Transcriptome Profiling Identifies Candidate Genes Contributing to Male and Female Gamete Development in Synthetic Brassica Allohexaploids
Abstract
1. Introduction
2. Results
2.1. Transcriptome Sequencing and Sequence Alignment
2.2. Anthers at Male Gamete Development Stage and Ovules at Female Gamete Development Stage Contained More Genes Than Leaves in Brassica Allohexaploids
2.3. Identification of Upregulated Genes Co-Expressed and Preferentially Expressed Genes in Male and Female Gamete Development of Brassica Allohexaploids
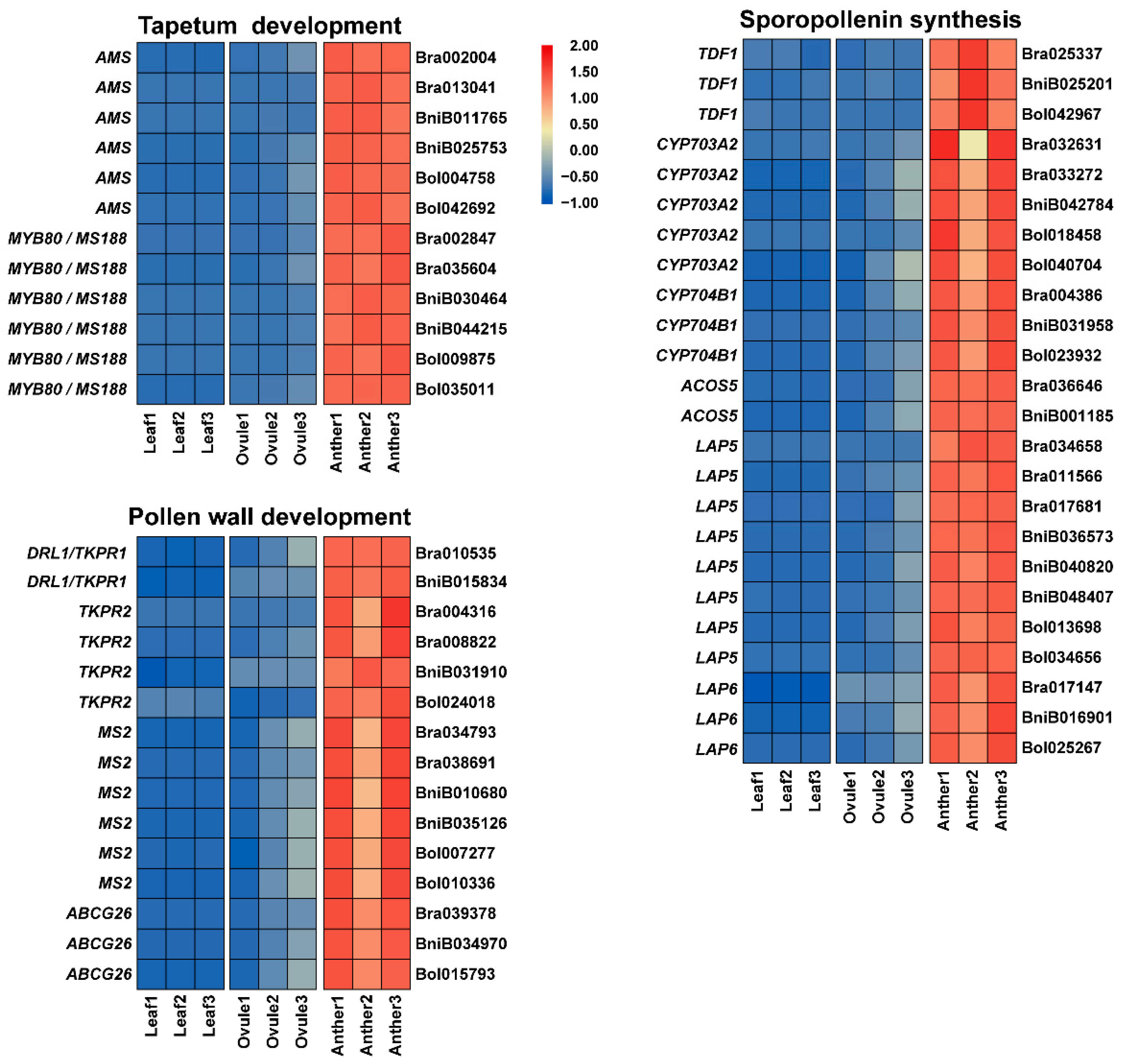
2.4. Identification of Preferentially Expressed Genes in Male Gamete Development of Brassica Allohexaploids
2.5. Identification of Preferentially Expressed Genes in Female Gamete Development of Brassica Allohexaploids
2.6. Validation of Expression Profiling by Real-Time Quantitative PCR (RT-qPCR)
3. Discussion
3.1. Meiosis-Related Genes May Affect Homologous Recombination during Male and Female Gamete Development in Brassica Allohexaploids
3.2. TDF1, AMS and MS188 May Influence Tapetum Development and Pollen Wall Formation in Brassica Allohexaploids
3.3. AUX1 and PIN1 May Regulate the Formation of the Seven-Cell Embryo Sac in Brassica Allohexaploids
4. Materials and Methods
4.1. Plant Materials, Tissue Collection and RNA Extraction
4.2. cDNA Library Construction, Filter and Alignment
4.3. FPKM and DEGs
4.4. Function Enrichment Analysis
4.5. RT-qPCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; et al. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Soltis, D.E.; Albert, V.A.; Leebens-Mack, J.; Bell, C.D.; Paterson, A.H.; Zheng, C.F.; Sankoff, D.; de Pamphilis, C.W.; Wall, P.K.; Soltis, P.S. Polyploidy and Angiosperm Diversification. Am. J. Bot. 2009, 96, 336–348. [Google Scholar] [CrossRef] [PubMed]
- Soltis, P.S.; Soltis, D.E. Ancient WGD events as drivers of key innovations in angiosperms. Curr. Opin. Plant Biol. 2016, 30, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Leitch, A.R.; Leitch, I.J. Genomic plasticity and the diversity of polyploid plants. Science 2008, 320, 481–483. [Google Scholar] [CrossRef]
- Soltis, P.S.; Soltis, D.E. The role of genetic and genomic attributes in the success of polyploids. Proc. Natl. Acad. Sci. USA 2000, 97, 7051–7057. [Google Scholar] [CrossRef]
- Ni, Z.F.; Kim, E.D.; Ha, M.S.; Lackey, E.; Liu, J.X.; Zhang, Y.R.; Sun, Q.X.; Chen, Z.J. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 2009, 457, 327–332. [Google Scholar] [CrossRef]
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 836–846. [Google Scholar] [CrossRef]
- Sattler, M.C.; Carvalho, C.R.; Clarindo, W.R. The polyploidy and its key role in plant breeding. Planta 2016, 243, 281–296. [Google Scholar] [CrossRef]
- Chen, Z.J. Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 2010, 15, 57–71. [Google Scholar] [CrossRef]
- Chen, S.; Nelson, M.N.; Chevre, A.M.; Jenczewski, E.; Li, Z.Y.; Mason, A.S.; Meng, J.L.; Plummer, J.A.; Pradhan, A.; Siddique, K.H.M.; et al. Trigenomic Bridges for Brassica Improvement. Crit. Rev. Plant Sci. 2011, 30, 524–547. [Google Scholar] [CrossRef]
- Gaebelein, R.; Mason, A.S. Allohexaploids in the Genus Brassica. Crit. Rev. Plant Sci. 2018, 37, 422–437. [Google Scholar] [CrossRef]
- Zou, J.; Zhu, J.L.; Huang, S.M.; Tian, E.T.; Xiao, Y.; Fu, D.H.; Tu, J.X.; Fu, T.D.; Meng, J.L. Broadening the avenue of intersubgenomic heterosis in oilseed Brassica. Theor. Appl. Genet. 2010, 120, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.N.; Mason, A.S.; Farooq, M.A.; Islam, F.; Quezada-Martinez, D.; Hu, D.D.; Yang, S.; Zou, J.; Zhou, W.J. Challenges and prospects for a potential allohexaploid Brassica crop. Theor. Appl. Genet. 2021, 134, 2711–2726. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Zou, J.; Meng, J.; Mei, S.; Wang, J. Tracing the transcriptomic changes in synthetic Trigenomic allohexaploids of Brassica using an RNA-Seq approach. PLoS ONE 2013, 8, e68883. [Google Scholar] [CrossRef]
- Gaebelein, R.; Schiessl, S.V.; Samans, B.; Batley, J.; Mason, A.S. Inherited allelic variants and novel karyotype changes influence fertility and genome stability in Brassica allohexaploids. New Phytol. 2019, 223, 965–978. [Google Scholar] [CrossRef]
- Zhou, J.; Tan, C.; Cui, C.; Ge, X.; Li, Z. Distinct subgenome stabilities in synthesized Brassica allohexaploids. Theor. Appl. Genet. 2016, 129, 1257–1271. [Google Scholar] [CrossRef]
- Ma, H.; Sundaresan, V. Development of flowering plant gametophytes. Curr. Top. Dev. Biol. 2010, 91, 379–412. [Google Scholar] [CrossRef]
- Hafidh, S.; Fila, J.; Honys, D. Male gametophyte development and function in angiosperms: A general concept. Plant Reprod. 2016, 29, 31–51. [Google Scholar] [CrossRef]
- Yadegari, R.; Drews, G.N. Female gametophyte development. Plant Cell 2004, 16, S133–S141. [Google Scholar] [CrossRef]
- Mccormick, S. Male Gametophyte Development. Plant Cell 1993, 5, 1265–1275. [Google Scholar] [CrossRef]
- Yang, W.C.; Shi, D.Q.; Chen, Y.H. Female gametophyte development in flowering plants. Annu. Rev. Plant Biol. 2010, 61, 89–108. [Google Scholar] [CrossRef] [PubMed]
- Nagalakshmi, U.; Waern, K.; Snyder, M. RNA-Seq: A method for comprehensive transcriptome analysis. Curr. Protoc. Mol. Biol. 2010, 4, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Yu, L.; Wei, L.; Yu, P.; Wang, J.; Zhao, H.; Zhang, Y.; Zhang, S.; Yang, Z.; Chen, G.; et al. BnTIR: An online transcriptome platform for exploring RNA-seq libraries for oil crop Brassica napus. Plant Biotechnol. J. 2021, 19, 1895–1897. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.J.; Zhang, P.P.; Lv, J.Y.; Cheng, Y.F.; Cui, J.M.; Zhao, H.X.; Hu, S.W. Global Dynamic Transcriptome Programming of Rapeseed (Brassica napus L.) Anther at Different Development Stages. PLoS ONE 2016, 11, e0154039. [Google Scholar] [CrossRef] [PubMed]
- Braynen, J.; Yang, Y.; Yuan, J.C.; Xie, Z.Q.; Cao, G.Q.; Wei, X.C.; Shi, G.Y.; Zhang, X.W.; Wei, F.; Tian, B.M. Comparative transcriptome analysis revealed differential gene expression in multiple signaling pathways at flowering in polyploid Brassica rapa. Cell Biosci. 2021, 11, 17. [Google Scholar] [CrossRef]
- Zhao, L.; He, J.; Cai, H.; Lin, H.; Li, Y.; Liu, R.; Yang, Z.; Qin, Y. Comparative expression profiling reveals gene functions in female meiosis and gametophyte development in Arabidopsis. Plant J. 2014, 80, 615–628. [Google Scholar] [CrossRef]
- Grelon, M.; Vezon, D.; Gendrot, G.; Pelletier, G. AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 2001, 20, 589–600. [Google Scholar] [CrossRef]
- De Muyt, A.; Vezon, D.; Gendrot, G.; Gallois, J.L.; Stevens, R.; Grelon, M. AtPRD1 is required for meiotic double strand break formation in Arabidopsis thaliana. EMBO J. 2007, 26, 4126–4137. [Google Scholar] [CrossRef]
- De Muyt, A.; Pereira, L.; Vezon, D.; Chelysheva, L.; Gendrot, G.; Chambon, A.; Laine-Choinard, S.; Pelletier, G.; Mercier, R.; Nogue, F.; et al. A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana. PLoS Genet. 2009, 5, e1000654. [Google Scholar] [CrossRef]
- Vrielynck, N.; Chambon, A.; Vezon, D.; Pereira, L.; Chelysheva, L.; De Muyt, A.; Mezard, C.; Mayer, C.; Grelon, M. A DNA topoisomerase VI-like complex initiates meiotic recombination. Science 2016, 351, 939–943. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, S.J.; Caryl, A.P.; Jones, G.H.; Franklin, F.C. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 2002, 115, 3645–3655. [Google Scholar] [CrossRef] [PubMed]
- Cuacos, M.; Lambing, C.; Pachon-Penalba, M.; Osman, K.; Armstrong, S.J.; Henderson, I.R.; Sanchez-Moran, E.; Franklin, F.C.H.; Heckmann, S. Meiotic chromosome axis remodelling is critical for meiotic recombination in Brassica rapa. J. Exp. Bot. 2021, 72, 3012–3027. [Google Scholar] [CrossRef] [PubMed]
- Ferdous, M.; Higgins, J.D.; Osman, K.; Lambing, C.; Roitinger, E.; Mechtler, K.; Armstrong, S.J.; Perry, R.; Pradillo, M.; Cunado, N.; et al. Inter-Homolog Crossing-Over and Synapsis in Arabidopsis Meiosis Are Dependent on the Chromosome Axis Protein AtASY3. PLoS Genet. 2012, 8, e1002507. [Google Scholar] [CrossRef]
- Higgins, J.D.; Sanchez-Moran, E.; Armstrong, S.J.; Jones, G.H.; Franklin, F.C.H. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 2005, 19, 2488–2500. [Google Scholar] [CrossRef]
- Chelysheva, L.; Vezon, D.; Chambon, A.; Gendrot, G.; Pereira, L.; Lemhemdi, A.; Vrielynck, N.; Le Guin, S.; Novatchkova, M.; Grelon, M. The Arabidopsis HEI10 Is a New ZMM Protein Related to Zip3. PLoS Genet. 2012, 8, e1002799. [Google Scholar] [CrossRef]
- Macaisne, N.; Novatchkova, M.; Peirera, L.; Vezon, D.; Jolivet, S.; Froger, N.; Chelysheva, L.; Grelon, M.; Mercier, R. SHOC1, an XPF endonuclease-related protein, is essential for the formation of class I meiotic crossovers. Curr. Biol. 2008, 18, 1432–1437. [Google Scholar] [CrossRef]
- Chelysheva, L.; Gendrot, G.; Vezon, D.; Doutriaux, M.P.; Mercier, R.; Grelon, M. Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet. 2007, 3, e83. [Google Scholar] [CrossRef]
- Higgins, J.D.; Vignard, J.; Mercier, R.; Pugh, A.G.; Franklin, F.C.; Jones, G.H. AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. Plant J. 2008, 55, 28–39. [Google Scholar] [CrossRef]
- Chelysheva, L.; Vezon, D.; Belcram, K.; Gendrot, G.; Grelon, M. The Arabidopsis BLAP75/Rmi1 Homologue Plays Crucial Roles in Meiotic Double-Strand Break Repair. PLoS Genet. 2008, 4, e1000309. [Google Scholar] [CrossRef][Green Version]
- Hartung, F.; Suer, S.; Knoll, A.; Wurz-Wildersinn, R.; Puchta, H. Topoisomerase 3alpha and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana. PLoS Genet. 2008, 4, e1000285. [Google Scholar] [CrossRef] [PubMed]
- Knoll, A.; Higgins, J.D.; Seeliger, K.; Reha, S.J.; Dangel, N.J.; Bauknecht, M.; Schropfer, S.; Franklin, F.C.H.; Puchta, H. The Fanconi Anemia Ortholog FANCM Ensures Ordered Homologous Recombination in Both Somatic and Meiotic Cells in Arabidopsis. Plant Cell 2012, 24, 1448–1464. [Google Scholar] [CrossRef] [PubMed]
- Hartung, F.; Suer, S.; Puchta, H. Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 104, 18836–18841. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, A.C.; Pearce, S.; Band, L.R.; Yang, C.Y.; Ferjentsikova, I.; King, J.; Yuan, Z.; Zhang, D.B.; Wilson, Z.A. Biphasic regulation of the transcription factor ABORTED MICROSPORES (AMS) is essential for tapetum and pollen development in Arabidopsis. New Phytol. 2017, 213, 778–790. [Google Scholar] [CrossRef] [PubMed]
- Phan, H.A.; Li, S.F.; Parish, R.W. MYB80, a regulator of tapetal and pollen development, is functionally conserved in crops. Plant Mol. Biol. 2012, 78, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Chen, H.; Li, H.; Gao, J.F.; Jiang, H.; Wang, C.; Guan, Y.F.; Yang, Z.N. Defective in Tapetal Development and Function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J. 2008, 55, 266–277. [Google Scholar] [CrossRef]
- Morant, M.; Jorgensen, K.; Schaller, H.; Pinot, F.; Moller, B.L.; Werck-Reichhart, D.; Bak, S. CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 2007, 19, 1473–1487. [Google Scholar] [CrossRef]
- Dobritsa, A.A.; Shrestha, J.; Morant, M.; Pinot, F.; Matsuno, M.; Swanson, R.; Moller, B.L.; Preuss, D. CYP704B1 Is a Long-Chain Fatty Acid omega-Hydroxylase Essential for Sporopollenin Synthesis in Pollen of Arabidopsis. Plant Physiol. 2009, 151, 574–589. [Google Scholar] [CrossRef]
- Souza, C.D.; Kim, S.S.; Koch, S.; Kienow, L.; Schneider, K.; McKim, S.M.; Haughn, G.; Kombrink, E.; Douglas, C.J. A Novel Fatty Acyl-CoA Synthetase Is Required for Pollen Development and Sporopollenin Biosynthesis in Arabidopsis. Plant Cell 2009, 21, 507–525. [Google Scholar] [CrossRef]
- Kim, S.S.; Grienenberger, E.; Lallemand, B.; Colpitts, C.C.; Kim, S.Y.; Souza, C.D.; Geoffroy, P.; Heintz, D.; Krahn, D.; Kaiser, M.; et al. LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B Encode Hydroxyalkyl alpha-Pyrone Synthases Required for Pollen Development and Sporopollenin Biosynthesis in Arabidopsis thaliana. Plant Cell 2010, 22, 4045–4066. [Google Scholar] [CrossRef]
- Grienenberger, E.; Kim, S.S.; Lallemand, B.; Geoffroy, P.; Heintz, D.; Souza, C.D.; Heitz, T.; Douglas, C.J.; Legrand, M. Analysis of TETRAKETIDE alpha-PYRONE REDUCTASE Function in Arabidopsis thaliana Reveals a Previously Unknown, but Conserved, Biochemical Pathway in Sporopollenin Monomer Biosynthesis. Plant Cell 2010, 22, 4067–4083. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.K.; Chu, H.; Yip, W.K.; Yeung, E.C.; Lo, C. An anther-specific dihydroflavonol 4-reductase-like gene (DRL1) is essential for male fertility in Arabidopsis. New Phytol. 2009, 181, 576–587. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.W.; Yu, X.H.; Zhang, K.S.; Shi, J.X.; De Oliveira, S.; Schreiber, L.; Shanklin, J.; Zhang, D.B. Male Sterile2 Encodes a Plastid-Localized Fatty Acyl Carrier Protein Reductase Required for Pollen Exine Development in Arabidopsis. Plant Physiol. 2011, 157, 842–853. [Google Scholar] [CrossRef]
- Quilichini, T.D.; Friedmann, M.C.; Samuels, A.L.; Douglas, C.J. ATP-Binding Cassette Transporter G26 Is Required for Male Fertility and Pollen Exine Formation in Arabidopsis. Plant Physiol. 2010, 154, 678–690. [Google Scholar] [CrossRef] [PubMed]
- Panoli, A.; Martin, M.V.; Alandete-Saez, M.; Simon, M.; Neff, C.; Swarup, R.; Bellido, A.; Yuan, L.; Pagnussat, G.C.; Sundaresan, V. Auxin Import and Local Auxin Biosynthesis Are Required for Mitotic Divisions, Cell Expansion and Cell Specification during Female Gametophyte Development in Arabidopsis thaliana. PLoS ONE 2015, 10, e0126164. [Google Scholar] [CrossRef] [PubMed]
- Ceccato, L.; Masiero, S.; Roy, D.S.; Bencivenga, S.; Roig-Villanova, I.; Ditengou, F.A.; Palme, K.; Simon, R.; Colombo, L. Maternal Control of PIN1 Is Required for Female Gametophyte Development in Arabidopsis. PLoS ONE 2013, 8, e66148. [Google Scholar] [CrossRef]
- Wang, Y.B.; Hou, Y.N.; Gu, H.Y.; Kang, D.M.; Chen, Z.L.; Liu, J.J.; Qu, L.J. The Arabidopsis Anaphase-Promoting Complex/Cyclosome Subunit 1 is Critical for Both Female Gametogenesis and Embryogenesis. J. Integr. Plant Biol. 2013, 55, 64–74. [Google Scholar] [CrossRef]
- Li, L.X.; Liao, H.Z.; Jiang, L.X.; Tan, Q.; Ye, D.; Zhang, X.Q. Arabidopsis thaliana NOP10 is required for gametophyte formation. J. Integr. Plant Biol. 2018, 60, 723–736. [Google Scholar] [CrossRef]
- Rodrigo-Peiris, T.; Xu, X.M.; Zhao, Q.; Wang, H.J.; Meier, I. RanGAP is required for post-meiotic mitosis in female gametophyte development in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 2705–2714. [Google Scholar] [CrossRef]
- Pastuglia, M.; Azimzadeh, J.; Goussot, M.; Camilleri, C.; Belcram, K.; Evrard, J.L.; Schmit, A.C.; Guerche, P.; Bouchez, D. gamma-tubulin is essential for microtubule organization and development in Arabidopsis. Plant Cell 2006, 18, 1412–1425. [Google Scholar] [CrossRef]
- Tanaka, H.; Ishikawa, M.; Kitamura, S.; Takahashi, Y.; Soyano, T.; Machida, C.; Machida, Y. The AtNACK1/HINKEL and STUD/TETRASPORE/AtNACK2 genes, which encode functionally redundant kinesins, are essential for cytokinesis in Arabidopsis. Genes Cells 2004, 9, 1199–1211. [Google Scholar] [CrossRef] [PubMed]
- Portereiko, M.F.; Sandaklie-Nikolova, L.; Lloyd, A.; Dever, C.A.; Otsuga, D.; Drews, G.N. Nuclear fusion defective1 encodes the Arabidopsis RPL21M protein and is required for karyogamy during female gametophyte development and fertilization. Plant Physiol. 2006, 141, 957–965. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Lau, S.; Jurgens, G. Twin plants from supernumerary egg cells in Arabidopsis. Curr. Biol. 2015, 25, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Cifuentes, M.; Eber, F.; Lucas, M.O.; Lode, M.; Chevre, A.M.; Jenczewski, E. Repeated Polyploidy Drove Different Levels of Crossover Suppression between Homoeologous Chromosomes in Brassica napus Allohaploids. Plant Cell 2010, 22, 2265–2276. [Google Scholar] [CrossRef]
- Page, S.L.; Hawley, R.S. Chromosome choreography: The meiotic ballet. Science 2003, 301, 785–789. [Google Scholar] [CrossRef]
- Youds, J.L.; Boulton, S.J. The choice in meiosis—Defining the factors that influence crossover or non-crossover formation. J. Cell Sci. 2011, 124, 501–513. [Google Scholar] [CrossRef]
- Li, D.D.; Xue, J.S.; Zhu, J.; Yang, Z.N. Gene Regulatory Network for Tapetum Development in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 1559. [Google Scholar] [CrossRef]
- Lou, Y.; Zhou, H.S.; Han, Y.; Zeng, Q.Y.; Zhu, J.; Yang, Z.N. Positive regulation of AMS by TDF1 and the formation of a TDF1-AMS complex are required for anther development in Arabidopsis thaliana. New Phytol. 2018, 217, 378–391. [Google Scholar] [CrossRef]
- Wang, K.; Guo, Z.L.; Zhou, W.T.; Zhang, C.; Zhang, Z.Y.; Lou, Y.; Xiong, S.X.; Yao, X.Z.; Fan, J.J.; Zhu, J.; et al. The Regulation of Sporopollenin Biosynthesis Genes for Rapid Pollen Wall Formation. Plant Physiol. 2018, 178, 283–294. [Google Scholar] [CrossRef]
- Serbes, I.E.; Palovaara, J.; Gross-Hardt, R. Development and function of the flowering plant female gametophyte. Plant Dev. Evol. 2019, 131, 401. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]







| Sample Name | Raw Reads Number | Clean Reads Number | Clean Reads Rate (%) | Raw Q30 Bases Rate (%) | Clean Q30 Bases Rate (%) |
|---|---|---|---|---|---|
| Leaf1 | 49,970,344 | 46,345,462 | 92.75 | 93.65 | 94.55 |
| Leaf2 | 45,415,696 | 40,202,856 | 88.52 | 93.22 | 94.63 |
| Leaf3 | 47,653,708 | 44,057,944 | 92.45 | 93.56 | 94.54 |
| Ovule1 | 51,752,802 | 45,556,284 | 88.03 | 92.76 | 94.86 |
| Ovule2 | 49,874,912 | 46,197,038 | 92.63 | 93.56 | 94.53 |
| Ovule3 | 48,619,388 | 45,120,190 | 92.8 | 93.36 | 94.36 |
| Anther1 | 55,572,362 | 47,450,126 | 85.38 | 91.81 | 94.29 |
| Anther2 | 50,943,546 | 45,328,282 | 88.98 | 92.73 | 94.42 |
| Anther3 | 50,431,536 | 46,943,730 | 93.08 | 93.67 | 94.51 |
| Sample Name | Total Reads Number | Mapped Reads Number | Mapping Rate (%) | MultiMap Reads Number | MultiMap Rate (%) |
|---|---|---|---|---|---|
| Leaf1 | 46,345,462 | 25,110,254 | 54.18 | 991,570 | 2.14 |
| Leaf2 | 40,202,856 | 21,502,559 | 53.49 | 868,719 | 2.16 |
| Leaf3 | 44,057,944 | 23,720,073 | 53.84 | 978,530 | 2.22 |
| Ovule1 | 45,556,284 | 38,811,219 | 85.19 | 1,467,965 | 3.22 |
| Ovule2 | 46,197,038 | 39,142,941 | 84.73 | 1,469,346 | 3.18 |
| Ovule3 | 45,120,190 | 38,282,041 | 84.84 | 1,488,352 | 3.30 |
| Anther1 | 47,450,126 | 39,735,180 | 83.74 | 1,558,104 | 3.28 |
| Anther2 | 45,328,282 | 38,359,513 | 84.63 | 1,487,356 | 3.28 |
| Anther3 | 46,943,730 | 39,794,184 | 84.77 | 1,508,397 | 3.21 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, C.; Tian, Z.; Liu, Y.; Shi, G.; Tian, B.; Chen, W.; Xie, Z.; Han, X.; Liang, N.; Wei, F.; et al. Transcriptome Profiling Identifies Candidate Genes Contributing to Male and Female Gamete Development in Synthetic Brassica Allohexaploids. Plants 2022, 11, 1556. https://doi.org/10.3390/plants11121556
Ji C, Tian Z, Liu Y, Shi G, Tian B, Chen W, Xie Z, Han X, Liang N, Wei F, et al. Transcriptome Profiling Identifies Candidate Genes Contributing to Male and Female Gamete Development in Synthetic Brassica Allohexaploids. Plants. 2022; 11(12):1556. https://doi.org/10.3390/plants11121556
Chicago/Turabian StyleJi, Chengyan, Zhaoran Tian, Yue Liu, Gongyao Shi, Baoming Tian, Weiwei Chen, Zhengqing Xie, Xingzhou Han, Niannian Liang, Fang Wei, and et al. 2022. "Transcriptome Profiling Identifies Candidate Genes Contributing to Male and Female Gamete Development in Synthetic Brassica Allohexaploids" Plants 11, no. 12: 1556. https://doi.org/10.3390/plants11121556
APA StyleJi, C., Tian, Z., Liu, Y., Shi, G., Tian, B., Chen, W., Xie, Z., Han, X., Liang, N., Wei, F., & Wei, X. (2022). Transcriptome Profiling Identifies Candidate Genes Contributing to Male and Female Gamete Development in Synthetic Brassica Allohexaploids. Plants, 11(12), 1556. https://doi.org/10.3390/plants11121556






