Viromes of Hungarian Peach Trees Identified by High-Throughput Sequencing of Small RNAs
Abstract
:1. Introduction
2. Results
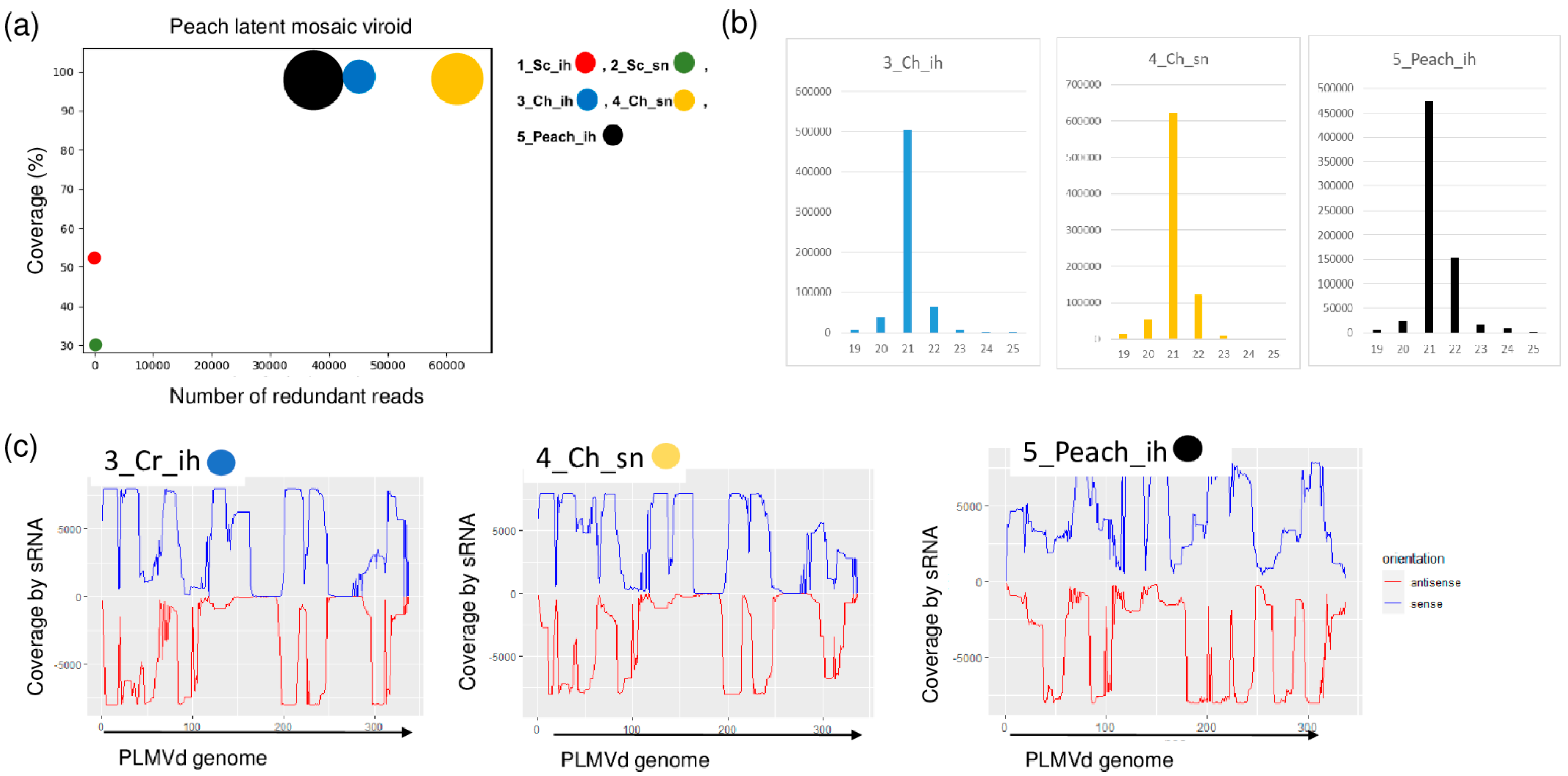
2.1. Small RNA HTS Revealed the Presence of Both Regulated and Non-Regulated Viruses
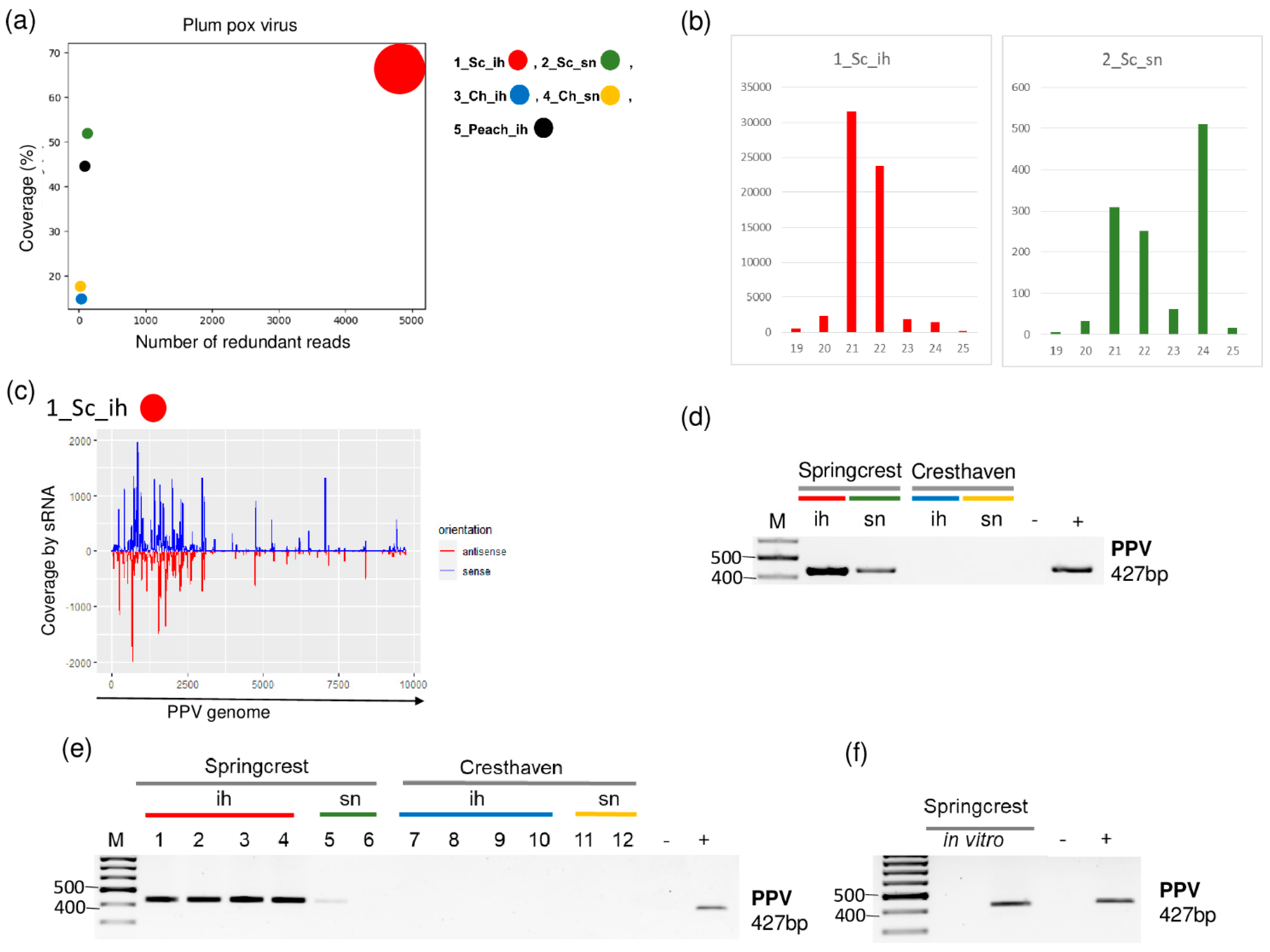
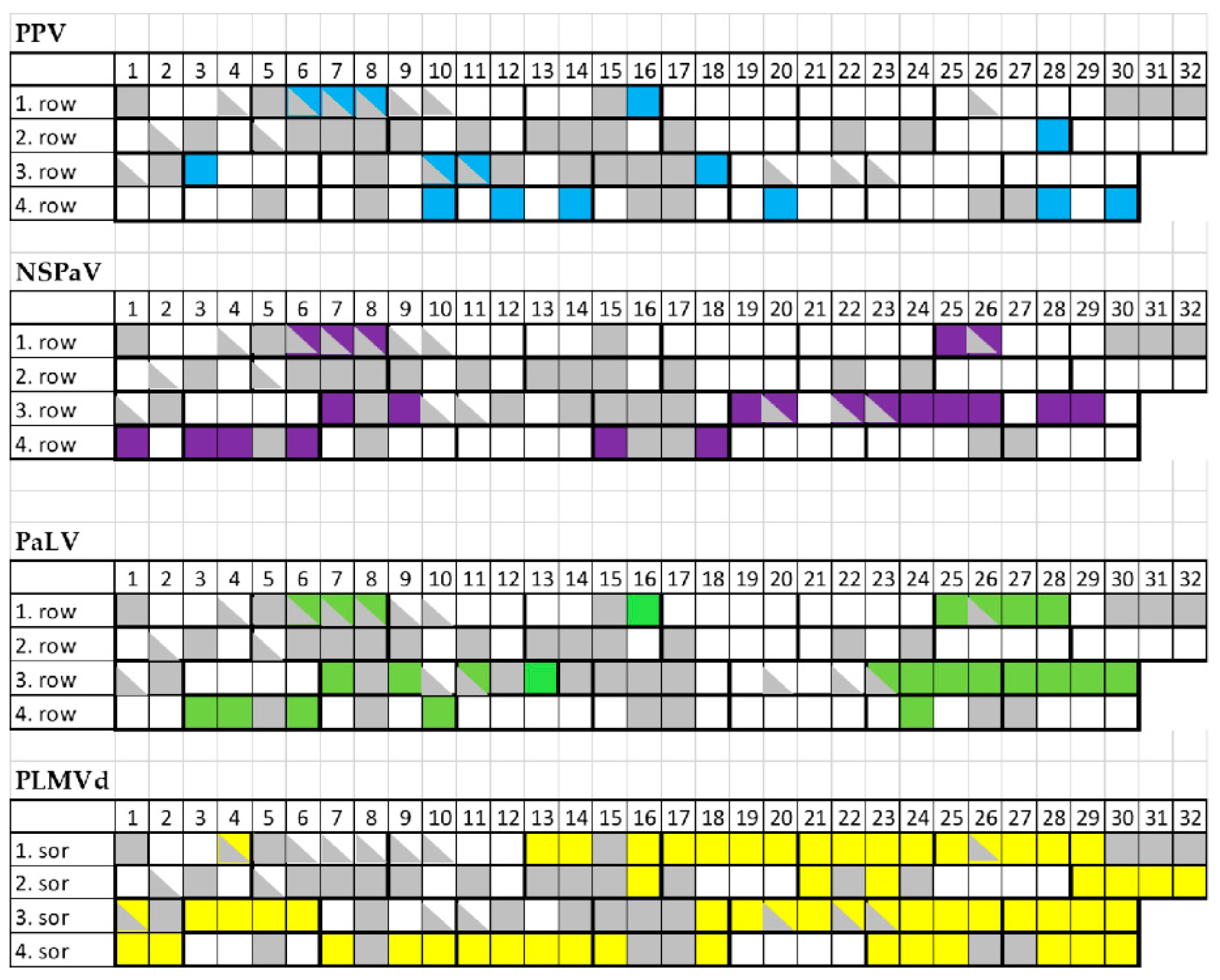
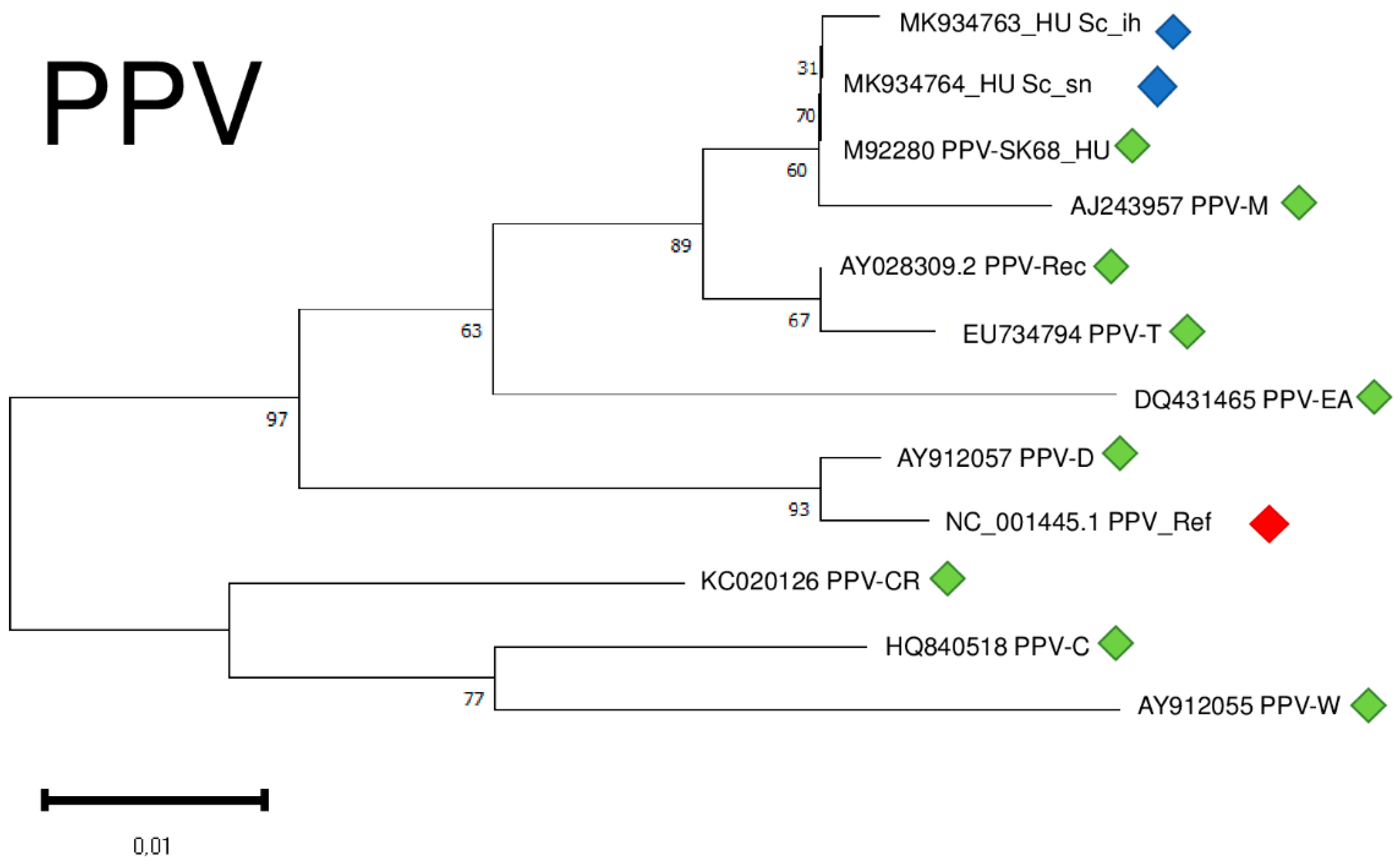
2.2. PPV Is Present in the Isolator
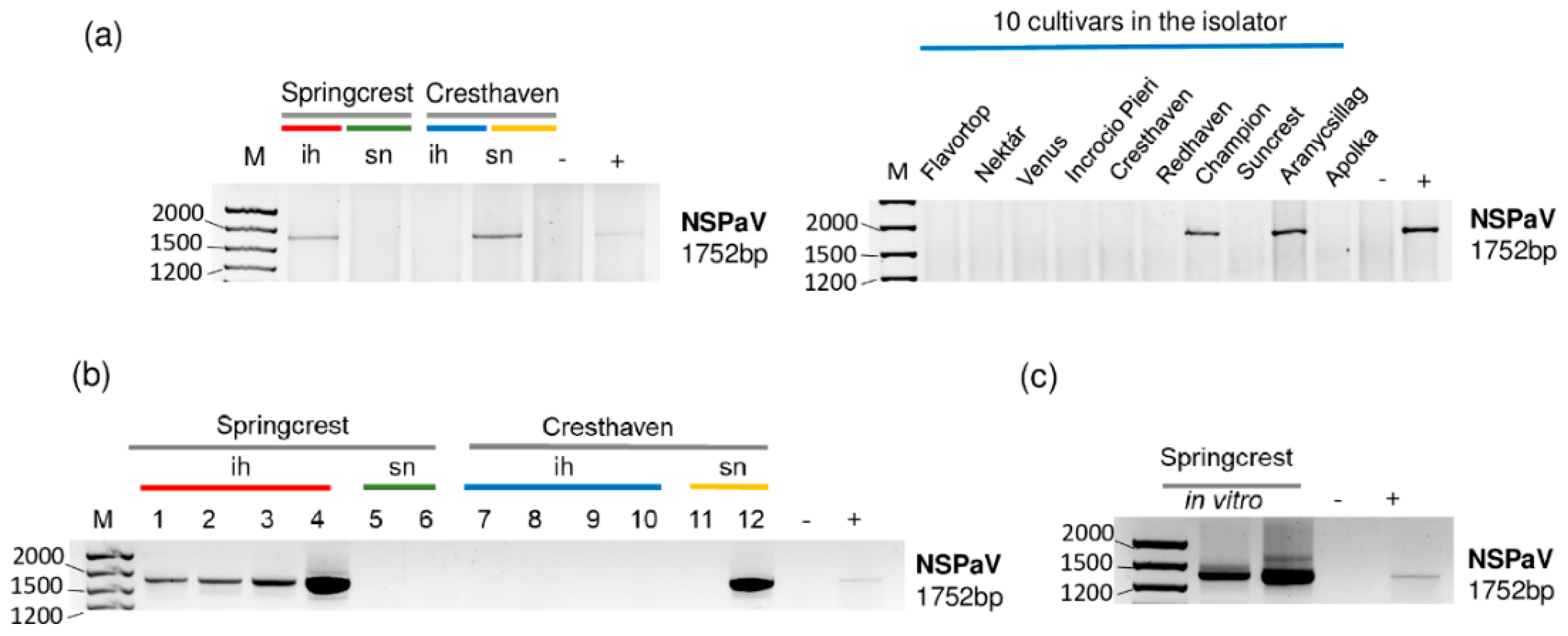
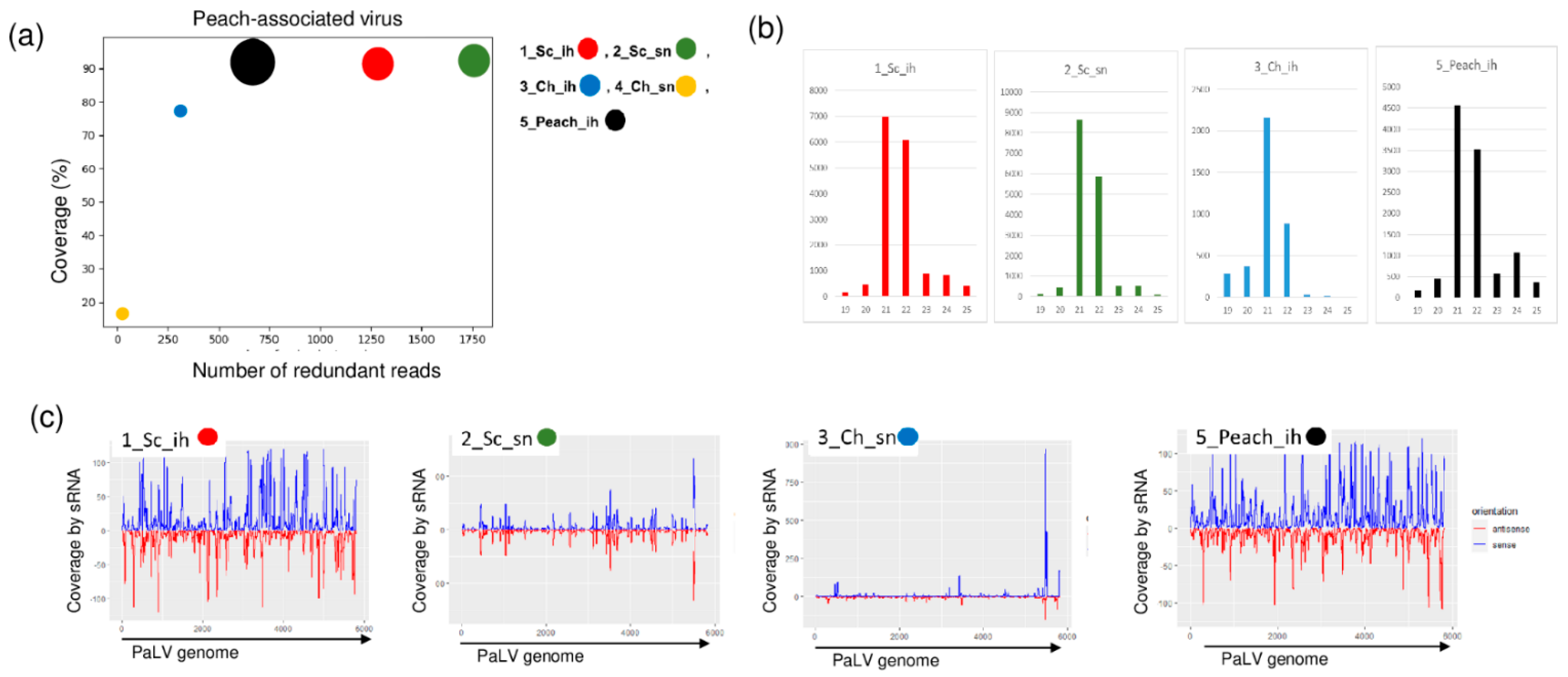
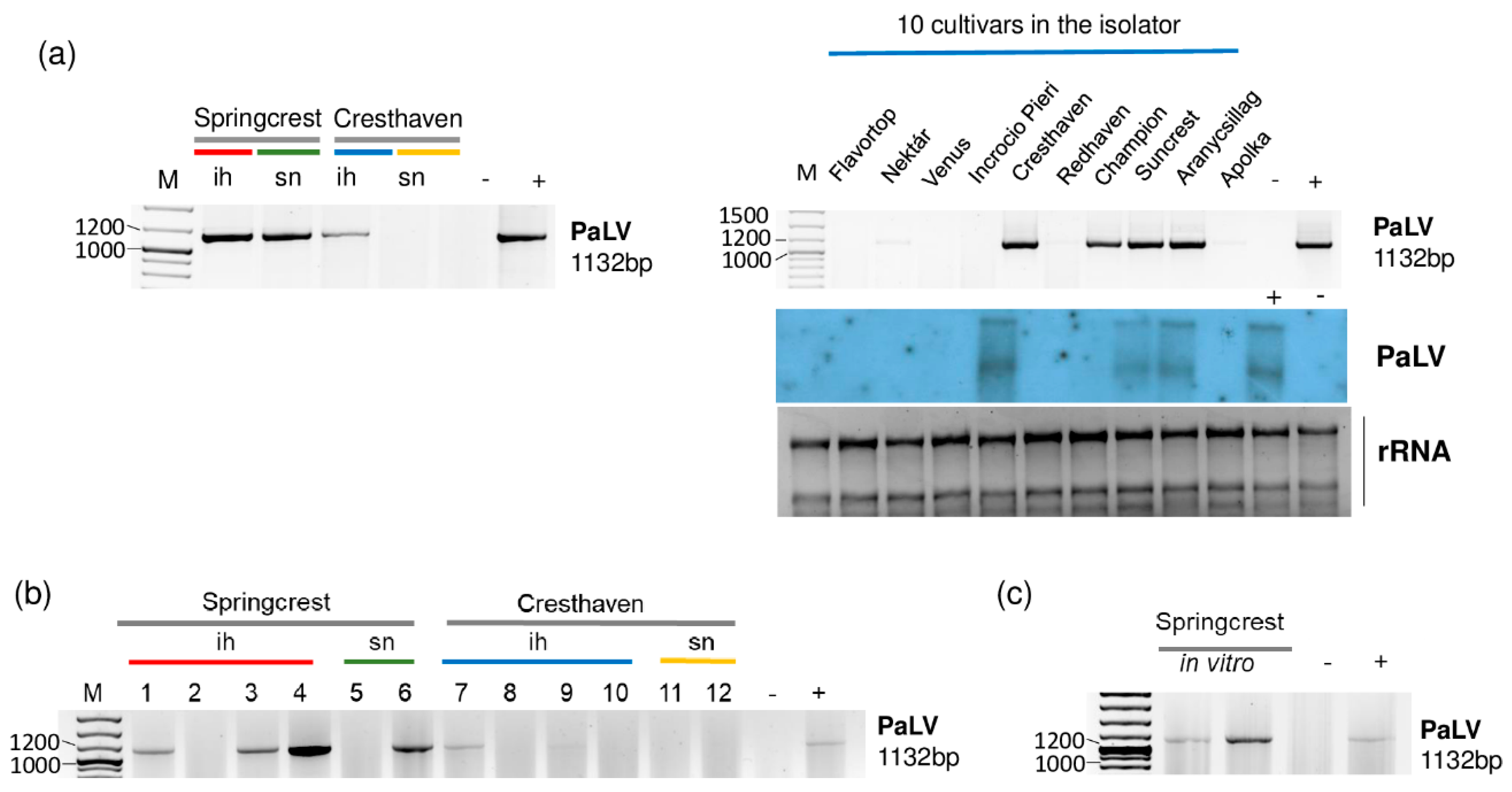
2.3. Luteoviruses: NSPaV and PaLV Are Present Both in the Isolator and in the Open Field
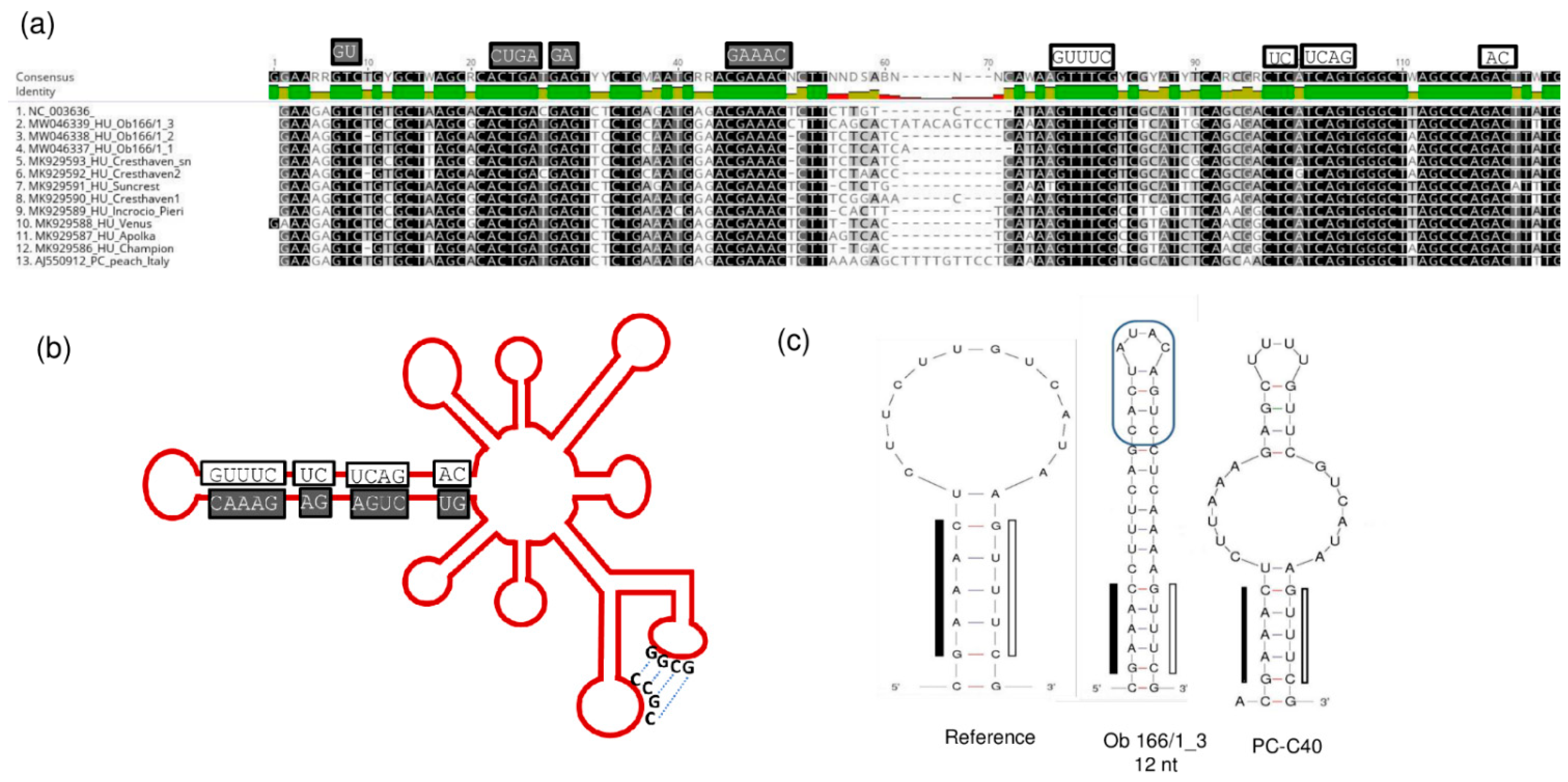
2.4. Frequent Presence of PLMVd in the Tested Trees Was Shown by sRNA HTS and RT-LAMP, but Not with Biological Assays on GF305
2.5. Infection with Luteoviruses and PLMVd Are Frequent in the Isolator, but Cannot Be Ascribed as the Only Cause of the Trees’ Decline
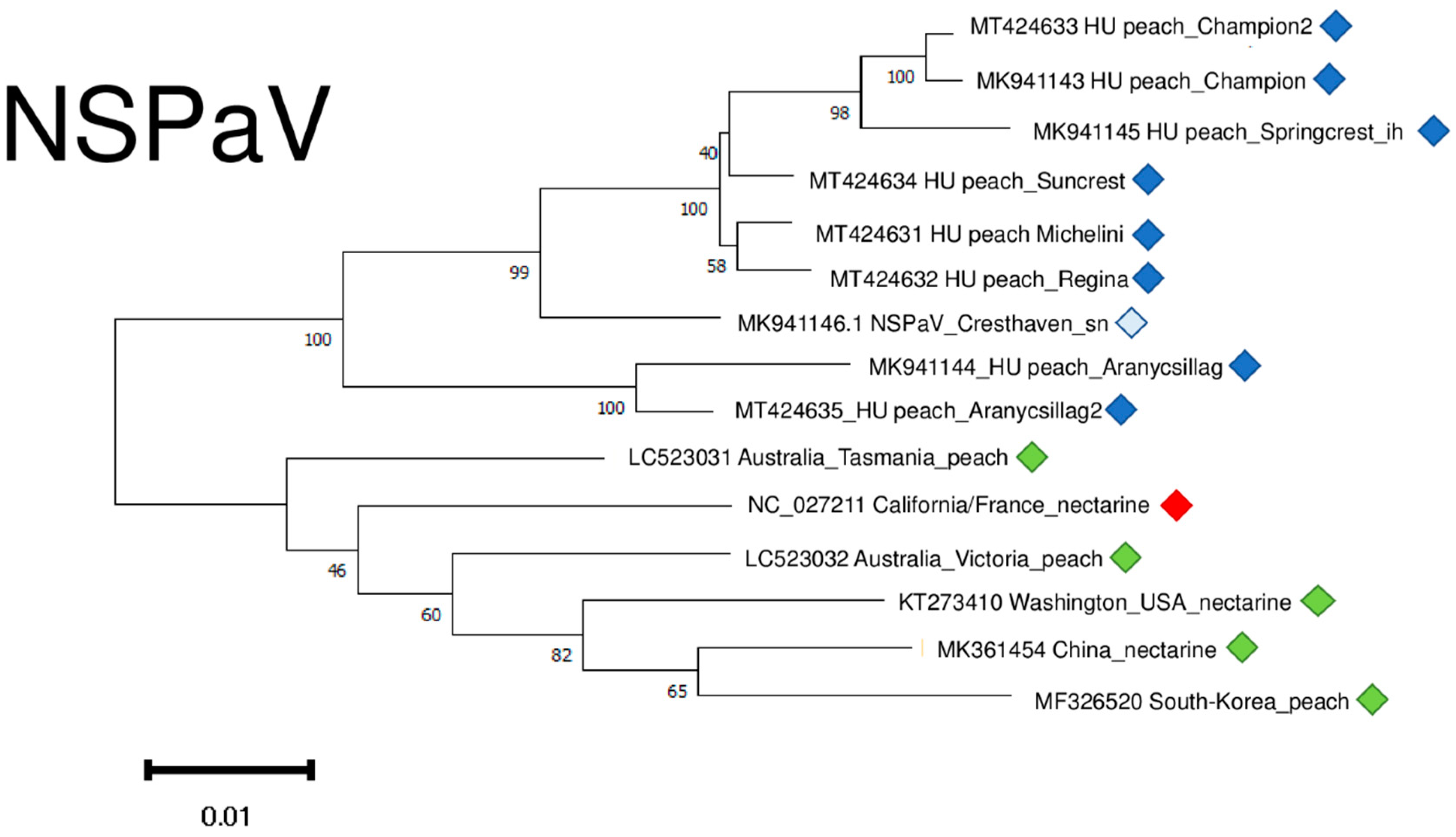
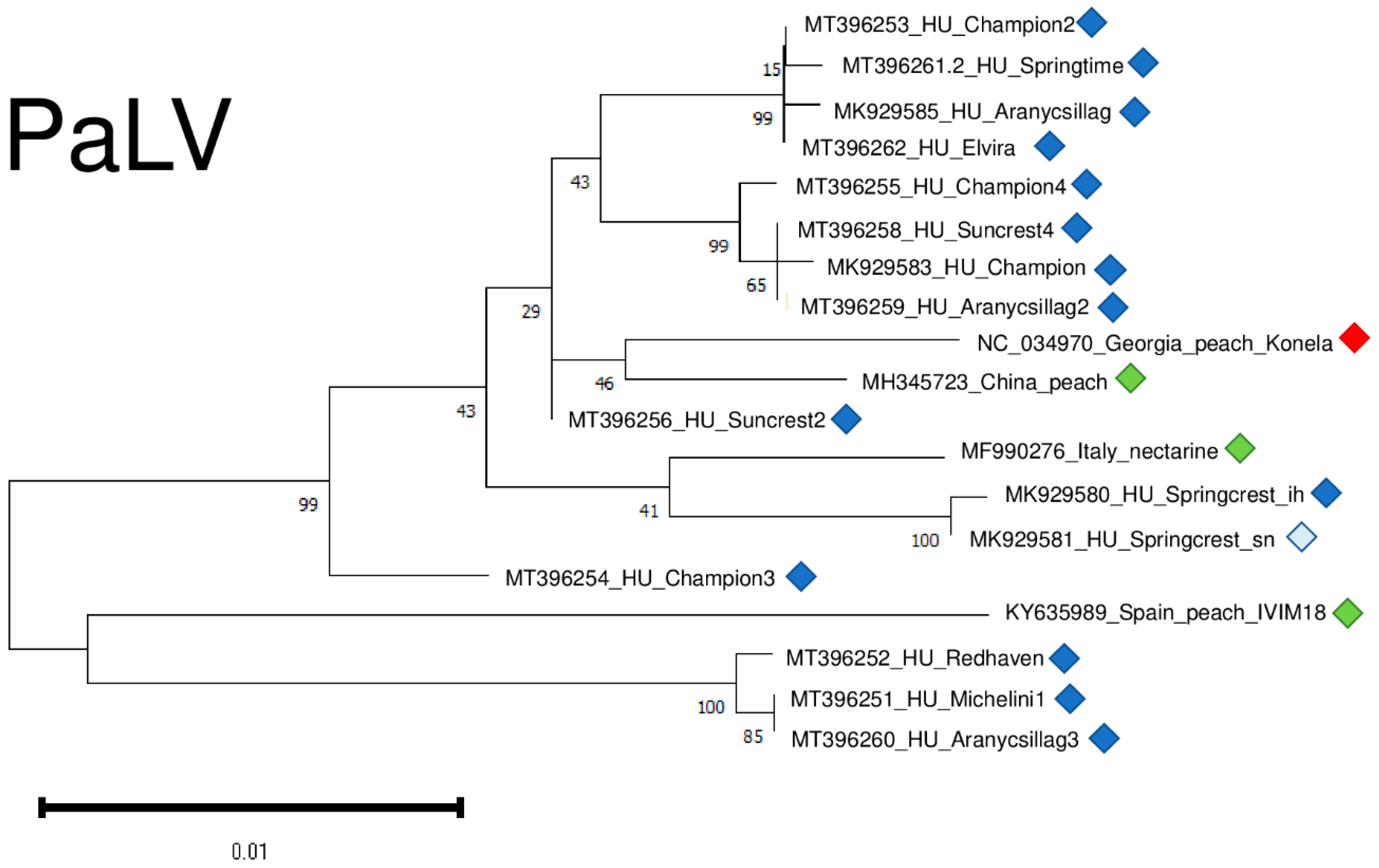
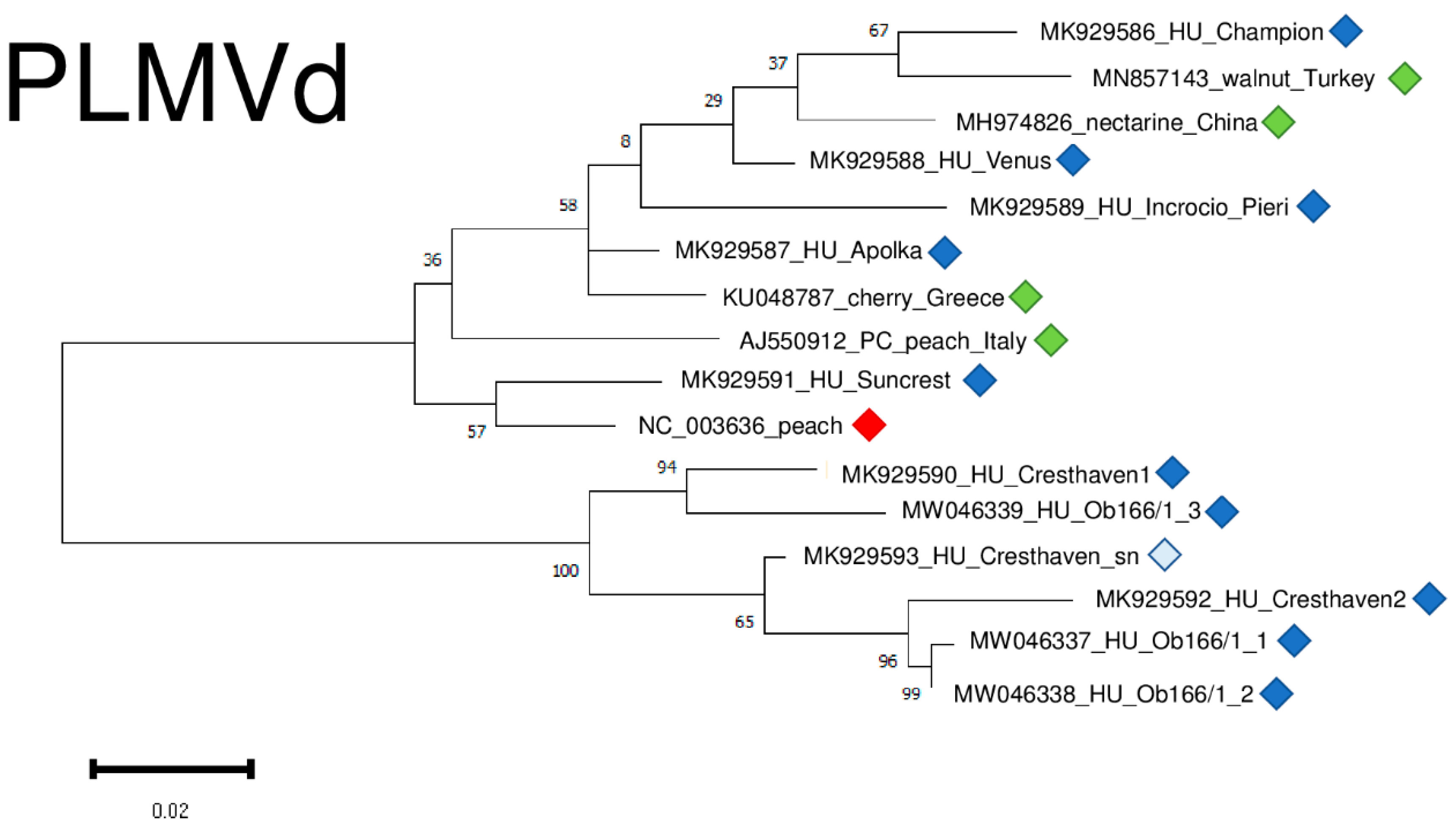
2.6. Phylogenetic Analysis of the Viral Strains
3. Discussion
4. Materials and Methods
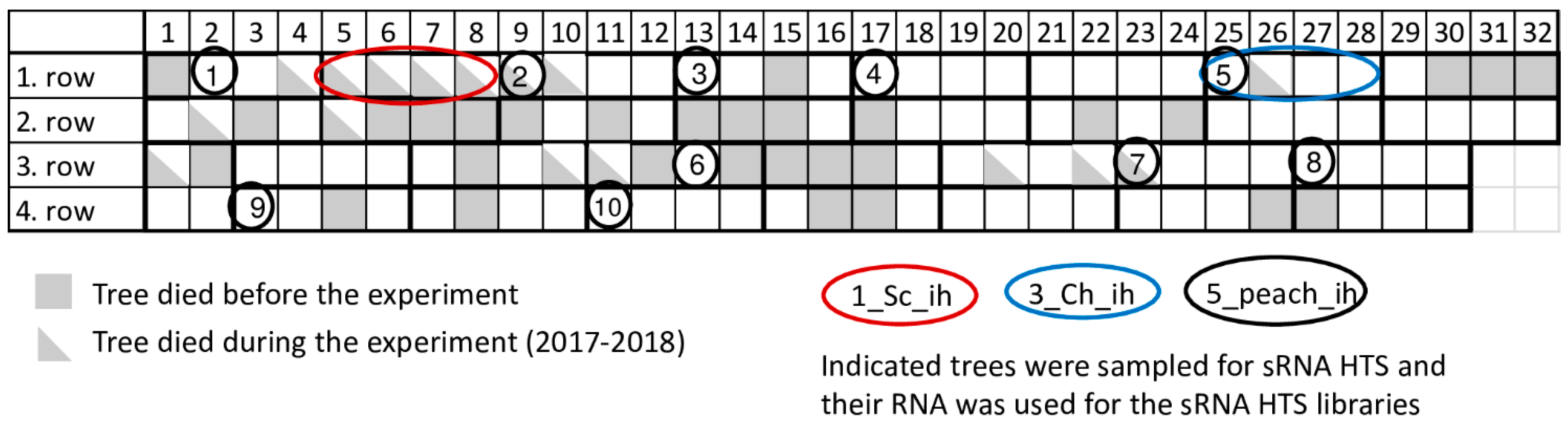
4.1. Plant Material and Sample Preparation
4.2. Pipeline for the Data Evaluation of the HTS Results (Bioinformatics)
4.3. Confirmation of the Obtained Results by RT-PCR, Northern Blot and Sanger Sequencing
4.4. RT—LAMP Assay for PLMVd Detection
4.5. Biological Assay
4.6. Phylogenetical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hadidi, A.; Barba, M. Chapter 1: Economic impact of pome and stone fruit viruses and viroids. In Virus and Virus-Like Diseases of Pome and Stone Fruits; The American Phytopathological Society: Saint Paul, MN, USA, 2011; pp. 1–7. [Google Scholar] [CrossRef]
- Barba, M.; Ilardi, V.; Pasquini, G. Control of pome and stone fruit virus diseases. Adv. Virus Res. 2015, 91, 47–83. [Google Scholar] [CrossRef] [PubMed]
- Roossinck, M.J.; Martin, D.P.; Roumagnac, P. Plant virus metagenomics: Advances in virus discovery. Phytopathology 2015, 105, 716–727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Studholme, D.J.; Glover, R.H.; Boonham, N. Application of high-throughput DNA sequencing in phytopathology. Annu. Rev. Phytopathol. 2011, 49, 87–105. [Google Scholar] [CrossRef] [PubMed]
- Massart, S.; Olmos, A.; Jijakli, H.; Candresse, T. Current impact and future directions of high throughput sequencing in plant virus diagnostics. Virus Res. 2014, 188, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Pooggin, M.M. Small rna-omics for plant virus identification, virome reconstruction, and antiviral defense characterization. Front. Microbiol. 2018, 9, 2779. [Google Scholar] [CrossRef]
- Maliogka, V.I.; Minafra, A.; Saldarelli, P.; Ruiz-García, A.B.; Glasa, M.; Katis, N.; Olmos, A. Recent advances on detection and characterization of fruit tree viruses using high-throughput sequencing technologies. Viruses 2018, 10, 436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maree, H.J.; Fox, A.; Al Rwahnih, M.; Boonham, N.; Candresse, T. Application of hts for routine plant virus diagnostics: State of the art and challenges. Front. Plant Sci. 2018, 9, 1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, W.; Li, S.; Massart, S. Is there a “biological desert” with the discovery of new plant viruses? A retrospective analysis for new fruit tree viruses. Front. Microbiol. 2020, 11, 592816. [Google Scholar] [CrossRef]
- Krizbai, L.; Kriston, E.; Kreuze, J.; Melika, G. Identification of nectarine stem pitting-associated virus infecting prunus persica in Hungary. New Dis. Rep. 2017, 35, 18. [Google Scholar] [CrossRef] [Green Version]
- García, J.A.; Glasa, M.; Cambra, M.; Candresse, T. Plum pox virus and sharka: A model potyvirus and a major disease. Mol. Plant Pathol. 2014, 15, 226–241. [Google Scholar] [CrossRef]
- Plum pox potyvirus. EPPO Bull. 2004, 34, 247–256. [CrossRef]
- Flores, R.; Delgado, S.; Rodio, M.-E.; Ambrós, S.; Hernández, C.; Di Serio, F. Peach latent mosaic viroid: Not so latent. Mol. Plant Pathol. 2006, 7, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Hernández, C.; Flores, R. Plus and minus rnas of peach latent mosaic viroid self-cleave in vitro via hammerhead structures. Proc. Natl. Acad. Sci. USA 1992, 89, 3711–3715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navarro, B.; Gisel, A.; Rodio, M.E.; Delgado, S.; Flores, R.; Di Serio, F. Small rnas containing the pathogenic determinant of a chloroplast-replicating viroid guide the degradation of a host mrna as predicted by rna silencing. Plant J. 2012, 70, 991–1003. [Google Scholar] [CrossRef] [PubMed]
- Boubourakas, I.N.; Fukuta, S.; Kyriakopoulou, P.E. Sensitive and rapid detection of peach latent mosaic viroid by the reverse transcription loop-mediated isothermal amplification. J. Virol. Methods 2009, 160, 63–68. [Google Scholar] [CrossRef]
- Lee, H.-J.; Jeong, R.-D. Reverse transcription loop-mediated isothermal amplification assay for detecting peach latent mosaic viroid in peach pollen. Hortic. Sci. Technol. 2020, 38, 830–839. [Google Scholar] [CrossRef]
- Bag, S.; Al Rwahnih, M.; Li, A.; Gonzalez, A.; Rowhani, A.; Uyemoto, J.K.; Sudarshana, M.R. Detection of a new luteovirus in imported nectarine trees: A case study to propose adoption of metagenomics in post-entry quarantine. Phytopathology 2015, 105, 840–846. [Google Scholar] [CrossRef] [Green Version]
- Villamor, D.E.V.; Mekuria, T.A.; Pillai, S.S.; Eastwell, K.C. High-throughput sequencing identifies novel viruses in nectarine: Insights to the etiology of stem-pitting disease. Phytopathology 2016, 106, 519–527. [Google Scholar] [CrossRef] [Green Version]
- Lu, M.G.; Zhang, C.; Zhang, Z.X.; Wang, C.A.; Li, S.F. Nectarine stem-pitting-associated virus detected in peach trees in China. Plant Dis. 2017, 101, 513. [Google Scholar] [CrossRef]
- Xu, Y.; Li, S.; Na, C.; Yang, L.; Lu, M. Analyses of virus/viroid communities in nectarine trees by next-generation sequencing and insight into viral synergisms implication in host disease symptoms. Sci. Rep. 2019, 9, 12261. [Google Scholar] [CrossRef] [Green Version]
- Jo, Y.; Lian, S.; Chu, H.; Cho, J.K.; Yoo, S.-H.; Choi, H.; Yoon, J.-Y.; Choi, S.-K.; Lee, B.C.; Cho, W.K. Peach RNA viromes in six different peach cultivars. Sci. Rep. 2018, 8, 1844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Igori, D.; Baek, D.; Kim San, Y.; Seo, E.; Lee, S.-H.; Jeong, R.-D.; Yi, S.-I.; Bong, J.-J.; Moon Jae, S. Complete genome sequence of nectarine stem pitting-associated virus, isolated from prunus persica in cheongdo county, south korea. Genome Announc. 2017, 5, e00908-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Candresse, T.; Faure, C.; Theil, S.; Marais, A. First report of nectarine stem pitting-associated virus infecting prunus mume in Japan. Plant Dis. 2017, 101, 393. [Google Scholar] [CrossRef]
- Kinoti, W.M.; Nancarrow, N.; Dann, A.; Rodoni, B.C.; Constable, F.E. Updating the quarantine status of prunus infecting viruses in australia. Viruses 2020, 12, 246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.P.; Liu, H.W.; Bateman, M.; Liu, Z.; Li, R. Molecular characterization of a novel luteovirus from peach identified by high-throughput sequencing. Arch. Virol. 2017, 162, 2903–2905. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, R.; Marais, A.; Faure, C.; Theil, S.; Alioto, D.; Candresse, T. First report of peach-associated luteovirus in nectarine (Prunus persica) in Italy. Plant Disease 2018, 102, 1465. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, Z.; Lu, M.; Xiao, H.; Li, S. First report of peach-associated luteovirus from flat peach and nectarine in China. Plant Dis. 2018, 102, 2669. [Google Scholar] [CrossRef]
- Lenz, O.; Přibylová, J.; Fránová, J.; Koloniuk, I.; Špak, J. Identification and characterization of a new member of the genus luteovirus from cherry. Arch. Virol. 2017, 162, 587–590. [Google Scholar] [CrossRef]
- Palkovics, L.; Burgyán, J.; Balázs, E. Comparative sequence analysis of four complete primary structures of plum pox virus strains. Virus Genes 1993, 7, 339–347. [Google Scholar] [CrossRef]
- Sihelská, N.; Glasa, M.; Šubr, Z.W. Host preference of the major strains of plum pox virus—Opinions based on regional and world-wide sequence data. J. Integr. Agric. 2017, 16, S2095–S3119. [Google Scholar] [CrossRef]
- Rubio, L.; Galipienso, L.; Ferriol, I. Detection of plant viruses and disease management: Relevance of genetic diversity and evolution. Front. Plant Sci. 2020, 11, 1092. [Google Scholar] [CrossRef] [PubMed]
- Massart, S.; Chiumenti, M.; De Jonghe, K.; Glover, R.; Haegeman, A.; Koloniuk, I.; Komínek, P.; Kreuze, J.; Kutnjak, D.; Lotos, L.; et al. Virus detection by high-throughput sequencing of small rnas: Large-scale performance testing of sequence analysis strategies. Phytopathology 2018, 109, 488–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navarro, L.; Llácer, G.; Cambra, M.; Arregui, J.M.; Juárez, J. Shoot-Tip Grafting In Vitro for Elimination of Viruses in Peach Plants (Prunus Persica Batsch); International Society for Horticultural Science (ISHS): Leuven, Belgium, 1983; pp. 185–192. [Google Scholar]
- Gambino, G.; Perrone, I.; Gribaudo, I. A rapid and effective method for rna extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 2008, 19, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Czotter, N.; Molnár, J.; Pesti, R.; Demián, E.; Baráth, D.; Varga, T.; Várallyay, É. Use of sirnas for diagnosis of viruses associated to woody plants in nurseries and stock collections. Methods Mol. Biol. 2018, 1746, 115–130. [Google Scholar] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. Mega11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evol. Int. J. Org. Evol. 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Olmos, A.; Boonham, N.; Candresse, T.; Gentit, P.; Giovani, B.; Kutnjak, D.; Liefting, L.; Maree, H.J.; Minafra, A.; Moreira, A.; et al. High-throughput sequencing technologies for plant pest diagnosis: Challenges and opportunities. EPPO Bull. 2018, 48, 219–224. [Google Scholar] [CrossRef] [Green Version]
- Massart, S.; Candresse, T.; Gil, J.; Lacomme, C.; Predajna, L.; Ravnikar, M.; Reynard, J.-S.; Rumbou, A.; Saldarelli, P.; Škorić, D.; et al. A framework for the evaluation of biosecurity, commercial, regulatory, and scientific impacts of plant viruses and viroids identified by ngs technologies. Front. Microbiol. 2017, 8, 45. [Google Scholar] [CrossRef] [Green Version]
- Rott, M.; Xiang, Y.; Boyes, I.; Belton, M.; Saeed, H.; Kesanakurti, P.; Hayes, S.; Lawrence, T.; Birch, C.; Bhagwat, B.; et al. Application of next generation sequencing for diagnostic testing of tree fruit viruses and viroids. Plant Dis. 2017, 101, 1489–1499. [Google Scholar] [CrossRef] [Green Version]


| Library code | Cultivar | Isolator House/Open Field Stock Nursery | Number of Tested Cultivars | Number of Tested Trees |
|---|---|---|---|---|
| 1_Sc_ih | Springcrest | isolator house | 1 | 4 |
| 2_Sc_sn | Springcrest | stock nursery/open field | 1 | 2 |
| 3_Ch_ih | Cresthaven | isolator house | 1 | 4 |
| 4_Ch_sn | Cresthaven | stock nursery/open field | 1 | 2 |
| 5_Peach_ih | 1_Flavortop | isolator house | 10 | 10 |
| 2_Nektár H | ||||
| 3_Venus | ||||
| 4_Incrocio Pieri | ||||
| 5_Cresthaven | ||||
| 6_Redhaven | ||||
| 7_Champion | ||||
| 8_Suncrest | ||||
| 9_Aranycsillag | ||||
| 10_Apolka |
| Library Code | Bioinformatics Analysis | Viruses | Viroid | |||
|---|---|---|---|---|---|---|
| PPV | NSPaV | PaLV | ChaLV | PLMVd | ||
| 1_Sc_ih | n of virus specific contigs | 94 | 2 | 17 | 0 | 0 |
| n of non-redundant reads | 3867 | 1353 | 2368 | 677 | 17 | |
| RPM | 4908 | 462 | 1296 | 197 | 3 | |
| % coverage | 68.3 | 89.7 | 93.9 | 48.3 | 51.6 | |
| 2_Sc_sn | n of virus specific contigs | 1 | 0 | 13 | 1 | 0 |
| n of non-redundant reads | 637 | 166 | 2547 | 714 | 6 | |
| RPM | 122 | 46 | 1759 | 312 | 1 | |
| % coverage | 52.0 | 32.4 | 94.3 | 51.2 | 291 | |
| 3_Ch_ih | n of virus specific contigs | 0 | 0 | 0 | 0 | 14 |
| n of non-redundant reads | 102 | 56 | 1012 | 244 | 2817 | |
| RPM | 34 | 9 | 307 | 47 | 43,915 | |
| % coverage | 14.2 | 13.5 | 76.9 | 29.4 | 99.1 | |
| 4_Ch_sn | n of virus specific contigs | 0 | 0 | 0 | 0 | 20 |
| n of non-redundant reads | 143 | 1291 | 95 | 66 | 3586 | |
| RPM | 33 | 350 | 16 | 14 | 63,289 | |
| % coverage | 17.1 | 81.5 | 16.5 | 12.1 | 100 | |
| 5_peach_ih | n of virus specific contigs | 0 | 16 | 41 | 1 | 89 |
| n of non-redundant reads | 509 | 1627 | 3076 | 848 | 5462 | |
| RPM | 79 | 239 | 670 | 125 | 38,197 | |
| % coverage | 43.9 | 93.6 | 98.5 | 61.1 | 100 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barath, D.; Jaksa-Czotter, N.; Varga, T.; Varallyay, E. Viromes of Hungarian Peach Trees Identified by High-Throughput Sequencing of Small RNAs. Plants 2022, 11, 1591. https://doi.org/10.3390/plants11121591
Barath D, Jaksa-Czotter N, Varga T, Varallyay E. Viromes of Hungarian Peach Trees Identified by High-Throughput Sequencing of Small RNAs. Plants. 2022; 11(12):1591. https://doi.org/10.3390/plants11121591
Chicago/Turabian StyleBarath, Daniel, Nikoletta Jaksa-Czotter, Tunde Varga, and Eva Varallyay. 2022. "Viromes of Hungarian Peach Trees Identified by High-Throughput Sequencing of Small RNAs" Plants 11, no. 12: 1591. https://doi.org/10.3390/plants11121591
APA StyleBarath, D., Jaksa-Czotter, N., Varga, T., & Varallyay, E. (2022). Viromes of Hungarian Peach Trees Identified by High-Throughput Sequencing of Small RNAs. Plants, 11(12), 1591. https://doi.org/10.3390/plants11121591








