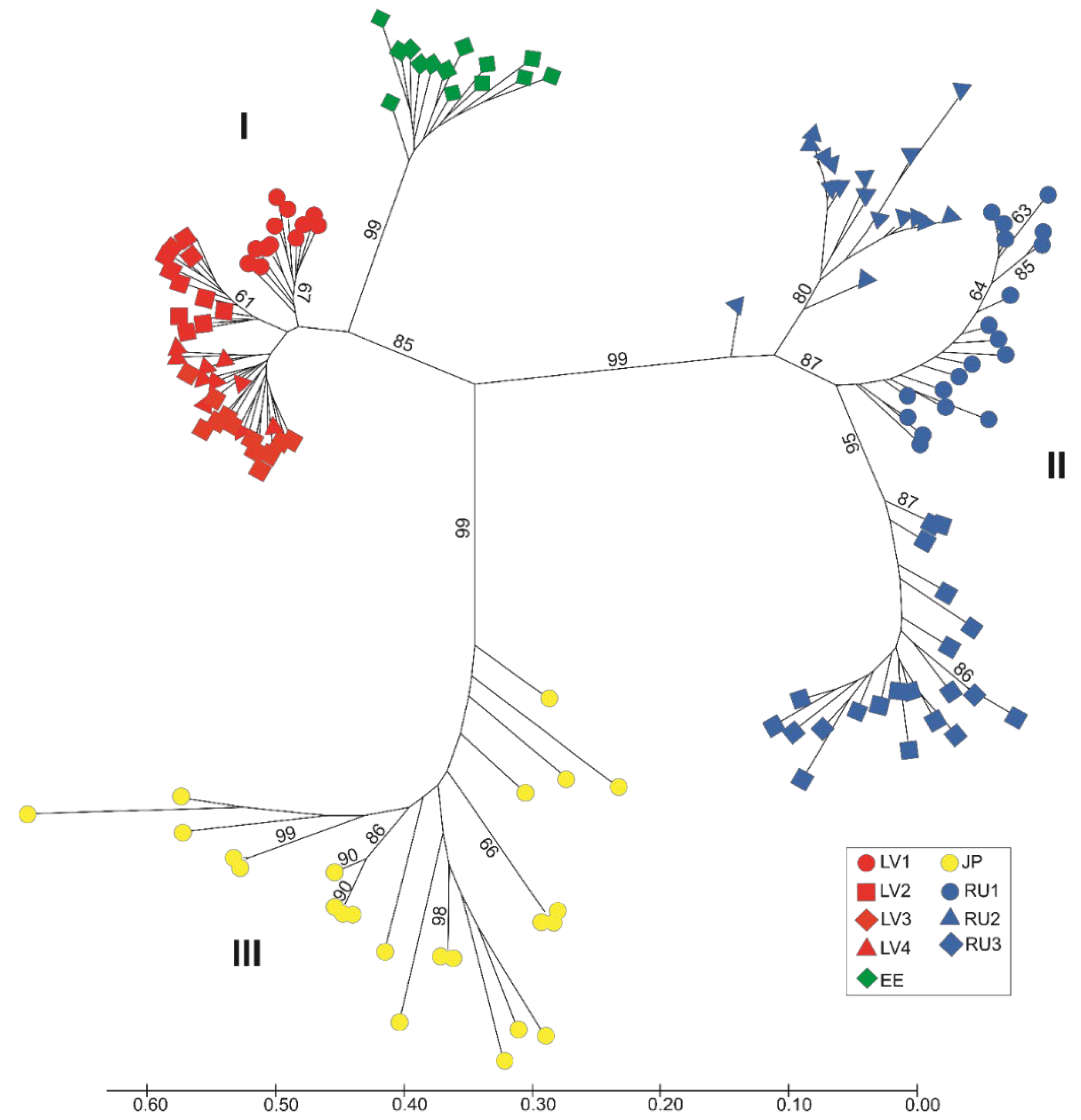
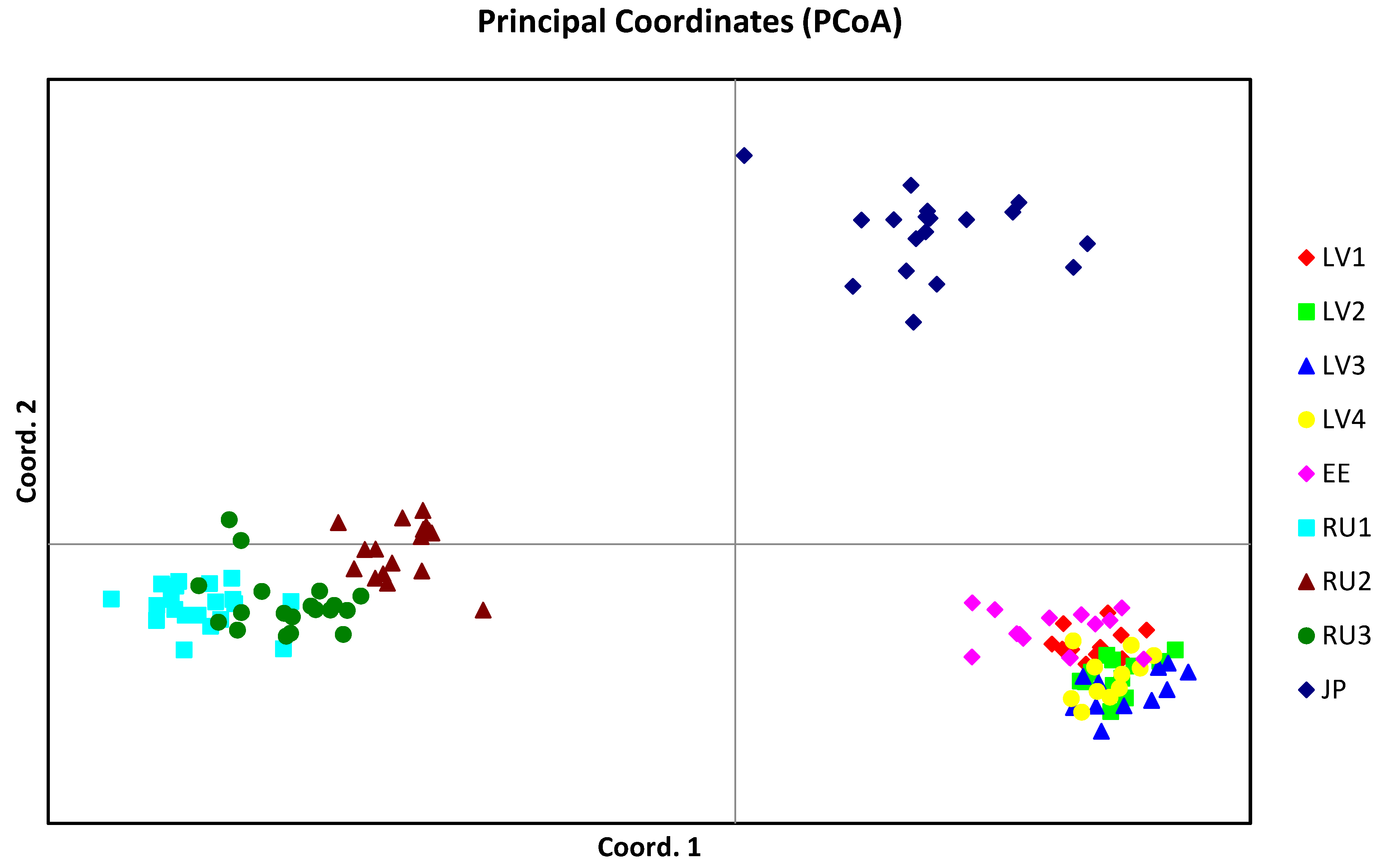
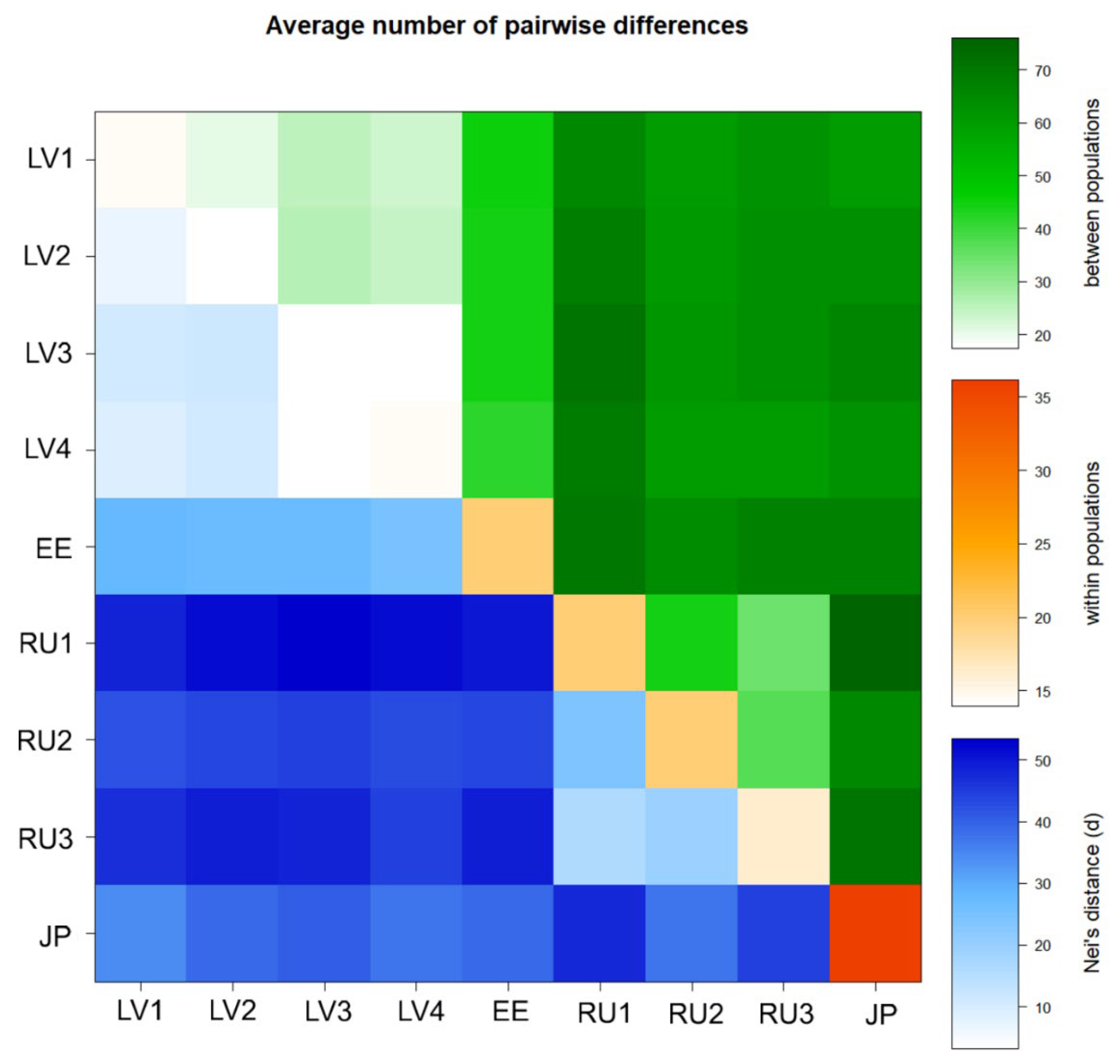
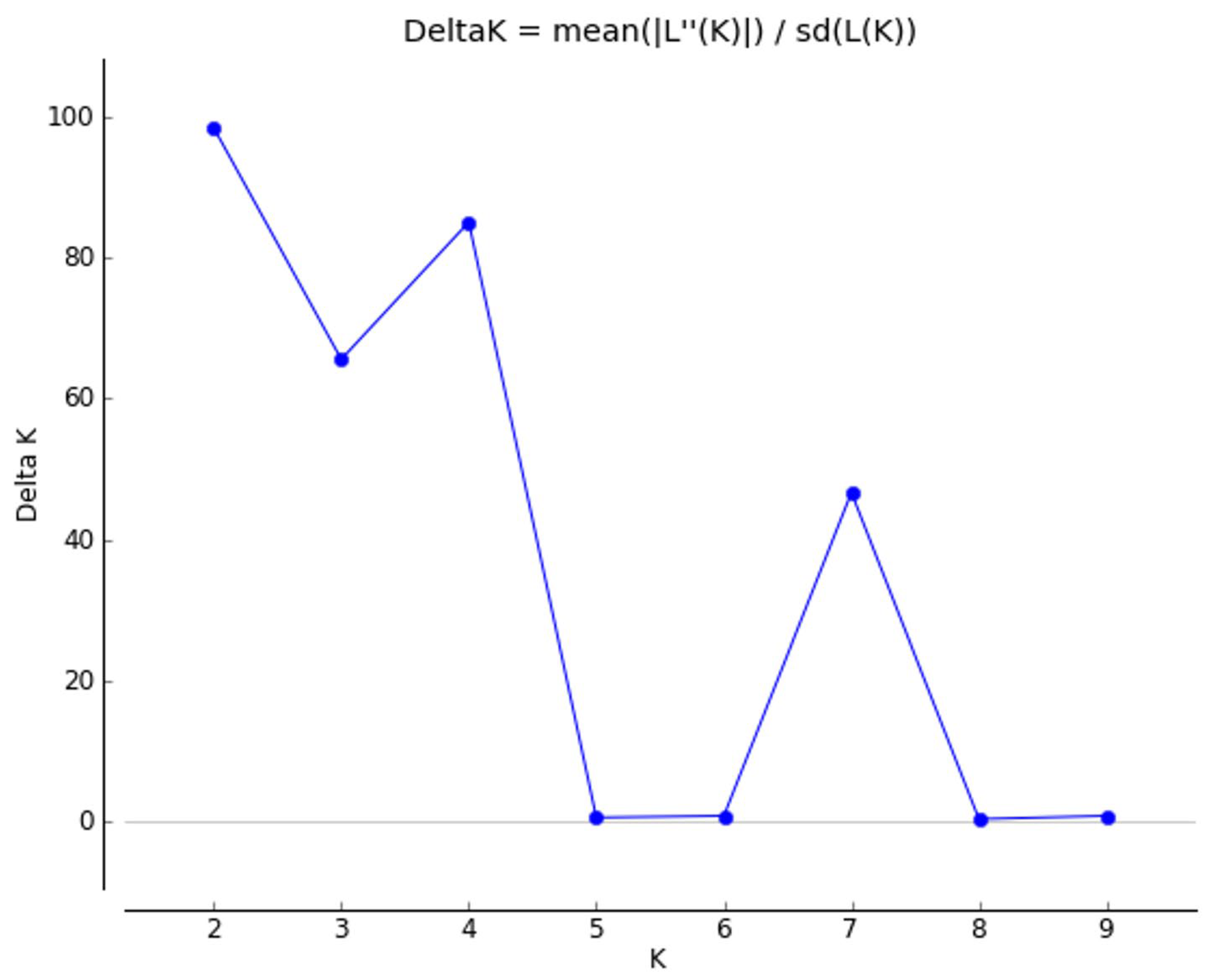
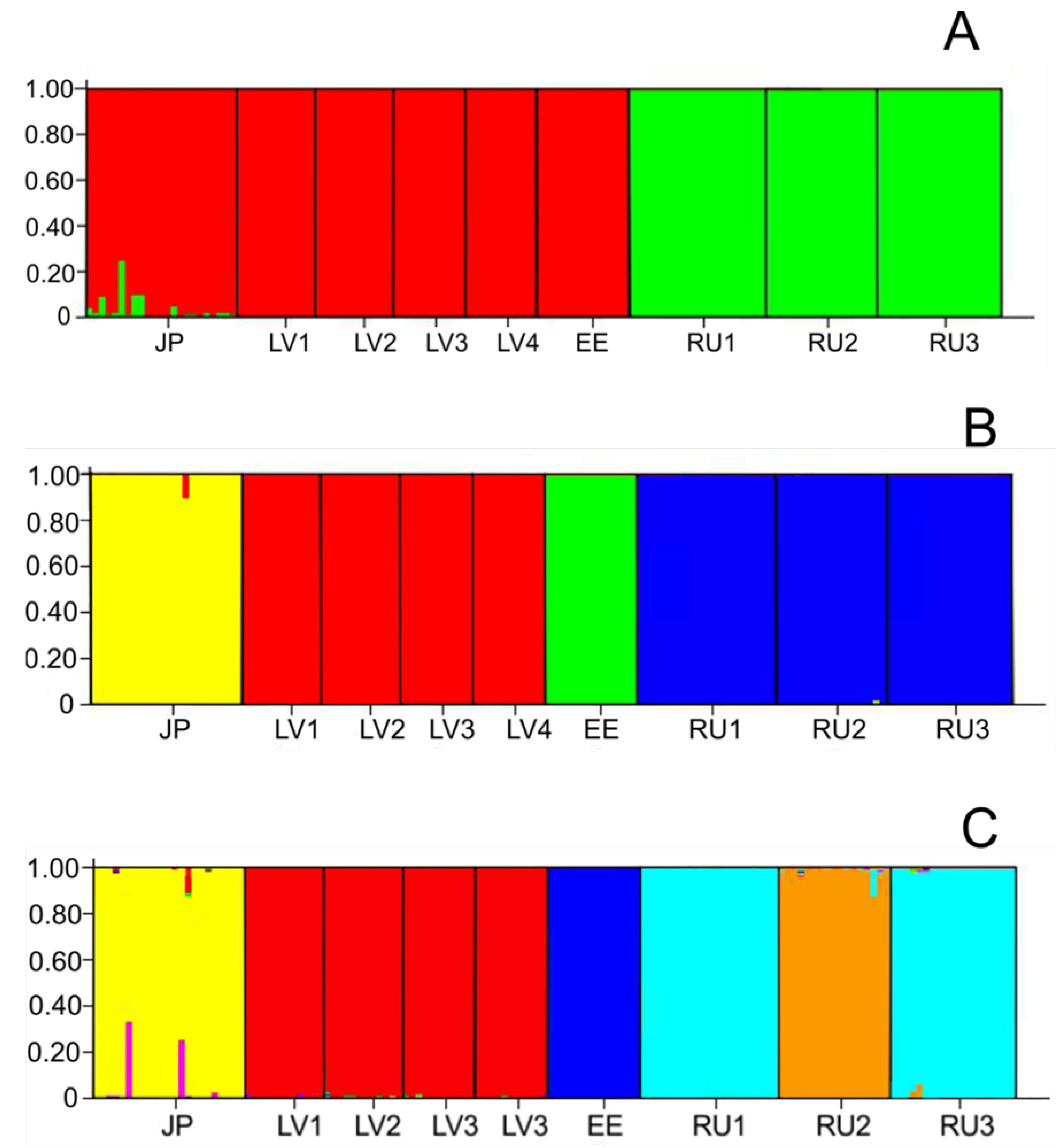
3. Results
4. Discussion
4.1. Genetic Diversity
4.2. Genetic Structure of Populations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Plekhanova, M.N. Blue honeysuckle (Lonicera caerulea L.) a new commercial berry crop for temperate climate: Genetic resources and breeding. Acta Hortic. 2000, 538, 159–164. [Google Scholar] [CrossRef]
- Lamoureux, D.; Sorokin, A.; Lefebre, I.; Alexanian, S.; Eyzaguirre, P.; Hausman, J.F. Investigation of genetic diversity in Russian collection of raspberry and blue honeysuckle. Plant Genet. Res. 2011, 9, 202–205. [Google Scholar] [CrossRef]
- Gerbrandt, E.M.; Bors, R.H.; Chibbar, R.N.; Baumann, T.E. Spring phenological adaptation of blue honeysuckle (Lonicera caerulea L.) foundation germplasm in a temperate climate. Can. J. Plant Sci. 2018, 98, 569–581. [Google Scholar] [CrossRef] [Green Version]
- Holubec, V.; Smekalova, T.; Leisova-Svobodova, L. Morphological and molecular evaluation of the Far East fruit genetic resources of Lonicera caerulea L.—Vegetation, ethnobotany, use and conservation. Genet. Resour. Crop Evol. 2019, 66, 121–141. [Google Scholar] [CrossRef]
- Kithma, A.B.; Rupasinghe, H.P.V. Polyphenols composition and anti-diabetic properties in vitro of haskap (Lonicera caerulea L.) berries in relation to cultivar and harvesting date. Food Compos. Anal. 2020, 88, 103402. [Google Scholar]
- Chaovanalikit, A.; Thompson, M.M.; Ronald, W.E. Characterization and quantification of anthocyanins and polyphenolics in blue honeysuckle (Lonicera caerulea L.). J. Agric. Food Chem. 2004, 52, 848–852. [Google Scholar] [CrossRef] [PubMed]
- Rupasinghe, H.P.V.; Arumuggam, N.; Amararathna, M.; De Silva, A.B.K.H. The potential health benefits of haskap (Lonicera caerulea L.): Role of cyanidin-3-O-glucoside. J. Funct. Foods 2018, 44, 24–39. [Google Scholar] [CrossRef]
- Gawroński, J.; Żebrowska, J.; Pabich, M.; Jackowska, I.; Kowalczyk, K.; Dyduch-Siemińska, M. Phytochemical characterization of blue honeysuckle in relation to the genotypic diversity of Lonicera sp. Appl. Sci. 2020, 10, 6545. [Google Scholar] [CrossRef]
- Smolik, M.; Ochmian, I.; Bobrowska-Chwat, A.; Chwat, G.; Arus, L.; Banaszczak, P.; Bocianowski, J.; Milczarski, P.; Ostrowska, K. Fingerprinting, structure, and genetic relationships among selected accessions of blue honeysuckle (Lonicera caerulea L.) from European collections. Biotechnol. Rep. 2022, 34, e00721. [Google Scholar] [CrossRef]
- Kuklina, A.G. The Nature of the Variability of Blue Honeysuckle in Natural and Cultural Populations. In New and Non-Traditional Plants and Prospects for Their Use, Proceedings of the V International Symposium, Pushchino, Russia, 9–14 June 2003; RAAS: Moscow-Pushchino, Russia, 2003; pp. 342–344. (In Russian) [Google Scholar]
- Miyashita, T.; Hoshino, Y. Interploid and intraploid hybridizations to produce polyploid Haskap (Lonicera caerulea var. emphyllocalyx) plants. Euphytica 2015, 201, 15–27. [Google Scholar] [CrossRef] [Green Version]
- Smith, S.A. Taking into account phylogenetic and divergence-time uncertainty in a parametric biogeographical analysis of the Northern Hemisphere plant clade Caprifolieae. J. Biogeogr. 2009, 36, 2324–2337. [Google Scholar] [CrossRef]
- Skvortsov, A.K. Blue honeysuckles (Lonicera subsect. Caeruleae) of Eurasia: Distribution, taxonomy, chromosome numbers, domestication. Acta Univ. Ups. Symb. Bot. Ups. 1986, 27, 95–105. [Google Scholar]
- Birkmane, K.Y.; Tabaka, L.V. Protected Plant Species of the Latvian SSR; Zinatne: Riga, Russia, 1974; p. 61. (In Russian) [Google Scholar]
- Kuklina, A.G. Rare Isolated Populations of Blue Honeysuckle (Lonicera caerulea L.) in Central Russia. In The Role of Botanical Gardens and Protected Natural Areas in the Study and Conservation of Plant and Fungal Diversity, Proceedings of the All-Russian Scientific Conference, Yaroslavl, Russia, 13–16 October 2011; YaSPU Publishing House: Yaroslavl, Russia, 2011; pp. 74–75. (In Russian) [Google Scholar]
- Brown, J.H. On the relationship between abundance and distribution of species. Am. Nat. 1984, 124, 255–279. [Google Scholar] [CrossRef]
- Beatty, G.E.; McEvoy, P.M.; Sweeney, O.; Provan, J. Range-edge effects promote clonal growth in peripheral populations of the one-sided wintergreen (Orthilia secunda). Divers. Distrib. 2008, 14, 546–555. [Google Scholar] [CrossRef]
- Hirsch, H.; Wagner, V.; Danihelka, J.; Ruprecht, E.; Sánchez-Gómez, P.; Seifert, M.; Hensen, I. High genetic diversity declines towards the geographic range periphery of Adonis vernalis, a Eurasian dry grassland plant. Plant Biol. 2015, 17, 1233–1241. [Google Scholar] [CrossRef]
- Naugžemys, D.; Žilinskaitė, S.; Denkovskij, J.; Patamsytė, J.; Literskis, J.; Žvingila, D. RAPD based study of genetic variation and relationships among Lonicera germplasm accession. Biologija 2007, 53, 34–39. [Google Scholar]
- Smolik, M.; Ochmian, I.; Grajkowski, J. Genetic variability of Polish and Russian accessions of cultivated blue honeysuckle (Lonicera caerulea). Rus. J. Genet. 2010, 46, 960–966. [Google Scholar] [CrossRef]
- Naugžemys, D.; Žilinskaitė, S.; Kleizaitė, V.; Skridaila, A.; Žvingila, D. Assessment of genetic variation among elite and wild germplasm of blue honeysuckle (Lonicera caerulea L.). Balt. For. 2011, 17, 8–16. [Google Scholar]
- Naugžemys, D.; Žilinskaitė, S.; Skridaila, A.; Žvingila, D. Phylogenetic analysis of the polymorphic 4x species complex Lonicera caerulea (Caprifoliaceae) using RAPD markers and noncoding chloroplast DNA sequences. Biologia 2014, 69, 585–593. [Google Scholar] [CrossRef]
- Weising, K.; Nybom, H.; Wolff, K.; Kahl, G. DNA Fingerprinting in Plants: Principles, Methods, and Applications, 2nd ed.; CRC Press; Taylor & Francis Group: Boca Raton, FL, USA, 2005; pp. 47–49. [Google Scholar]
- Krokaitė, E.; Janulionienė, R.; Jocienė, L.; Rekašius, T.; Rajackaitė, G.; Paulauskas, A.; Marozas, V.; Kupčinskienė, E. Relating invasibility and invasiveness: Case study of Impatiens parviflora. Front. Ecol. Evol. 2022, 10, 845947. [Google Scholar] [CrossRef]
- Zhang, Y.; Iaffaldano, B.J.; Zhuang, X.; Cardina, J.; Cornish, K. Chloroplast genome resources and molecular markers differentiate rubber dandelion species from weedy relatives. BMC Plant Biol. 2017, 17, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyashita, T.; Araki, H.; Hoshino, Y. Ploidy distribution and DNA content variations of Lonicera caerulea (Caprifoliaceae) in Japan. J. Plant Res. 2011, 124, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Beck, H.; Zimmermann, N.; McVicar, T.; Vergopolan, N.; Berg, A.; Wood, E. Present and future Köppen-Geiger climate classification maps at 1-km resolution. Sci. Data 2018, 5, 180214. [Google Scholar] [CrossRef] [Green Version]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 39–40. [Google Scholar]
- Butkuvienė, J.; Sinkevičienė, Z.; Naugžemys, D.; Patamsytė, J.; Žvingila, D. Genetic diversity of Batrachium (Ranunculaceae) species reveals the necessity of their protection in Lithuanian rivers. Aquat. Bot. 2017, 142, 61–70. [Google Scholar] [CrossRef]
- Patamsytė, J.; Rančelis, V.; Čėsnienė, T.; Kleizaitė, V.; Tunaitienė, V.; Naugžemys, D.; Vaitkūnienė, V.; Žvingila, D. Clonal structure and reduced diversity of the invasive alien plant Erigeron annuus in Lithuania. Cent. Eur. J. Biol. 2013, 8, 898–911. [Google Scholar] [CrossRef]
- Shaw, J.; Lickey, E.B.; Beck, J.T.; Farmer, S.B.; Liu, W.S.; Miller, J.; Siripun, K.C.; Winder, C.T.; Schilling, E.E.; Small, R.L. The tortoise and the hare II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 2005, 92, 142–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coart, E.; Van Glabeke, S.; Petit, R.J.; Van Bockstaele, E.; Roldan-Ruiz, I. Range wide versus local patterns of genetic diversity in hornbeam (Carpinus betulus L.). Conserv. Genet. 2005, 6, 259–273. [Google Scholar] [CrossRef]
- Yeh, F.C.; Yang, R.; Boyle, T. POPGENE: Microsoft Windows-Based Freeware for Population Genetic Analysis: Release 1.31; University of Alberta: Edmonton, AB, Canada, 1999. [Google Scholar]
- Vekemans, X.; Beauwens, T.; Lemaire, M.; Roldan-Ruiz, I. Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Mol. Ecol. 2002, 11, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef]
- Evanno, G.; Regnault, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, N.A. Distruct: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Ge, X.J.; Zhang, L.B.; Yuan, Y.M.; Hao, G.; Chiang, T.Y. Strong genetic differentiation of the East-Himalayan Megacodon stylophorus (Gentianaceae) detected by Inter-Simple Sequence Repeats (ISSR). Biodivers. Conserv. 2005, 14, 849–861. [Google Scholar] [CrossRef] [Green Version]
- Shrestha, M.K.; Golan-Goldhirsh, A.; Ward, D. Population genetic structure and the conservation of isolated populations of Acacia raddiana in the Negev Desert. Biol. Conserv. 2002, 108, 119–127. [Google Scholar] [CrossRef]
- Nybom, H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol. 2004, 13, 1143–1155. [Google Scholar] [CrossRef]
- Zeng, X.; Michalski, S.G.; Fischer, M.; Durka, W. Species diversity and population density affect genetic structure and gene dispersal in a subtropical understory shrub. J. Plant Ecol. 2012, 5, 270–278. [Google Scholar] [CrossRef] [Green Version]
- Vashishtha, A.; Jehan, T.; Lakhanpaul, S. Genetic diversity and population structure of Butea monosperma (Lam.) Taub.—A potential medicinal legume tree. Physiol. Mol. Biol. Plants 2013, 19, 389–397. [Google Scholar] [CrossRef] [Green Version]
- Abbasi, S.; Afsharzadeh, S.; Dinarvand, M. Genetic structure of Zannichellia, L. populations according to mountain barriers in Iran as revealed by ISSR and SRAP markers. Turk. J. Bot. 2021, 45, 328–339. [Google Scholar] [CrossRef]
- Vilcinskas, R.; Jociene, L.; Rekasius, T.; Marozas, V.; Paulauskas, A.; Kupcinskiene, E. Genetic diversity of Lithuanian populations of Juniperus communis L. in relation to abiotic and biotic factors. Dendrobiology 2016, 76, 61–71. [Google Scholar] [CrossRef]
- Lavergne, S.; Molofsky, J. Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proc. Natl. Acad. Sci. USA 2007, 104, 3883–3888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, B.; Nakamura, K.; Tamura, S.; Lee, B.Y.; Kwak, M. Genetic diversity and population structure of Lychnis wilfordii (Caryophyllaceae) with newly developed 17 microsatellite markers. Genes Genom. 2019, 41, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, Z.; Su, Y.; Wang, T. Population transcriptomic sequencing reveals allopatric divergence and local adaptation in Pseudotaxus chienii (Taxaceae). BMC Genom. 2021, 22, 388. [Google Scholar] [CrossRef]
- Gillies, A.C.M.; Cornelius, J.P.; Newton, A.C.; Navarro, C.; Hernández, M.; Wilson, J. Genetic variation in Costa Rican populations of the tropical timber species Cedrela odorata L., assessed using RAPDs. Mol. Ecol. 1997, 6, 1133–1145. [Google Scholar] [CrossRef]
- Patamsytė, J.; Kleizaitė, V.; Čėsnienė, T.; Rančelis, V.; Žvingila, D. The genetic structure of red raspberry (Rubus idaeus L.) populations in Lithuania. Cent. Eur. J. Biol. 2010, 5, 496–506. [Google Scholar] [CrossRef]
- Bochkarnikova, N.M. A new species of honeysuckle. Proc. Appl. Bot. Genet. Breed. 1975, 54, 41–45. (In Russian) [Google Scholar]
- Poyarkova, A.I. Caprifoliaceae Juss.—Honeysuckle. In Flora of the European Part of the USSR; Fedorov, A.A., Ed.; Nauka: Leningrad, Russia, 1978; Volume 3, pp. 10–21. (In Russian) [Google Scholar]
- Nedoluzhko, V.A. Systematic and geographical review of honeysuckle in the North East of Eurasia. Komar. Read. 1986, 33, 54–109. (In Russian) [Google Scholar]
- Voroshilov, V.I. Stage-chorological analysis of the honeysuckle Lonicera, L. (Caprifoliaceae) from the subsection Caerulea Rehd. sections (Adans) Rehd. Bull. MOIP Div. Biol. 1992, 97, 89–94. (In Russian) [Google Scholar]
- Rehder, A. Synopsis of the genus Lonicera. Mo. Bot. Gard. Ann. Rep. 1903, 14, 27–232. [Google Scholar] [CrossRef]
- Plekhanova, M.N.; Rostova, N.S. The analysis of variability of morphological, anatomical and biochemical characters of Lonicera in subsection caerulea (Caprifoliaceae) with the help of the method of principal components. Bot. Zhurnal 1994, 79, 45–64. (In Russian) [Google Scholar]
- Skvortsov, A.K.; Kuklina, A.G. Problems of the formation of a new cultivated plant. Bull. Main Bot. Gard. 2002, 184, 3–7. [Google Scholar]
- Plekhanova, M.N. On the specific composition of the blue honeysuckle Lonicera subsect. Caeruleae (fam. Caprifoliaceae). Genetic resources of fruit, small fruit crops and grape: Keeping and study. Bull. Appl. Bot. Genet. Plant Breed. 2007, 161, 57–68. (In Russian) [Google Scholar]
- Ben-Meni Schuler, S.; Picazo-Aragonés, J.; Rumsey, F.J.; Romero-García, A.T.; Suárez-Santiago, V.N. Macaronesia acts as a museum of genetic diversity of relict ferns: The case of Diplazium caudatum (Athyriaceae). Plants 2021, 10, 2425. [Google Scholar] [CrossRef]
- Boltenkov, E.V.; Artyukova, E.V.; Trias-Blasi, A. Taxonomic composition of Iris Subser. Chrysographes (Iridaceae) inferred from chloroplast DNA and morphological analyses. Plants 2021, 10, 2232. [Google Scholar] [CrossRef]
- Kuklina, A.; Brindza, J.; Stehlikova, B. Opportunities for the development of genetic resources of blue honeysuckle in Slovakia. Fruit Grow. Berry Grow. Russ. 2012, 34, 414–419. [Google Scholar]
- Grygorieva, O.; Klymenko, S.; Kuklina, A.; Vinogradova, Y.; Vergun, O.; Sedláčková, V.H.; Brindza, J. Evaluation of Lonicera caerulea L. genotypes based on morphological characteristics of fruits germplasm collection. Turk. J. Agric. For. 2021, 45, 850–860. [Google Scholar] [CrossRef]
- Reddy, M.P.; Sarla, N.; Siddiq, E.A. Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica 2002, 128, 9–17. [Google Scholar] [CrossRef]
- Garshasbi, S.; Iranbakhsh, A.; Asri, Y.; Bostanabad, S.Z. Genetic diversity and population structure analysis in Lonicera L. (Caprifoliaceae) with the use of ISSR molecular markers. Genetika 2021, 53, 1273–1286. [Google Scholar] [CrossRef]

| Pop | N | PLP (5%) [11] | Na | Ne | I | Hj | Br [11] |
|---|---|---|---|---|---|---|---|
| JP | 23 | 0.558 | 1.425 | 1.348 | 0.300 | 0.199 | 1.512 |
| LV1 | 12 | 0.232 | 1.022 | 1.137 | 0.119 | 0.079 | 1.226 |
| LV2 | 12 | 0.210 | 0.989 | 1.123 | 0.108 | 0.076 | 1.206 |
| LV3 | 11 | 0.210 | 0.983 | 1.136 | 0.114 | 0.078 | 1.210 |
| LV4 | 11 | 0.204 | 0.989 | 1.134 | 0.111 | 0.079 | 1.204 |
| EE | 14 | 0.271 | 1.044 | 1.185 | 0.154 | 0.111 | 1.263 |
| RU1 | 21 | 0.315 | 1.110 | 1.195 | 0.168 | 0.110 | 1.284 |
| RU2 | 17 | 0.293 | 1.094 | 1.179 | 0.158 | 0.110 | 1.274 |
| RU3 | 19 | 0.254 | 1.000 | 1.167 | 0.141 | 0.090 | 1.234 |
| Eastern region (JP + RU1 + RU2 + RU3) | |||||||
| Mean ± SE | 80 | 0.355 ± 0.062 | 1.157 ± 0.093 | 1.222 ± 0.042 | 0.192 ± 0.036 | 0.127 ± 0.022 | 1.326 ± 0.056 |
| Baltic region (LV1 + LV2 + LV3 + LV4 + EE) | |||||||
| Mean ± SE | 60 | 0.225 ± 0.014 | 1.006 ± 0.012 | 1.143 ± 0.011 | 0.121 ± 0.008 | 0.085 ± 0.008 | 1.222 ± 0.013 |
| Overall | |||||||
| Mean ± SE | 140 | 0.283 ± 0.077 | 1.073 ± 0.047 | 1.178 ± 0.023 | 0.153 ± 0.020 | 0.104 ± 0.026 | 1.268 ± 0.068 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naugžemys, D.; Patamsytė, J.; Žilinskaitė, S.; Hoshino, Y.; Skridaila, A.; Žvingila, D. Genetic Structure of Native Blue Honeysuckle Populations in the Western and Eastern Eurasian Ranges. Plants 2022, 11, 1480. https://doi.org/10.3390/plants11111480
Naugžemys D, Patamsytė J, Žilinskaitė S, Hoshino Y, Skridaila A, Žvingila D. Genetic Structure of Native Blue Honeysuckle Populations in the Western and Eastern Eurasian Ranges. Plants. 2022; 11(11):1480. https://doi.org/10.3390/plants11111480
Chicago/Turabian StyleNaugžemys, Donatas, Jolanta Patamsytė, Silva Žilinskaitė, Yoichiro Hoshino, Audrius Skridaila, and Donatas Žvingila. 2022. "Genetic Structure of Native Blue Honeysuckle Populations in the Western and Eastern Eurasian Ranges" Plants 11, no. 11: 1480. https://doi.org/10.3390/plants11111480
APA StyleNaugžemys, D., Patamsytė, J., Žilinskaitė, S., Hoshino, Y., Skridaila, A., & Žvingila, D. (2022). Genetic Structure of Native Blue Honeysuckle Populations in the Western and Eastern Eurasian Ranges. Plants, 11(11), 1480. https://doi.org/10.3390/plants11111480






