A Comprehensive Study of the WRKY Transcription Factor Family in Strawberry
Abstract
1. Introduction
2. Results and Discussion
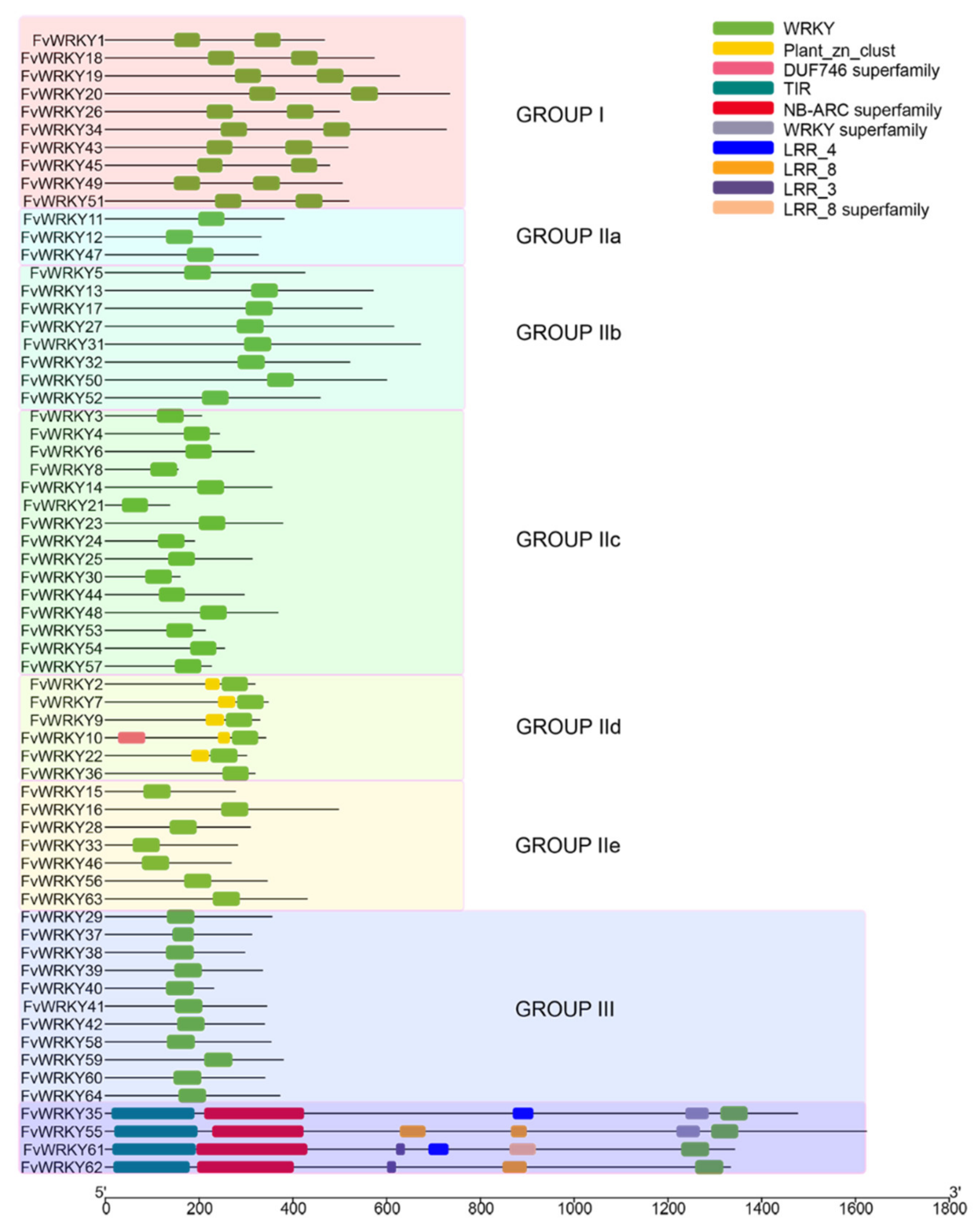
2.1. WRKY Members of F. vesca and F. ananassa
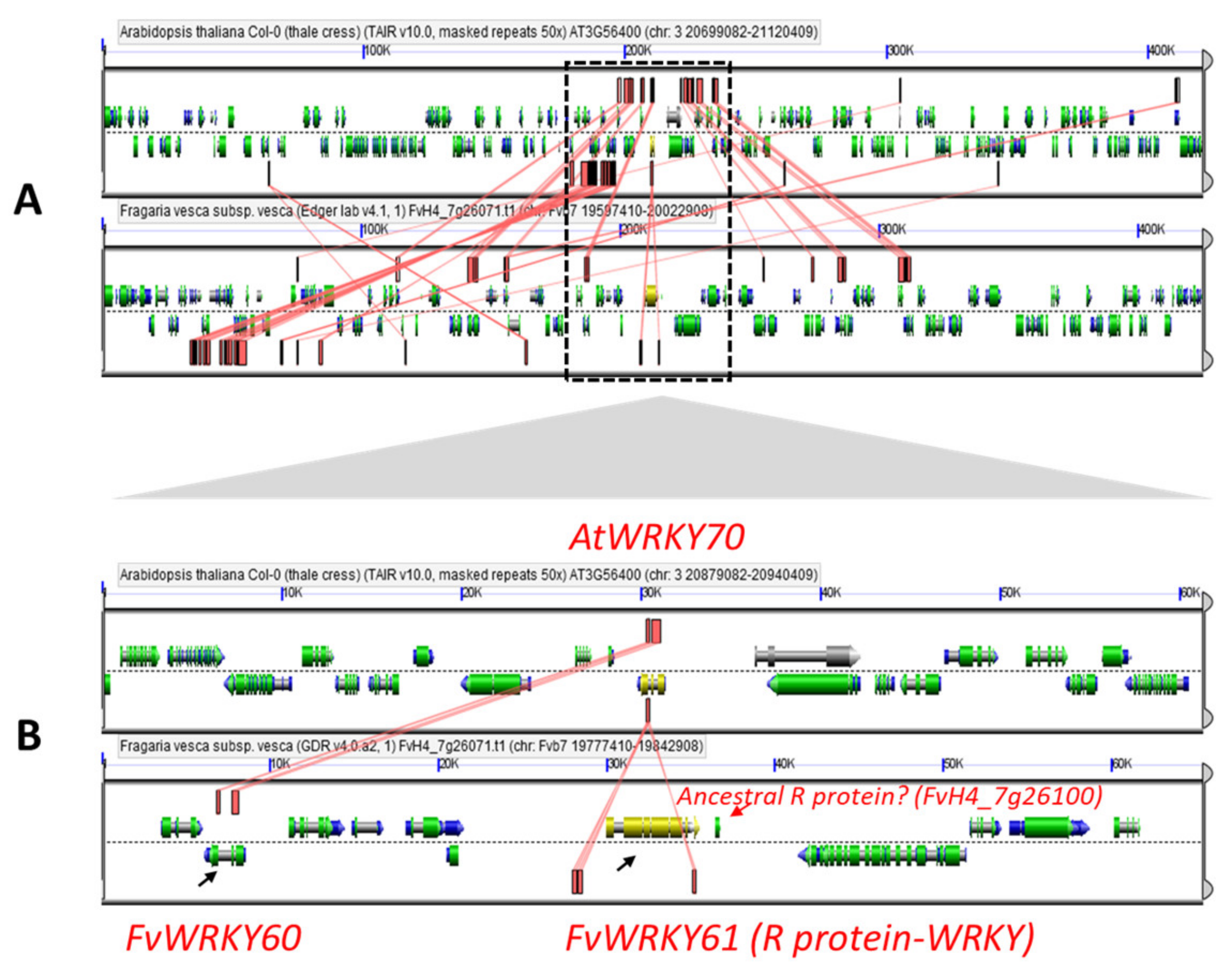
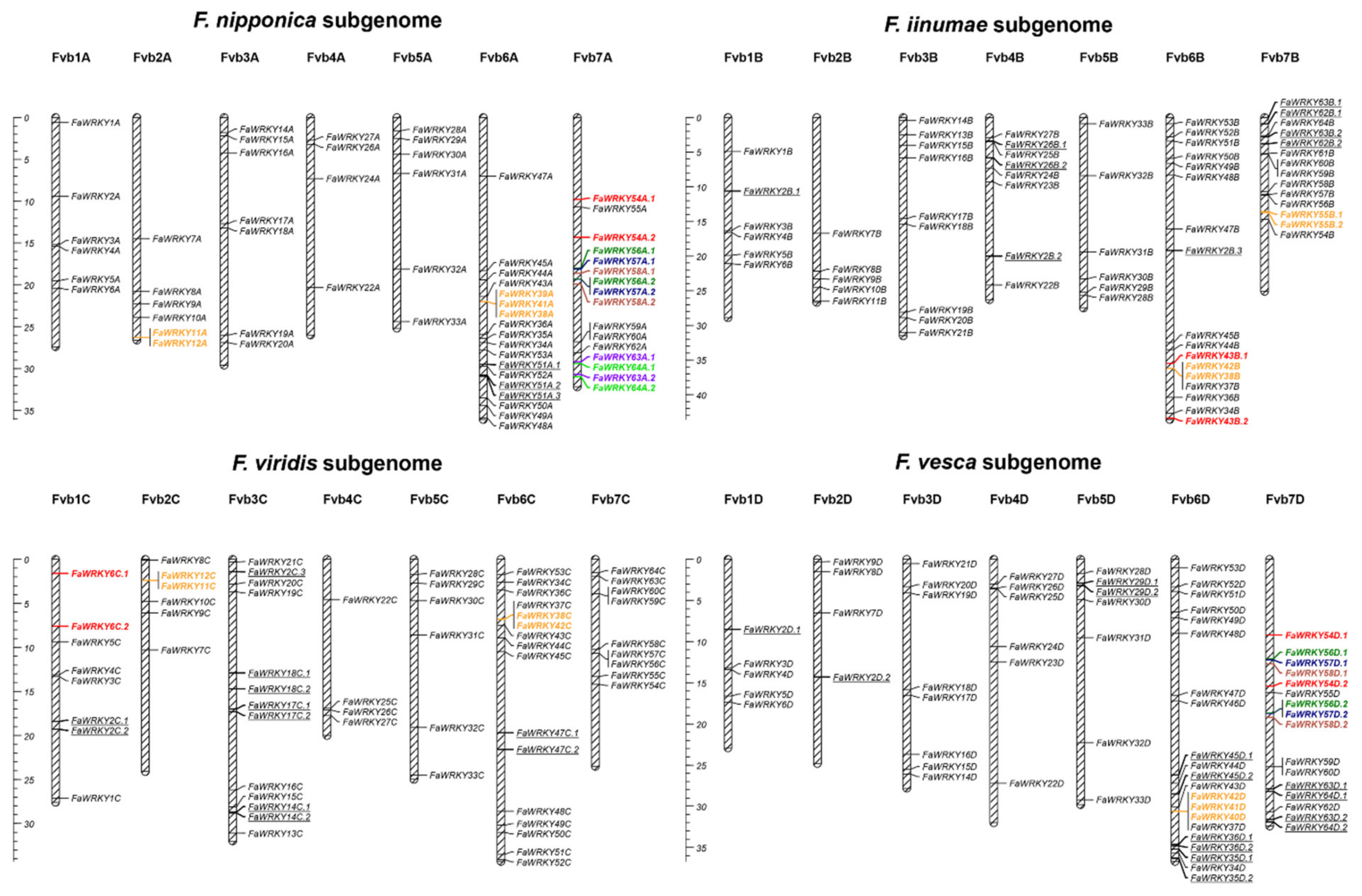
2.2. Genome-Wide Distribution and Gene Duplications of the Strawberry WRKY Family
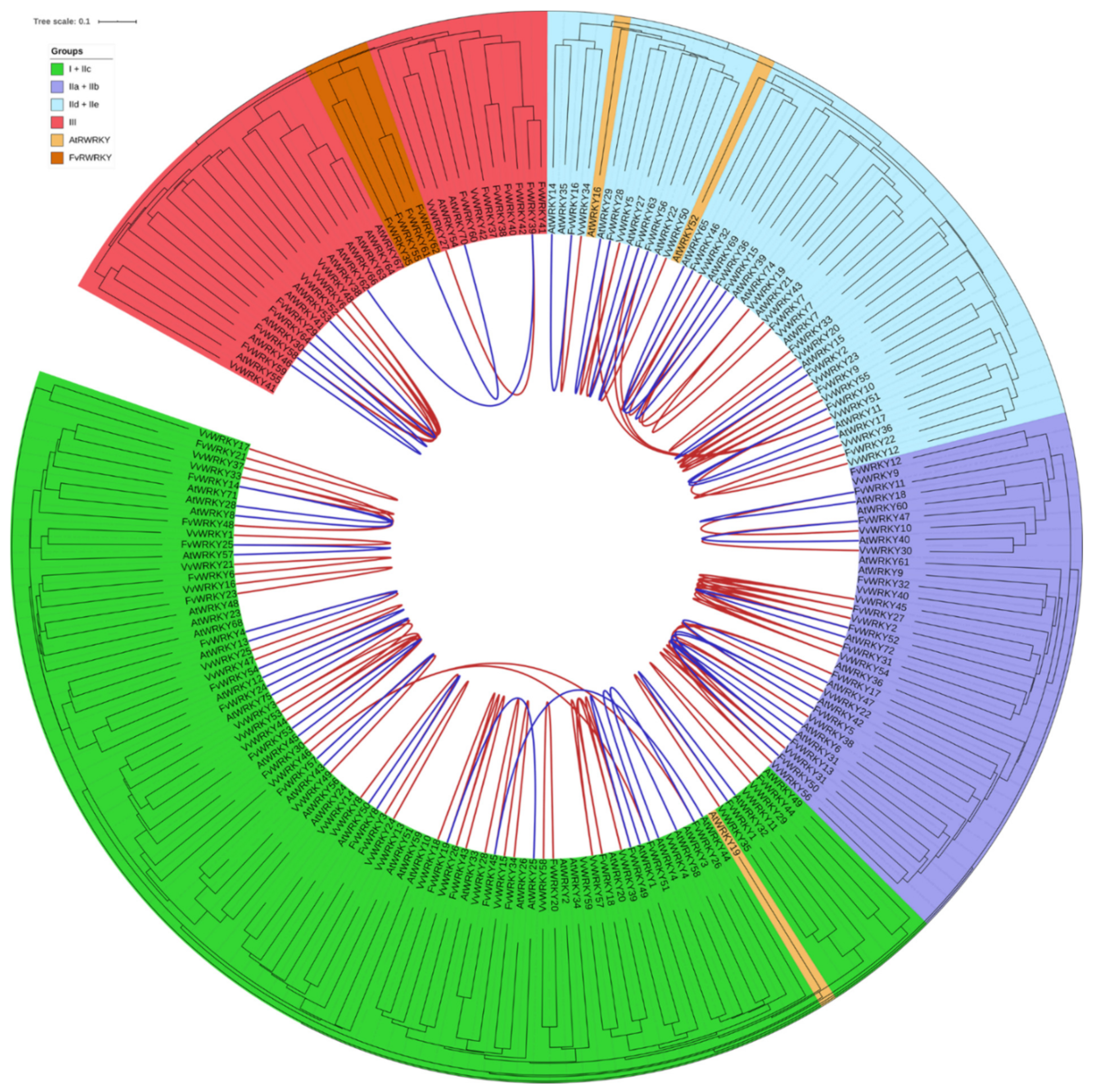
2.3. Phylogenomic Analysis of the Strawberry WRKY Family
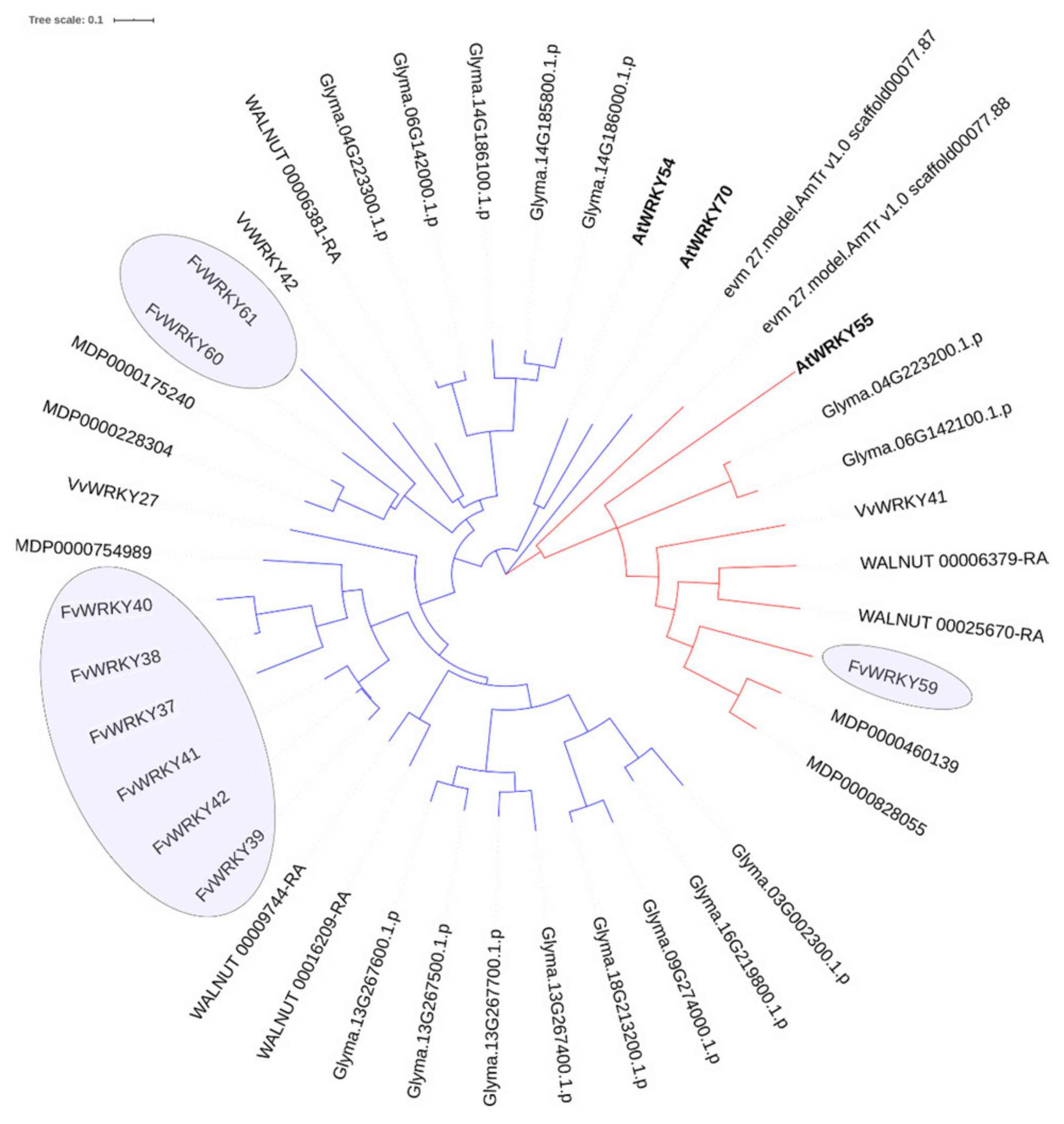
2.4. Orthology Relationships and Annotation of the Strawberry WRKYs
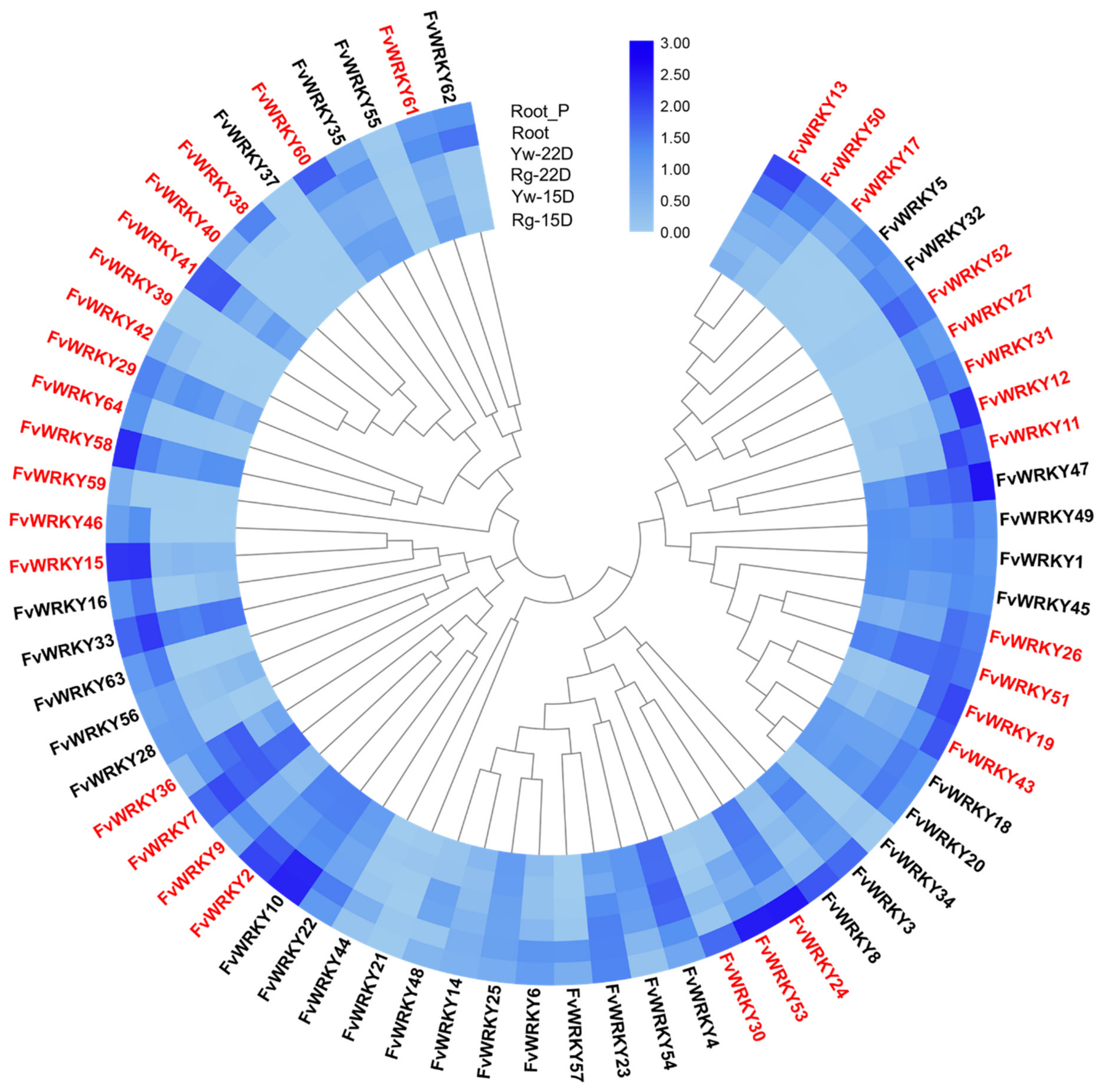
2.5. Expression Analysis of FvWRKY Genes in Different Tissues, Development Stages and Growth Conditions
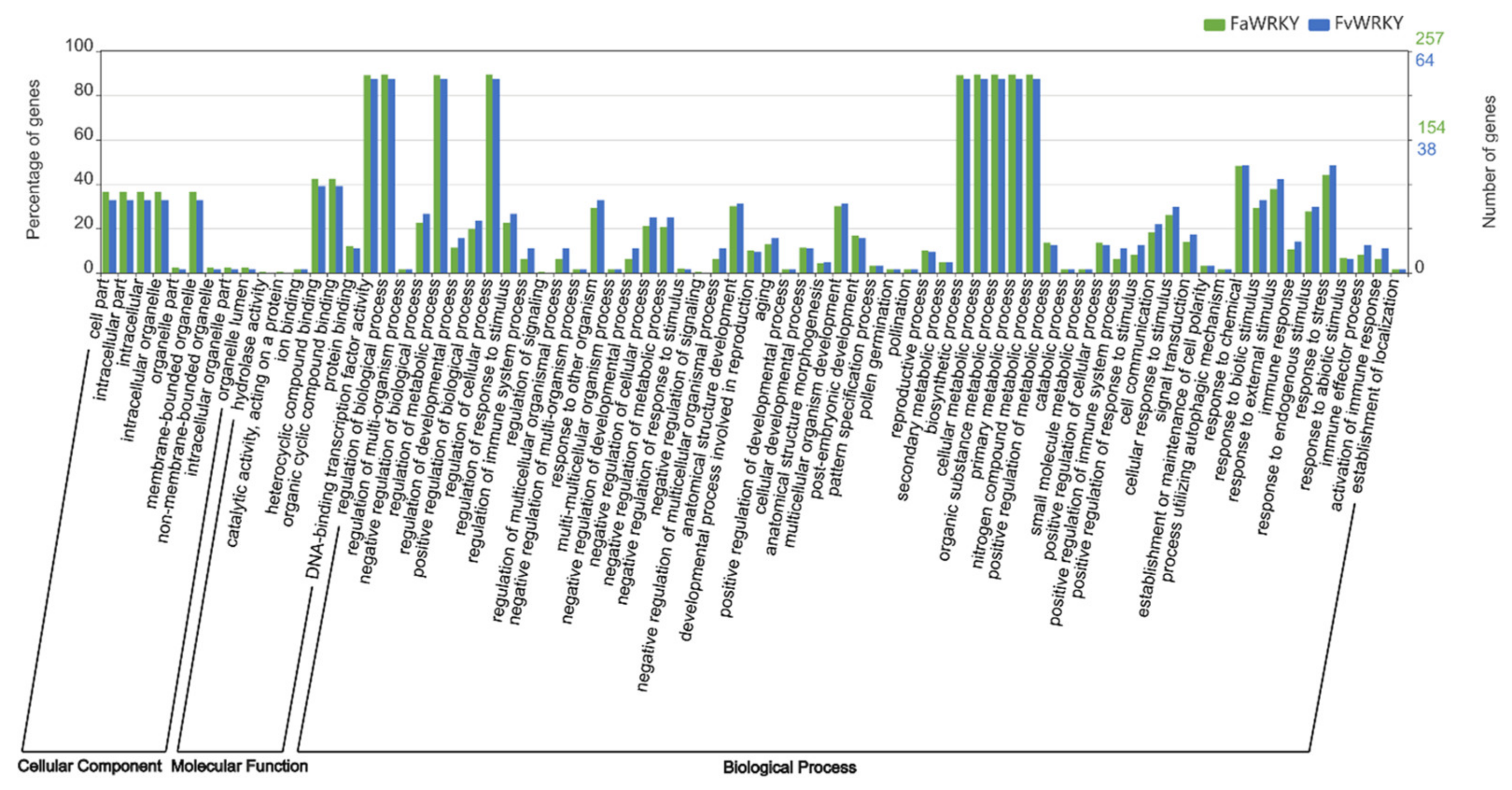
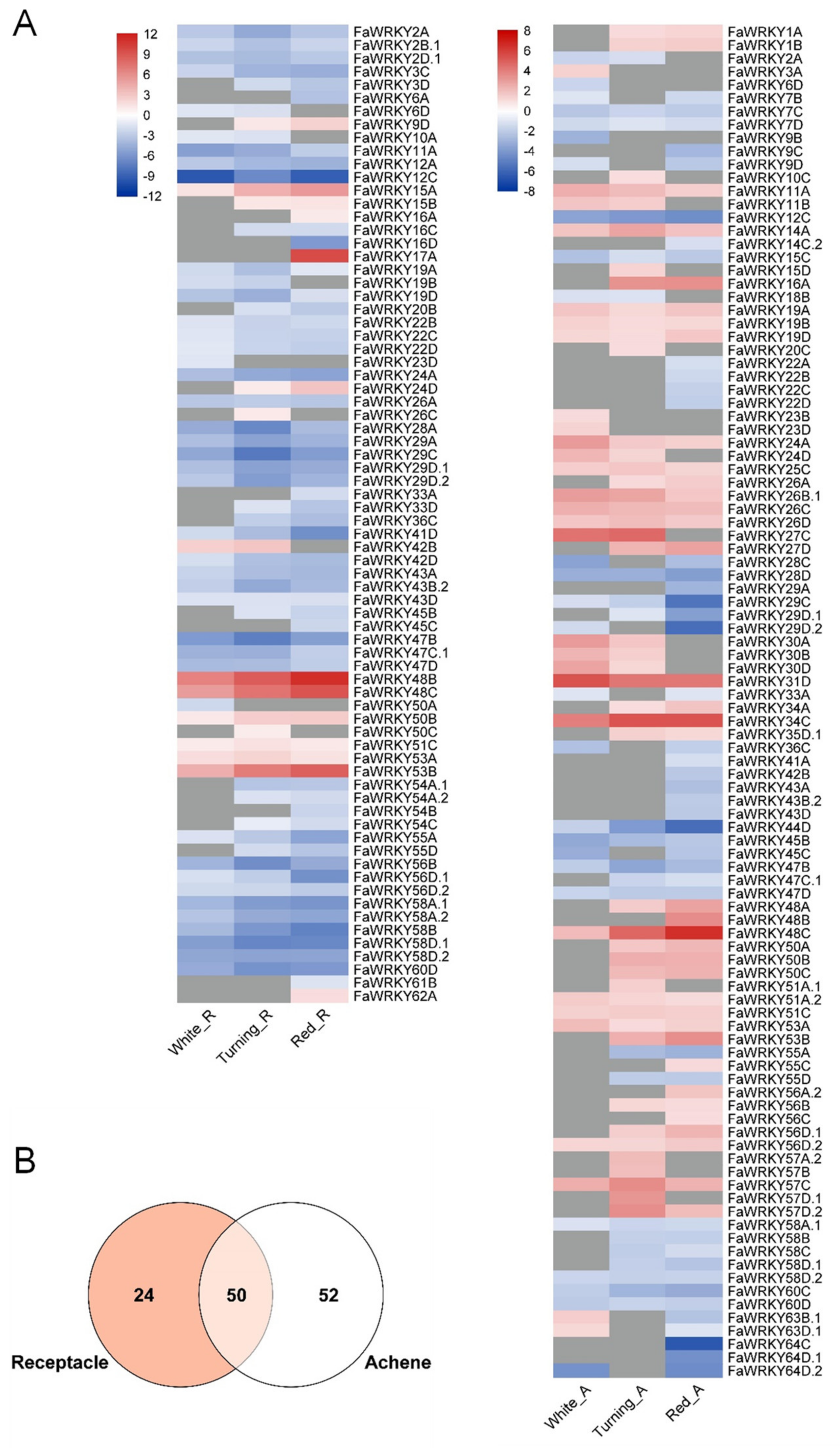
2.6. Expression Analysis of the FaWRKY Gene Family in Strawberry Tissues, the Fruit Ripening Process and Defense Responses
3. Conclusions
4. Materials and Methods
4.1. Identification of WRKY Family Members
4.2. Phylogenomics
4.3. Identification of Cis-Acting Element Analysis of WRKY Genes
4.4. Expression Analyses of Diploid and Octoploid Strawberry WRKYs
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Bakshi, M.; Oelmuller, R. WRKY transcription factors: Jack of many trades in plants. Plant Signal. Behav. 2014, 9, e27700. [Google Scholar] [CrossRef] [PubMed]
- Mohanta, T.K.; Park, Y.H.; Bae, H. Novel Genomic and Evolutionary Insight of WRKY Transcription Factors in Plant Lineage. Sci. Rep. 2016, 6, 37309. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, L. The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 2005, 5, 1. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Besseau, S.; Li, J.; Palva, E.T. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot 2012, 63, 2667–2679. [Google Scholar] [CrossRef]
- Chen, L.; Xiang, S.; Chen, Y.; Li, D.; Yu, D. Arabidopsis WRKY45 Interacts with the DELLA Protein RGL1 to Positively Regulate Age-Triggered Leaf Senescence. Mol. Plant 2017, 10, 1174–1189. [Google Scholar] [CrossRef]
- Doll, J.; Muth, M.; Riester, L.; Nebel, S.; Bresson, J.; Lee, H.C.; Zentgraf, U. Arabidopsis thaliana WRKY25 Transcription Factor Mediates Oxidative Stress Tolerance and Regulates Senescence in a Redox-Dependent Manner. Front. Plant Sci. 2019, 10, 1734. [Google Scholar] [CrossRef]
- Zhang, C.Q.; Xu, Y.; Lu, Y.; Yu, H.X.; Gu, M.H.; Liu, Q.Q. The WRKY transcription factor OsWRKY78 regulates stem elongation and seed development in rice. Planta 2011, 234, 541–554. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, L.; Yu, D. Transcription Factor WRKY75 Interacts with DELLA Proteins to Affect Flowering. Plant Physiol. 2018, 176, 790–803. [Google Scholar] [CrossRef]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 2012, 1819, 120–128. [Google Scholar] [CrossRef]
- Tsuda, K.; Somssich, I.E. Transcriptional networks in plant immunity. New Phytol. 2015, 206, 932–947. [Google Scholar] [CrossRef]
- Alves, M.S.; Dadalto, S.P.; Goncalves, A.B.; de Souza, G.B.; Barros, V.A.; Fietto, L.G. Transcription Factor Functional Protein-Protein Interactions in Plant Defense Responses. Proteomes 2014, 2, 85–106. [Google Scholar] [CrossRef]
- Ciolkowski, I.; Wanke, D.; Birkenbihl, R.P.; Somssich, I.E. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 2008, 68, 81–92. [Google Scholar] [CrossRef]
- Maeo, K.; Hayashi, S.; Kojima-Suzuki, H.; Morikami, A.; Nakamura, K. Role of conserved residues of the WRKY domain in the DNA-binding of tobacco WRKY family proteins. Biosci. Biotechnol. Biochem. 2001, 65, 2428–2436. [Google Scholar] [CrossRef]
- Cheng, X.; Zhao, Y.; Jiang, Q.; Yang, J.; Zhao, W.; Taylor, I.A.; Peng, Y.L.; Wang, D.; Liu, J. Structural basis of dimerization and dual W-box DNA recognition by rice WRKY domain. Nucleic Acids Res. 2019, 47, 4308–4318. [Google Scholar] [CrossRef]
- Singh, A.; Sharma, A.K.; Singh, N.K.; Sonah, H.; Deshmukh, R.; Sharma, T.R. Understanding the Effect of Structural Diversity in WRKY Transcription Factors on DNA Binding Efficiency through Molecular Dynamics Simulation. Biology 2019, 8, 83. [Google Scholar] [CrossRef]
- Van Verk, M.C.; Pappaioannou, D.; Neeleman, L.; Bol, J.F.; Linthorst, H.J. A Novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 2008, 146, 1983–1995. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, Y.; Chi, Y.; Fan, B.; Chen, Z. Characterization of Soybean WRKY Gene Family and Identification of Soybean WRKY Genes that Promote Resistance to Soybean Cyst Nematode. Sci. Rep. 2017, 7, 17804. [Google Scholar] [CrossRef]
- Chen, X.; Li, C.; Wang, H.; Guo, Z. WRKY transcription factors: Evolution, binding, and action. Phytopathol. Res. 2019, 1, 13. [Google Scholar] [CrossRef]
- Machens, F.; Becker, M.; Umrath, F.; Hehl, R. Identification of a novel type of WRKY transcription factor binding site in elicitor-responsive cis-sequences from Arabidopsis thaliana. Plant Mol. Biol. 2014, 84, 371–385. [Google Scholar] [CrossRef]
- Rinerson, C.I.; Rabara, R.C.; Tripathi, P.; Shen, Q.J.; Rushton, P.J. The evolution of WRKY transcription factors. BMC Plant Biol. 2015, 15, 66. [Google Scholar] [CrossRef]
- DiMeglio, L.M.; Staudt, G.; Yu, H.; Davis, T.M. A phylogenetic analysis of the genus Fragaria (strawberry) using intron-containing sequence from the ADH-1 gene. PLoS ONE 2014, 9, e102237. [Google Scholar] [CrossRef]
- Shulaev, V.; Sargent, D.J.; Crowhurst, R.N.; Mockler, T.C.; Folkerts, O.; Delcher, A.L.; Jaiswal, P.; Mockaitis, K.; Liston, A.; Mane, S.P.; et al. The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 2011, 43, 109–116. [Google Scholar] [CrossRef]
- Darwish, O.; Shahan, R.; Liu, Z.; Slovin, J.P.; Alkharouf, N.W. Re-annotation of the woodland strawberry (Fragaria vesca) genome. BMC Genom. 2015, 16, 29. [Google Scholar] [CrossRef]
- Tennessen, J.A.; Govindarajulu, R.; Ashman, T.L.; Liston, A. Evolutionary origins and dynamics of octoploid strawberry subgenomes revealed by dense targeted capture linkage maps. Genome Biol. Evol. 2014, 6, 3295–3313. [Google Scholar] [CrossRef]
- Li, Y.; Wei, W.; Feng, J.; Luo, H.; Pi, M.; Liu, Z.; Kang, C. Genome re-annotation of the wild strawberry Fragaria vesca using extensive Illumina- and SMRT-based RNA-seq datasets. DNA Res. 2018, 25, 61–70. [Google Scholar] [CrossRef]
- Edger, P.P.; VanBuren, R.; Colle, M.; Poorten, T.J.; Wai, C.M.; Niederhuth, C.E.; Alger, E.I.; Ou, S.; Acharya, C.B.; Wang, J.; et al. Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. Gigascience 2018, 7, gix124. [Google Scholar] [CrossRef]
- Li, Y.; Pi, M.; Gao, Q.; Liu, Z.; Kang, C. Updated annotation of the wild strawberry Fragaria vesca V4 genome. Hortic. Res. 2019, 6, 61. [Google Scholar] [CrossRef]
- Edger, P.P.; Poorten, T.J.; VanBuren, R.; Hardigan, M.A.; Colle, M.; McKain, M.R.; Smith, R.D.; Teresi, S.J.; Nelson, A.D.L.; Wai, C.M.; et al. Origin and evolution of the octoploid strawberry genome. Nat. Genet. 2019, 51, 541–547. [Google Scholar] [CrossRef]
- Edger, P.P.; McKain, M.R.; Yocca, A.E.; Knapp, S.J.; Qiao, Q.; Zhang, T. Reply to: Revisiting the origin of octoploid strawberry. Nat. Genet. 2020, 52, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Wang, J.; Harris, A.J.; Folta, K.M.; Zhao, M.; Kang, M. Tracing the diploid ancestry of the cultivated octoploid strawberry. Mol. Biol. Evol. 2020, 38, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Liston, A.; Wei, N.; Tennessen, J.A.; Li, J.; Dong, M.; Ashman, T.L. Revisiting the origin of octoploid strawberry. Nat. Genet. 2020, 52, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Medina-Puche, L.; Blanco-Portales, R.; Molina-Hidalgo, F.J.; Cumplido-Laso, G.; Garcia-Caparros, N.; Moyano-Canete, E.; Caballero-Repullo, J.L.; Munoz-Blanco, J.; Rodriguez-Franco, A. Extensive transcriptomic studies on the roles played by abscisic acid and auxins in the development and ripening of strawberry fruits. Funct. Integr. Genom. 2016, 16, 671–692. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Sevilla, J.F.; Vallarino, J.G.; Osorio, S.; Bombarely, A.; Pose, D.; Merchante, C.; Botella, M.A.; Amaya, I.; Valpuesta, V. Gene expression atlas of fruit ripening and transcriptome assembly from RNA-seq data in octoploid strawberry (Fragaria × ananassa). Sci. Rep. 2017, 7, 13737. [Google Scholar] [CrossRef] [PubMed]
- Amil-Ruiz, F.; Garrido-Gala, J.; Blanco-Portales, R.; Folta, K.M.; Munoz-Blanco, J.; Caballero, J.L. Identification and validation of reference genes for transcript normalization in strawberry (Fragaria × ananassa) defense responses. PLoS ONE 2013, 8, e70603. [Google Scholar] [CrossRef] [PubMed]
- Amil-Ruiz, F.; Garrido-Gala, J.; Gadea, J.; Blanco-Portales, R.; Munoz-Merida, A.; Trelles, O.; de Los Santos, B.; Arroyo, F.T.; Aguado-Puig, A.; Romero, F.; et al. Partial Activation of SA- and JA-Defensive Pathways in Strawberry upon Colletotrichum acutatum Interaction. Front. Plant Sci. 2016, 7, 1036. [Google Scholar] [CrossRef]
- Wei, W.; Hu, Y.; Han, Y.T.; Zhang, K.; Zhao, F.L.; Feng, J.Y. The WRKY transcription factors in the diploid woodland strawberry Fragaria vesca: Identification and expression analysis under biotic and abiotic stresses. Plant Physiol. Biochem. 2016, 105, 129–144. [Google Scholar] [CrossRef]
- Zhou, H.; Li, Y.; Zhang, Q.; Ren, S.; Shen, Y.; Qin, L.; Xing, Y. Genome-Wide Analysis of the Expression of WRKY Family Genes in Different Developmental Stages of Wild Strawberry (Fragaria vesca) Fruit. PLoS ONE 2016, 11, e0154312. [Google Scholar] [CrossRef]
- Solovyev, V.; Kosarev, P.; Seledsov, I.; Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006, 7 (Suppl. 1), S10. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Lyons, E.; Freeling, M. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 2008, 53, 661–673. [Google Scholar] [CrossRef]
- Glover, N.M.; Redestig, H.; Dessimoz, C. Homoeologs: What Are They and How Do We Infer Them? Trends Plant Sci. 2016, 21, 609–621. [Google Scholar] [CrossRef]
- Haas, B.J.; Delcher, A.L.; Wortman, J.R.; Salzberg, S.L. DAGchainer: A tool for mining segmental genome duplications and synteny. Bioinformatics 2004, 20, 3643–3646. [Google Scholar] [CrossRef]
- Haug-Baltzell, A.; Stephens, S.A.; Davey, S.; Scheidegger, C.E.; Lyons, E. SynMap2 and SynMap3D: Web-based whole-genome synteny browsers. Bioinformatics 2017, 33, 2197–2198. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, M.; Zhang, X.; Hao, B.; Kaushik, S.K.; Pan, Y. WRKY gene family evolution in Arabidopsis thaliana. Genetica 2011, 139, 973–983. [Google Scholar] [CrossRef]
- Chanderbali, A.S.; Berger, B.A.; Howarth, D.G.; Soltis, D.E.; Soltis, P.S. Evolution of floral diversity: Genomics, genes and gamma. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20150509. [Google Scholar] [CrossRef]
- Sahebi, M.; Hanafi, M.M.; van Wijnen, A.J.; Rice, D.; Rafii, M.Y.; Azizi, P.; Osman, M.; Taheri, S.; Bakar, M.F.A.; Isa, M.N.M.; et al. Contribution of transposable elements in the plant’s genome. Gene 2018, 665, 155–166. [Google Scholar] [CrossRef]
- Bailey, P.C.; Schudoma, C.; Jackson, W.; Baggs, E.; Dagdas, G.; Haerty, W.; Moscou, M.; Krasileva, K.V. Dominant integration locus drives continuous diversification of plant immune receptors with exogenous domain fusions. Genome Biol. 2018, 19, 23. [Google Scholar] [CrossRef]
- Moore, R.C.; Purugganan, M.D. The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 2005, 8, 122–128. [Google Scholar] [CrossRef]
- Vicient, C.M.; Casacuberta, J.M. Impact of transposable elements on polyploid plant genomes. Ann. Bot. 2017, 120, 195–207. [Google Scholar] [CrossRef]
- Bashir, T.; Chandra Mishra, R.; Hasan, M.M.; Mohanta, T.K.; Bae, H. Effect of Hybridization on Somatic Mutations and Genomic Rearrangements in Plants. Int. J. Mol. Sci. 2018, 19, 3758. [Google Scholar] [CrossRef]
- Hughes, A.L.; Friedman, R.; Ekollu, V.; Rose, J.R. Non-random association of transposable elements with duplicated genomic blocks in Arabidopsis thaliana. Mol. Phylogenet. Evol. 2003, 29, 410–416. [Google Scholar] [CrossRef]
- Liu, D.; Hunt, M.; Tsai, I.J. Inferring synteny between genome assemblies: A systematic evaluation. BMC Bioinform. 2018, 19, 26. [Google Scholar] [CrossRef]
- Cooper, G.M.; Brown, C.D. Qualifying the relationship between sequence conservation and molecular function. Genome Res. 2008, 18, 201–205. [Google Scholar] [CrossRef]
- Jiao, Y.; Leebens-Mack, J.; Ayyampalayam, S.; Bowers, J.E.; McKain, M.R.; McNeal, J.; Rolf, M.; Ruzicka, D.R.; Wafula, E.; Wickett, N.J.; et al. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 2012, 13, R3. [Google Scholar] [CrossRef]
- Murat, F.; Armero, A.; Pont, C.; Klopp, C.; Salse, J. Reconstructing the genome of the most recent common ancestor of flowering plants. Nat. Genet. 2017, 49, 490–496. [Google Scholar] [CrossRef]
- Xiang, Y.; Huang, C.H.; Hu, Y.; Wen, J.; Li, S.; Yi, T.; Chen, H.; Xiang, J.; Ma, H. Evolution of Rosaceae Fruit Types Based on Nuclear Phylogeny in the Context of Geological Times and Genome Duplication. Mol. Biol. Evol. 2017, 34, 262–281. [Google Scholar] [CrossRef]
- Koonin, E.V. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 2005, 39, 309–338. [Google Scholar] [CrossRef]
- Nehrt, N.L.; Clark, W.T.; Radivojac, P.; Hahn, M.W. Testing the ortholog conjecture with comparative functional genomic data from mammals. PLoS Comput. Biol. 2011, 7, e1002073. [Google Scholar] [CrossRef]
- APG (The Angiosperm Phylogeny Group). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef]
- Tiley, G.P.; Ane, C.; Burleigh, J.G. Evaluating and Characterizing Ancient Whole-Genome Duplications in Plants with Gene Count Data. Genome Biol. Evol. 2016, 8, 1023–1037. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.D.; El Baidouri, M.; Abernathy, B.; Iwata-Otsubo, A.; Chavarro, C.; Gonzales, M.; Libault, M.; Grimwood, J.; Jackson, S.A. A Comparative Epigenomic Analysis of Polyploidy-Derived Genes in Soybean and Common Bean. Plant Physiol. 2015, 168, 1433–1447. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef]
- Velasco, R.; Zharkikh, A.; Affourtit, J.; Dhingra, A.; Cestaro, A.; Kalyanaraman, A.; Fontana, P.; Bhatnagar, S.K.; Troggio, M.; Pruss, D.; et al. The genome of the domesticated apple (Malus x domestica Borkh.). Nat. Genet. 2010, 42, 833–839. [Google Scholar] [CrossRef]
- Luo, M.C.; You, F.M.; Li, P.; Wang, J.R.; Zhu, T.; Dandekar, A.M.; Leslie, C.A.; Aradhya, M.; McGuire, P.E.; Dvorak, J. Synteny analysis in Rosids with a walnut physical map reveals slow genome evolution in long-lived woody perennials. BMC Genom. 2015, 16, 707. [Google Scholar] [CrossRef]
- Chen, J.; Nolan, T.M.; Ye, H.; Zhang, M.; Tong, H.; Xin, P.; Chu, J.; Chu, C.; Li, Z.; Yin, Y. Arabidopsis WRKY46, WRKY54, and WRKY70 Transcription Factors Are Involved in Brassinosteroid-Regulated Plant Growth and Drought Responses. Plant Cell 2017, 29, 1425–1439. [Google Scholar] [CrossRef]
- Li, J.; Zhong, R.; Palva, E.T. WRKY70 and its homolog WRKY54 negatively modulate the cell wall-associated defenses to necrotrophic pathogens in Arabidopsis. PLoS ONE 2017, 12, e0183731. [Google Scholar] [CrossRef]
- Amborella Genome, P. The Amborella genome and the evolution of flowering plants. Science 2013, 342, 1241089. [Google Scholar] [CrossRef]
- Lei, Y.; Sun, Y.; Wang, B.; Yu, S.; Dai, H.; Li, H.; Zhang, Z.; Zhang, J. Woodland strawberry WRKY71 acts as a promoter of flowering via a transcriptional regulatory cascade. Hortic. Res. 2020, 7, 137. [Google Scholar] [CrossRef]
- Wei, W.; Cui, M.Y.; Hu, Y.; Gao, K.; Xie, Y.G.; Jiang, Y.; Feng, J.Y. Ectopic expression of FvWRKY42, a WRKY transcription factor from the diploid woodland strawberry (Fragaria vesca), enhances resistance to powdery mildew, improves osmotic stress resistance, and increases abscisic acid sensitivity in Arabidopsis. Plant Sci. An. Int. J. Exp. Plant Biol. 2018, 275, 60–74. [Google Scholar] [CrossRef]
- Zhang, W.W.; Zhao, S.Q.; Gu, S.; Cao, X.Y.; Zhang, Y.; Niu, J.F.; Liu, L.; Li, A.R.; Jia, W.S.; Qi, B.X.; et al. FvWRKY48 binds to the pectate lyase FvPLA promoter to control fruit softening in Fragaria vesca. Plant Physiol. 2022, 189, 1037–1049. [Google Scholar] [CrossRef]
- Hawkins, C.; Caruana, J.; Li, J.; Zawora, C.; Darwish, O.; Wu, J.; Alkharouf, N.; Liu, Z. An eFP browser for visualizing strawberry fruit and flower transcriptomes. Hortic. Res. 2017, 4, 17029. [Google Scholar] [CrossRef]
- Hollender, C.A.; Geretz, A.C.; Slovin, J.P.; Liu, Z. Flower and early fruit development in a diploid strawberry, Fragaria vesca. Planta 2012, 235, 1123–1139. [Google Scholar] [CrossRef]
- Toljamo, A.; Blande, D.; Karenlampi, S.; Kokko, H. Reprogramming of Strawberry (Fragaria vesca) Root Transcriptome in Response to Phytophthora cactorum. PLoS ONE 2016, 11, e0161078. [Google Scholar] [CrossRef]
- Hussain, R.M.F.; Sheikh, A.H.; Haider, I.; Quareshy, M.; Linthorst, H.J.M. Arabidopsis WRKY50 and TGA Transcription Factors Synergistically Activate Expression of PR1. Front. Plant Sci. 2018, 9, 930. [Google Scholar] [CrossRef]
- Gao, Q.M.; Venugopal, S.; Navarre, D.; Kachroo, A. Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 2011, 155, 464–476. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, L.; Xiang, S.; Chen, Y.; Zhang, H.; Yu, D. The transcription factor WRKY75 positively regulates jasmonate-mediated plant defense to necrotrophic fungal pathogens. J. Exp. Bot. 2021, 72, 1473–1489. [Google Scholar] [CrossRef]
- Encinas-Villarejo, S.; Maldonado, A.M.; Amil-Ruiz, F.; de los Santos, B.; Romero, F.; Pliego-Alfaro, F.; Muñoz-Blanco, J.; Caballero, J.L. Evidence for a positive regulatory role of strawberry (Fragaria × ananassa) Fa WRKY1 and Arabidopsis At WRKY75 proteins in resistance. J. Exp. Bot. 2009, 60, 3043–3065. [Google Scholar] [CrossRef]
- Higuera, J.J.; Garrido-Gala, J.; Lekhbou, A.; Arjona-Girona, I.; Amil-Ruiz, F.; Mercado, J.A.; Pliego-Alfaro, F.; Munoz-Blanco, J.; Lopez-Herrera, C.J.; Caballero, J.L. The Strawberry FaWRKY1 Transcription Factor Negatively Regulates Resistance to Colletotrichum acutatum in Fruit Upon Infection. Front. Plant Sci. 2019, 10, 480. [Google Scholar] [CrossRef]
- Eulgem, T. Dissecting the WRKY web of plant defense regulators. PLoS Pathog. 2006, 2, e126. [Google Scholar] [CrossRef][Green Version]
- Wang, D.; Amornsiripanitch, N.; Dong, X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2006, 2, e123. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Kracher, B.; Ross, A.; Kramer, K.; Finkemeier, I.; Somssich, I.E. Principles and characteristics of the Arabidopsis WRKY regulatory network during early MAMP-triggered immunity. Plant J. 2018, 96, 487–502. [Google Scholar] [CrossRef]
- Zheng, Z.; Mosher, S.L.; Fan, B.; Klessig, D.F.; Chen, Z. Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol. 2007, 7, 2. [Google Scholar] [CrossRef]
- Roulin, A.; Auer, P.L.; Libault, M.; Schlueter, J.; Farmer, A.; May, G.; Stacey, G.; Doerge, R.W.; Jackson, S.A. The fate of duplicated genes in a polyploid plant genome. Plant J. 2013, 73, 143–153. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, X.; He, C.; Zhang, Q.Y.; Zou, X.; Duan, K.; Gao, Q. Novel Fungal Pathogenicity and Leaf Defense Strategies Are Revealed by Simultaneous Transcriptome Analysis of Colletotrichum fructicola and Strawberry Infected by This Fungus. Front. Plant Sci. 2018, 9, 434. [Google Scholar] [CrossRef]
- Devaiah, B.N.; Karthikeyan, A.S.; Raghothama, K.G. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 2007, 143, 1789–1801. [Google Scholar] [CrossRef]
- Garrido-Gala, J.; Higuera, J.J.; Munoz-Blanco, J.; Amil-Ruiz, F.; Caballero, J.L. The VQ motif-containing proteins in the diploid and octoploid strawberry. Sci. Rep. 2019, 9, 4942. [Google Scholar] [CrossRef]
- Medina-Puche, L.; Martinez-Rivas, F.J.; Molina-Hidalgo, F.J.; Mercado, J.A.; Moyano, E.; Rodriguez-Franco, A.; Caballero, J.L.; Munoz-Blanco, J.; Blanco-Portales, R. An atypical HLH transcriptional regulator plays a novel and important role in strawberry ripened receptacle. BMC Plant Biol. 2019, 19, 586. [Google Scholar] [CrossRef]
- Zhao, F.; Li, G.; Hu, P.; Zhao, X.; Li, L.; Wei, W.; Feng, J.; Zhou, H. Identification of basic/helix-loop-helix transcription factors reveals candidate genes involved in anthocyanin biosynthesis from the strawberry white-flesh mutant. Sci. Rep. 2018, 8, 2721. [Google Scholar] [CrossRef]
- Bhattarai, K.K.; Atamian, H.S.; Kaloshian, I.; Eulgem, T. WRKY72-type transcription factors contribute to basal immunity in tomato and Arabidopsis as well as gene-for-gene resistance mediated by the tomato R gene Mi-1. Plant J. 2010, 63, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Van Verk, M.C.; Bol, J.F.; Linthorst, H.J. WRKY transcription factors involved in activation of SA biosynthesis genes. BMC Plant Biol. 2011, 11, 89. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Dong, Q.; Yu, D. Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. Int. J. Exp. Plant Biol. 2012, 185–186, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Brader, G.; Kariola, T.; Palva, E.T. WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 2006, 46, 477–491. [Google Scholar] [CrossRef]
- Li, J.; Brader, G.; Palva, E.T. The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 2004, 16, 319–331. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Kracher, B.; Roccaro, M.; Somssich, I.E. Induced Genome-Wide Binding of Three Arabidopsis WRKY Transcription Factors during Early MAMP-Triggered Immunity. Plant Cell 2017, 29, 20–38. [Google Scholar] [CrossRef]
- Pandey, S.P.; Roccaro, M.; Schon, M.; Logemann, E.; Somssich, I.E. Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. Plant J. 2010, 64, 912–923. [Google Scholar] [CrossRef]
- Zheng, Z.; Qamar, S.A.; Chen, Z.; Mengiste, T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006, 48, 592–605. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, F.; Chen, M.; Liu, Z.; Zhang, Z.; Fu, J.; Ma, Y. Comparative Transcriptomics Reveals Differential Gene Expression Related to Colletotrichum gloeosporioides Resistance in the Octoploid Strawberry. Front. Plant Sci. 2017, 8, 779. [Google Scholar] [CrossRef]
- Mukhi, N.; Brown, H.; Gorenkin, D.; Ding, P.; Bentham, A.R.; Stevenson, C.E.M.; Jones, J.D.G.; Banfield, M.J. Perception of structurally distinct effectors by the integrated WRKY domain of a plant immune receptor. Proc. Natl. Acad. Sci. USA 2021, 118, e2113996118. [Google Scholar] [CrossRef]
- Narusaka, M.; Shirasu, K.; Noutoshi, Y.; Kubo, Y.; Shiraishi, T.; Iwabuchi, M.; Narusaka, Y. RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 2009, 60, 218–226. [Google Scholar] [CrossRef]
- Jung, S.; Lee, T.; Cheng, C.H.; Buble, K.; Zheng, P.; Yu, J.; Humann, J.; Ficklin, S.P.; Gasic, K.; Scott, K.; et al. 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 2019, 47, D1137–D1145. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; Ugene Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Guo, C.; Guo, R.; Xu, X.; Gao, M.; Li, X.; Song, J.; Zheng, Y.; Wang, X. Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J. Exp. Bot. 2014, 65, 1513–1528. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Sperschneider, J.; Catanzariti, A.M.; DeBoer, K.; Petre, B.; Gardiner, D.M.; Singh, K.B.; Dodds, P.N.; Taylor, J.M. LOCALIZER: Subcellular localization prediction of both plant and effector proteins in the plant cell. Sci. Rep. 2017, 7, 44598. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; van den Beek, M.; Blankenberg, D.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Eberhard, C.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016, 44, W3–W10. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Suyama, M.; Torrents, D.; Bork, P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006, 34, W609–W612. [Google Scholar] [CrossRef]
- Yang, Z.; Nielsen, R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 2002, 19, 908–917. [Google Scholar] [CrossRef]
- Villanueva-Canas, J.L.; Laurie, S.; Alba, M.M. Improving genome-wide scans of positive selection by using protein isoforms of similar length. Genome Biol. Evol. 2013, 5, 457–467. [Google Scholar] [CrossRef]
- De La Torre, A.R.; Li, Z.; Van de Peer, Y.; Ingvarsson, P.K. Contrasting Rates of Molecular Evolution and Patterns of Selection among Gymnosperms and Flowering Plants. Mol. Biol. Evol. 2017, 34, 1363–1377. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; von Mering, C.; Bork, P. Fast Genome-Wide Functional Annotation through Orthology Assignment by eggNOG-Mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernandez-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Ye, J.; Zhang, Y.; Cui, H.; Liu, J.; Wu, Y.; Cheng, Y.; Xu, H.; Huang, X.; Li, S.; Zhou, A.; et al. WEGO 2.0: A web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 2018, 46, W71–W75. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2020, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Kopylova, E.; Noe, L.; Touzet, H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]


| Gene 1 | Gene 2 | Gene 3 | Gene 4 | Gene 5 | Kn | Ks | ω |
|---|---|---|---|---|---|---|---|
| Segmental duplications | |||||||
| FvWRKY2 | FvWRKY9 | 0.4583 | 6.7082 | 0.0683 | |||
| FvWRKY7 | FvWRKY36 | 0.3183 | 1.5265 | 0.2085 | |||
| FvWRKY13 | FvWRKY50 * | 0.3550 | 2.6663 | 0.1331 | |||
| FvWRKY14 | FvWRKY48 | 0.4566 | 2.6947 | 0.1694 | |||
| FvWRKY15 | FvWRKY46 | 0.4185 | 5.7462 | 0.0728 | |||
| FvWRKY17 | FvWRKY50 * | 0.3908 | 5.7974 | 0.0674 | |||
| FvWRKY19 | FvWRKY43 | 0.3680 | 1.8100 | 0.2033 | |||
| FvWRKY24 * | FvWRKY53 | 0.4702 | 3.1511 | 0.1492 | |||
| FvWRKY24 * | FvWRKY30 | 0.4020 | 5.0096 | 0.0802 | |||
| FvWRKY26 | FvWRKY51 | 0.3295 | 3.0282 | 0.1088 | |||
| FvWRKY27 | FvWRKY31 | 0.7246 | 4.1163 | 0.1760 | |||
| FvWRKY27 | FvWRKY52 | 0.5880 | 2.9760 | 0.1976 | |||
| FvWRKY29 | FvWRKY58 * | 0.4700 | 2.2460 | 0.2093 | |||
| FvWRKY30 | FvWRKY53 | 0.4628 | 77.4216 | 0.0060 | |||
| FvWRKY39 * | FvWRKY61 * | 0.7849 | 2.7159 | 0.2890 | |||
| FvWRKY58 * | FvWRKY64 | 0.5863 | 63.4312 | 0.0092 | |||
| Tandem repeats | |||||||
| FvWRKY11 | FvWRKY12 | NA | NA | NA | |||
| FvWRKY38 | FvWRKY39 * | FvWRKY40 | FvWRKY41 | FvWRKY42 | NA | NA | NA |
| FvWRKY59 | FvWRKY60 | FvWRKY61 * | NA | NA | NA | ||
| Gene 1 | Gene 2 | Gene 3 | Kn | Ks | ω |
|---|---|---|---|---|---|
| Segmental duplications | |||||
| FaWRKY6C.1 | FaWRKY6C.2 | 0.0026 | 0.0000 | NA | |
| FaWRKY43B.1 | FaWRKY43B.2 | 0.0017 | 0.0055 | NA | |
| FaWRKY54A.1 | FaWRKY54A.2 | 0.0000 | 0.0039 | NA | |
| FaWRKY54D.1 | FaWRKY54D.2 | 0.0016 | 0.0000 | NA | |
| FaWRKY56A.1 | FaWRKY56A.2 | 0.0083 | 0.0109 | 0.7615 | |
| FaWRKY56D.1 | FaWRKY56D.2 | 0.0013 | 0.0047 | NA | |
| FaWRKY57A.1 | FaWRKY57A.2 | 0.0000 | 0.0087 | NA | |
| FaWRKY57D.1 | FaWRKY57D.2 | 0.0022 | 0.0000 | NA | |
| FaWRKY58A.1 | FaWRKY58A.2 | 0.0064 | 0.0111 | 0.5766 | |
| FaWRKY58D.1 | FaWRKY58D.2 | 0.0012 | 0.0043 | NA | |
| FaWRKY63A.1 | FaWRKY63A.2 | 0.0024 | 0.0000 | NA | |
| FaWRKY64A.1 | FaWRKY64A.2 | 0.0026 | 0.0063 | NA | |
| Segmental, non-syntenic | |||||
| FaWRKY2D.1 | FaWRKY2D.2 | 0.0131 | 0.0153 | 0.8562 | |
| FaWRKY2C.1 | FaWRKY2C.2 | 0.0000 | 0.0000 | NA | |
| FaWRKY2C.1 | FaWRKY2C.3 | 0.0124 | 0.0147 | 0.8482 | |
| FaWRKY2C.2 | FaWRKY2C.3 | 0.0124 | 0.0147 | 0.8482 | |
| FaWRKY2B.1 | FaWRKY2B.2 | 0.0134 | 0.0153 | 0.8758 | |
| FaWRKY2B.1 | FaWRKY2B.3 | 0.0125 | 0.0145 | 0.8621 | |
| FaWRKY2B.2 | FaWRKY2B.3 | 0.0133 | 0.0103 | 1.2913 | |
| FaWRKY14C.1 | FaWRKY14C.2 | 0.0071 | 0.0279 | 0.2545 | |
| FaWRKY17C.1 | FaWRKY17C.2 | 0.0021 | 0.0084 | NA | |
| FaWRKY18C.1 | FaWRKY18C.2 | 0.0072 | 0.0069 | NA | |
| FaWRKY26B.1 | FaWRKY26B.2 | 0.3425 | 0.5631 | 0.6082 | |
| FaWRKY29D.1 | FaWRKY29D.2 | 0.0011 | 0.0000 | NA | |
| FaWRKY35D.1 | FaWRKY35D.2 | 0.0024 | 0.0101 | 0.2376 | |
| FaWRKY36D.1 | FaWRKY36D.2 | 0.0190 | 0.0322 | 0.5901 | |
| FaWRKY45D.1 | FaWRKY45D.2 | 0.0000 | 0.0000 | NA | |
| FaWRKY47C.1 | FaWRKY47C.2 | 0.1398 | 0.2113 | 0.6616 | |
| FaWRKY51A.1 | FaWRKY51A.2 | 0.0400 | 0.0457 | 0.8753 | |
| FaWRKY51A.1 | FaWRKY51A.3 | 0.0965 | 0.1091 | 0.8845 | |
| FaWRKY51A.2 | FaWRKY51A.3 | 0.0630 | 0.0586 | 1.0751 | |
| FaWRKY62B.1 | FaWRKY62B.2 | 0.0298 | 0.0269 | 1.1078 | |
| FaWRKY63D.1 | FaWRKY63D.2 | 0.0022 | 0.0053 | NA | |
| FaWRKY63B.1 | FaWRKY63B.2 | 0.0147 | 0.0132 | 1.1136 | |
| FaWRKY64D.1 | FaWRKY64D.2 | 0.0025 | 0.0032 | NA | |
| Tandem duplications | |||||
| FaWRKY11C | FaWRKY12C | NA | NA | NA | |
| FaWRKY11A | FaWRKY12A | NA | NA | NA | |
| FaWRKY55B.1 | FaWRKY55B.2 | NA | NA | NA | |
| FaWRKY38A | FaWRKY39A | FaWRKY41A | NA | NA | NA |
| FaWRKY38B | FaWRKY42B | NA | NA | NA | |
| FaWRKY38C | FaWRKY42C | NA | NA | NA | |
| FaWRKY40D | FaWRKY41D | FaWRKY42D | NA | NA | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garrido-Gala, J.; Higuera, J.-J.; Rodríguez-Franco, A.; Muñoz-Blanco, J.; Amil-Ruiz, F.; Caballero, J.L. A Comprehensive Study of the WRKY Transcription Factor Family in Strawberry. Plants 2022, 11, 1585. https://doi.org/10.3390/plants11121585
Garrido-Gala J, Higuera J-J, Rodríguez-Franco A, Muñoz-Blanco J, Amil-Ruiz F, Caballero JL. A Comprehensive Study of the WRKY Transcription Factor Family in Strawberry. Plants. 2022; 11(12):1585. https://doi.org/10.3390/plants11121585
Chicago/Turabian StyleGarrido-Gala, José, José-Javier Higuera, Antonio Rodríguez-Franco, Juan Muñoz-Blanco, Francisco Amil-Ruiz, and José L. Caballero. 2022. "A Comprehensive Study of the WRKY Transcription Factor Family in Strawberry" Plants 11, no. 12: 1585. https://doi.org/10.3390/plants11121585
APA StyleGarrido-Gala, J., Higuera, J.-J., Rodríguez-Franco, A., Muñoz-Blanco, J., Amil-Ruiz, F., & Caballero, J. L. (2022). A Comprehensive Study of the WRKY Transcription Factor Family in Strawberry. Plants, 11(12), 1585. https://doi.org/10.3390/plants11121585






