Salt Priming as a Smart Approach to Mitigate Salt Stress in Faba Bean (Vicia faba L.)
Abstract
:1. Introduction
2. Results
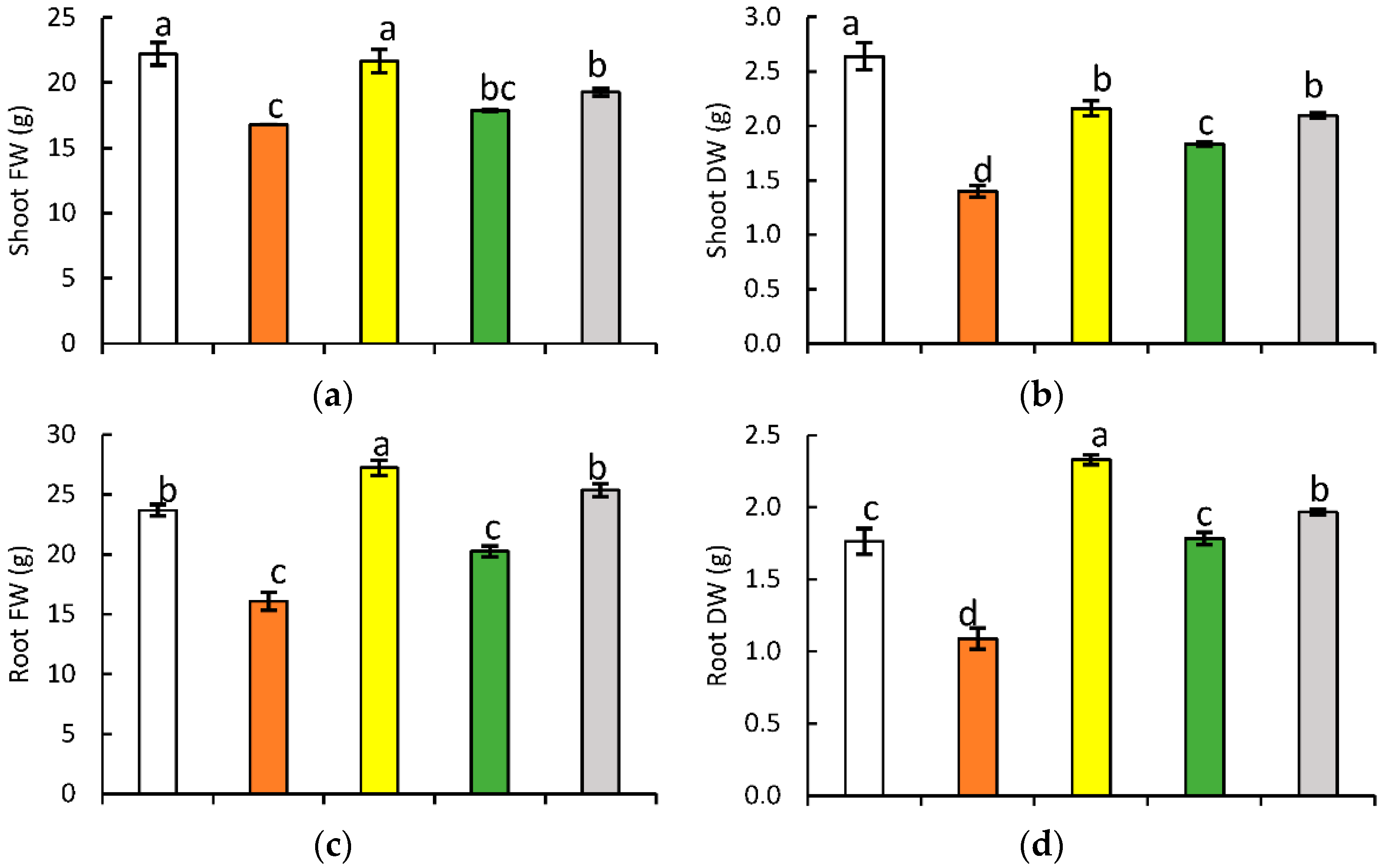
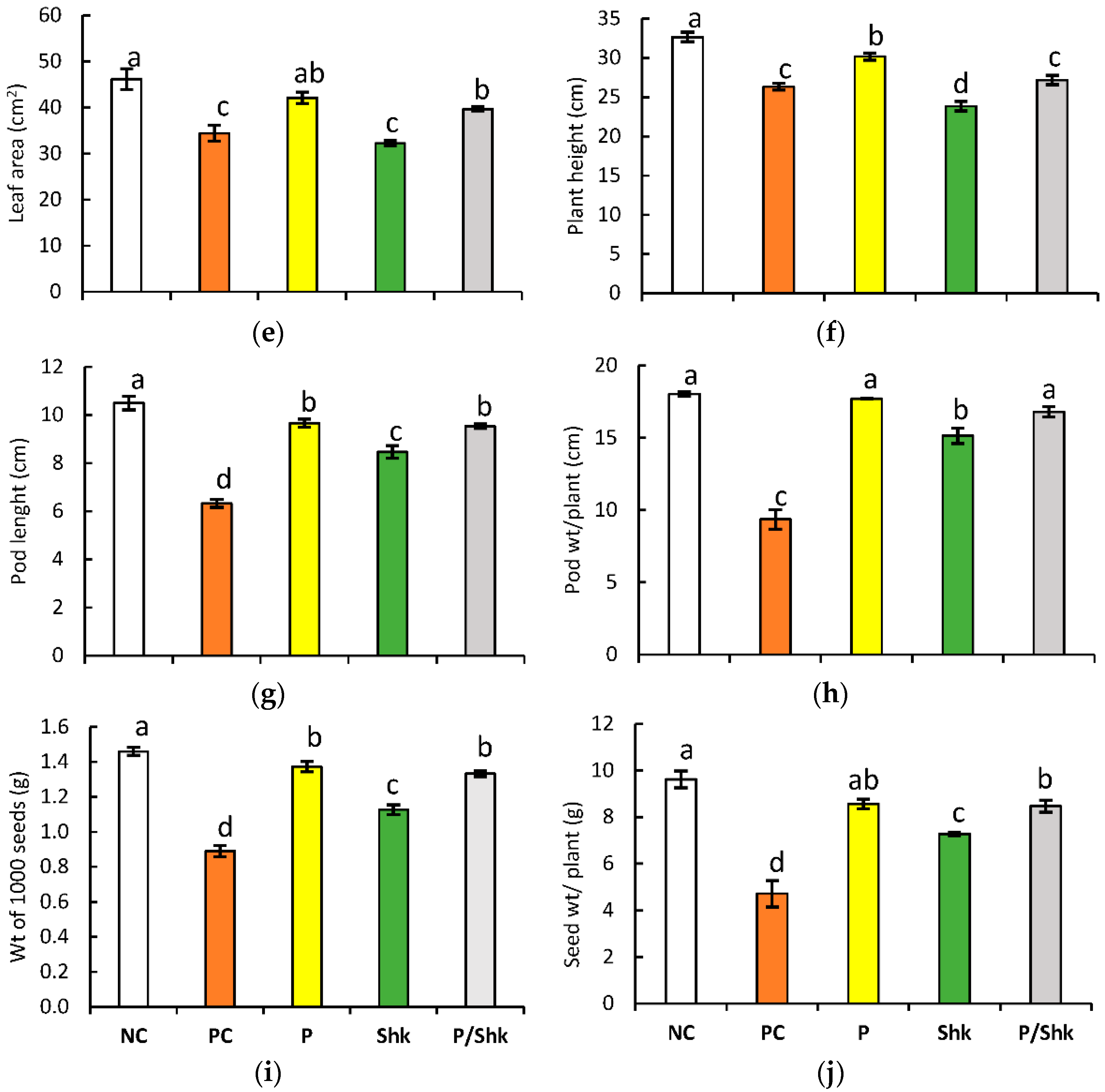
2.1. Plant Growth and Yield
2.2. Chlorophyll and Carotenoids
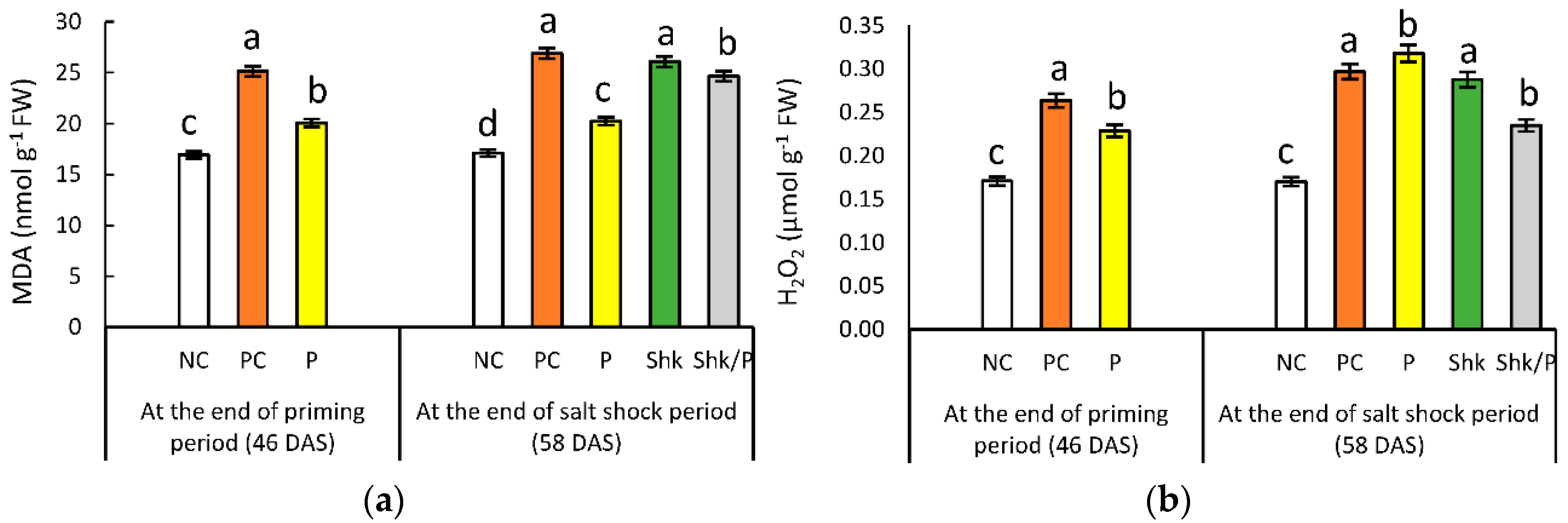
2.3. Lipid Peroxidation
2.4. Hydrogen Peroxide Content
2.5. Membrane Stability Index (MSI)
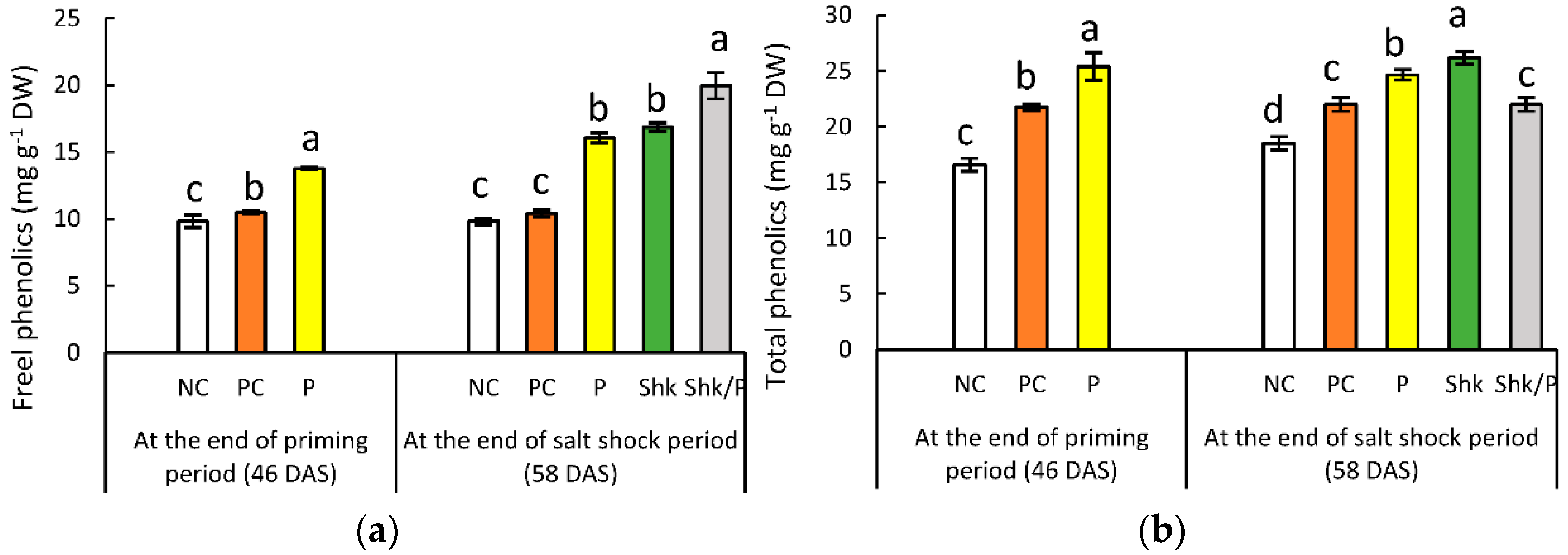
2.6. Phenolic Content
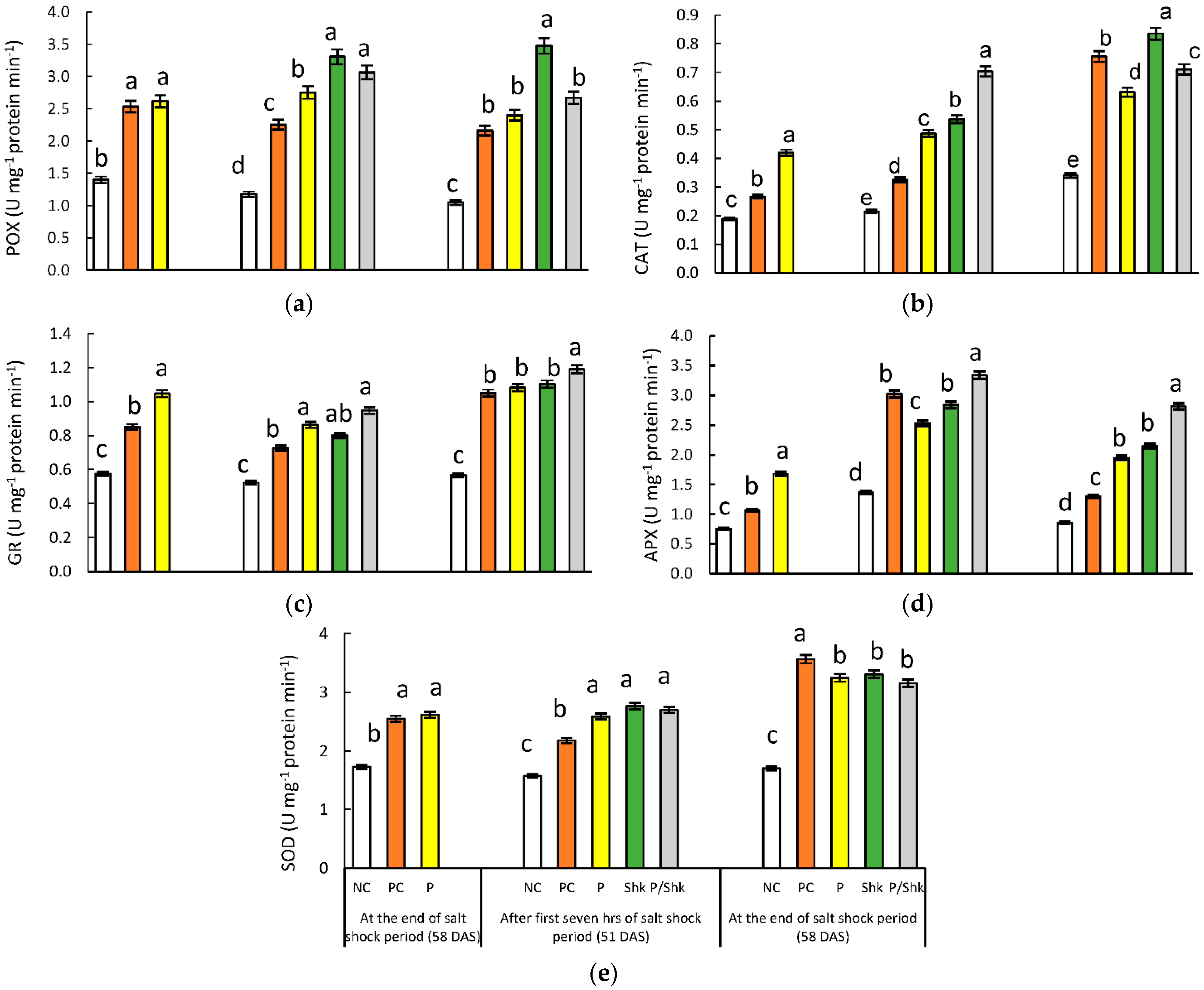
2.7. Enzymatic Antioxidant Activities
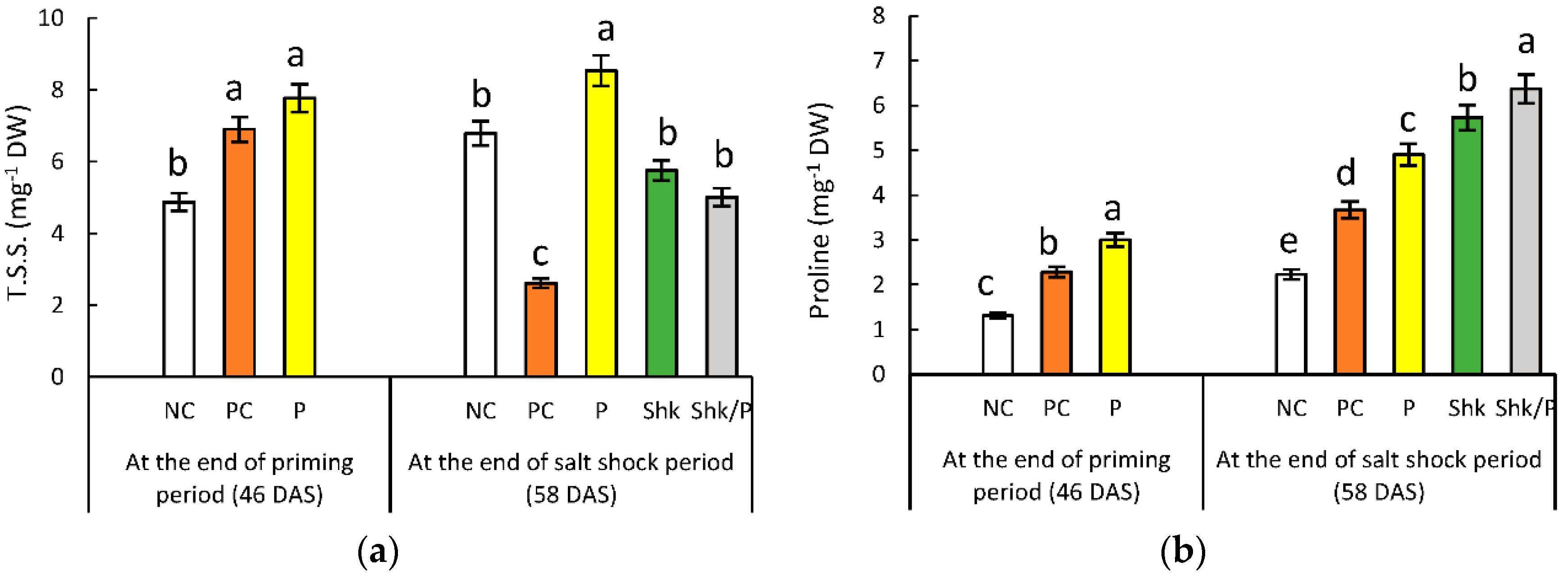
2.8. Total Soluble Sugars
2.9. Free Proline Content
2.10. Expression of Glutathione Metabolism Genes
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
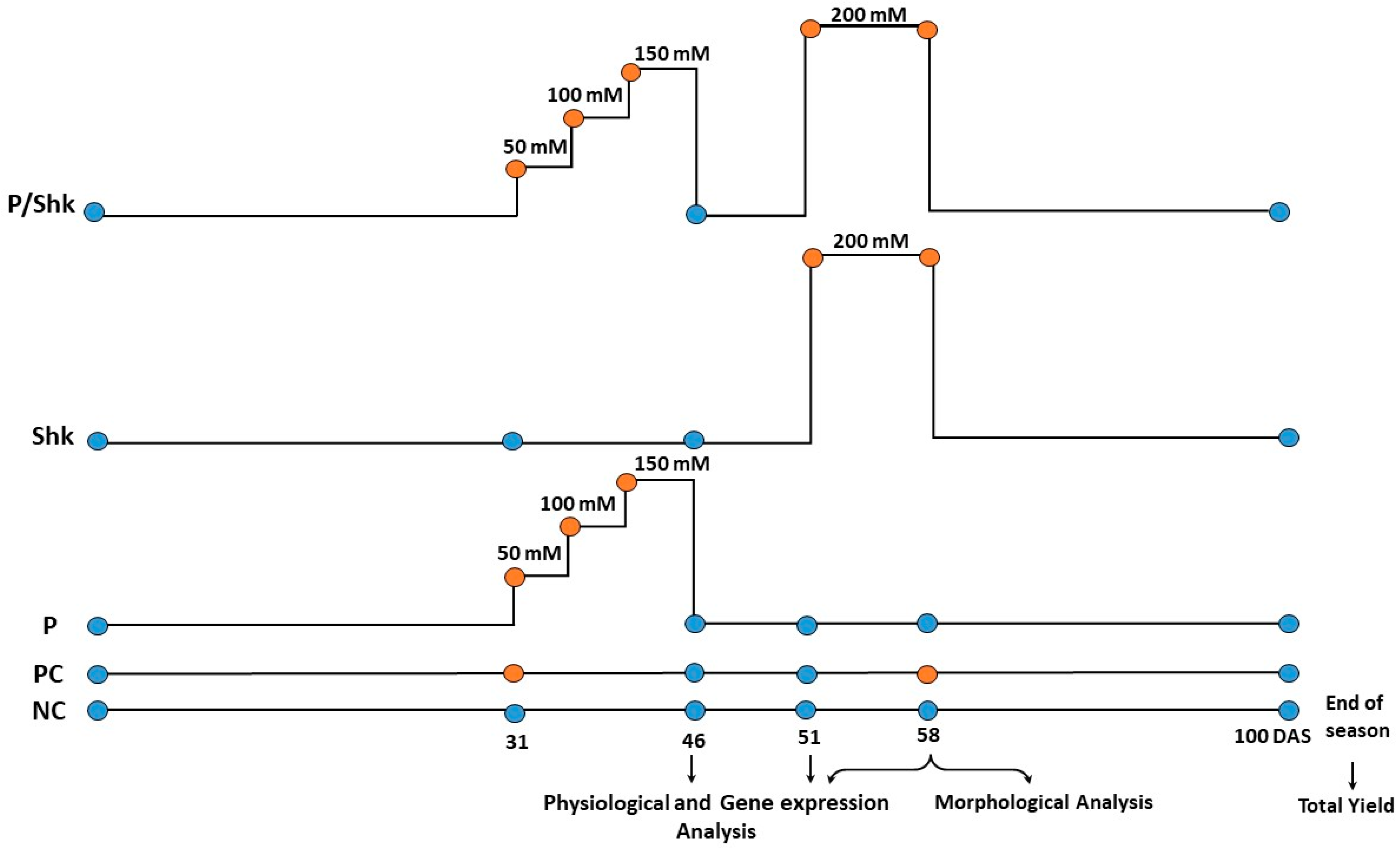
4.2. Salt Treatment
- NC: irrigated with tap water (negative control).
- PC: irrigated with 150 mM NaCl solution (positive control).
- P: salt primed for 15 days divided into three equal intervals; plants were irrigated with 50 mM NaCl solution in the first interval (5 days), then the salt concentration was increased by 50 mM in each of the next intervals. Thereafter, the salt solution was replaced with tap water till the end of the season.
- Shk: Exposed to 200 mM NaCl for 7 days starting 51 days after sowing (DAS).
- P/Shk: Primed as detailed in P, then shocked as described in Shk.
4.3. Plant Sampling
4.4. Physiological Analysis
4.4.1. Photosynthetic Pigments
4.4.2. Determination of Peroxide
4.4.3. Malondialdehyde (MDA)
4.4.4. Membrane Stability Index (MSI)
4.4.5. Phenolic Content
4.4.6. Antioxidant Enzymes Assay
4.4.7. Total Soluble Sugar (TSS) Content
4.4.8. Proline Content
4.5. Gene Expression Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mateos-Aparicio, I.; Redondo-Cuenca, A.; Villanueva-Suárez, M.-J.; Zapata-Revilla, M.-A.; Tenorio-Sanz, M.-D. Pea Pod, Broad Bean Pod and Okara, Potential Sources of Functional Compounds. LWT Food Sci. Technol. 2010, 43, 1467–1470. [Google Scholar] [CrossRef]
- Abdelkader, A.A.; Khalil, M.S.; Mohamed, M.S.M. Simultaneous biodegradation of λ-cyhalothrin pesticide and Vicia faba growth promotion under greenhouse conditions. AMB Expr. 2022, 12, 1–17. [Google Scholar] [CrossRef]
- Ofuya, Z.; Akhidue, V. The Role of Pulses in Human Nutrition: A Review. J. Appl. Sci. Environ. Manag. 2005, 9, 99–104. [Google Scholar] [CrossRef] [Green Version]
- Dhull, S.B.; Kidwai, M.K.; Noor, R.; Chawla, P.; Rose, P.K. A review of nutritional profile and processing of faba bean (Vicia faba L.). Legume Sci. 2021, e129. [Google Scholar] [CrossRef]
- Ramya, K.B.; Thaakur, S. Herbs Containing L-Dopa: An Update. Anc. Sci. Life 2007, 27, 50. [Google Scholar] [PubMed]
- Barton, L.; Thamo, T.; Engelbrecht, D.; Biswas, W.K. Does Growing Grain Legumes or Applying Lime Cost Effectively Lower Greenhouse Gas Emissions from Wheat Production in a Semi-Arid Climate? J. Clean. Prod. 2014, 83, 194–203. [Google Scholar] [CrossRef] [Green Version]
- El-Mogy, M.M.; Garchery, C.; Stevens, R. Irrigation with Salt Water Affects Growth, Yield, Fruit Quality, Storability and Marker-Gene Expression in Cherry Tomato. Acta Agric. Scand. Sect. B Soil Plant Sci. 2018, 68, 727–737. [Google Scholar] [CrossRef]
- Machado, R.M.A.; Serralheiro, R.P. Soil Salinity: Effect on Vegetable Crop Growth. Management Practices to Prevent and Mitigate Soil Salinization. Horticulturae 2017, 3, 30. [Google Scholar] [CrossRef] [Green Version]
- Lhissoui, R.; El Harti, A.; Chokmani, K. Mapping Soil Salinity in Irrigated Land Using Optical Remote Sensing Data. Eurasian J. Soil Sci. 2014, 3, 82–88. [Google Scholar] [CrossRef] [Green Version]
- Scudiero, E.; Skaggs, T.H.; Corwin, D.L. Comparative Regional-Scale Soil Salinity Assessment with near-Ground Apparent Electrical Conductivity and Remote Sensing Canopy Reflectance. Ecol. Indic. 2016, 70, 276–284. [Google Scholar] [CrossRef] [Green Version]
- Alzahrani, O.; Abouseadaa, H.; Abdelmoneim, T.K.; Alshehri, M.A.; Mohamed, E.-M.; El-Beltagi, H.S.; Atia, M.A. Agronomical, Physiological and Molecular Evaluation Reveals Superior Salt-Tolerance in Bread Wheat through Salt-Induced Priming Approach. Not. Bot. Horti Agrobot. Cluj Napoca 2021, 49, 12310. [Google Scholar] [CrossRef]
- Abdeldym, E.A.; El-Mogy, M.M.; Abdellateaf, H.R.; Atia, M.A. Genetic Characterization, Agro-Morphological and Physiological Evaluation of Grafted Tomato under Salinity Stress Conditions. Agronomy 2020, 10, 1948. [Google Scholar] [CrossRef]
- Abdelgawad, K.F.; El-Mogy, M.M.; Mohamed, M.I.A.; Garchery, C.; Stevens, R.G. Increasing Ascorbic Acid Content and Salinity Tolerance of Cherry Tomato Plants by Suppressed Expression of the Ascorbate Oxidase Gene. Agronomy 2019, 9, 51. [Google Scholar] [CrossRef] [Green Version]
- Almeida, D.M.; Oliveira, M.M.; Saibo, N.J. Regulation of Na+ and K+ Homeostasis in Plants: Towards Improved Salt Stress Tolerance in Crop Plants. Genet. Mol. Biol. 2017, 40, 326–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, N.; Bano, A.; Babar, M. The Stimulatory Effects of Plant Growth Promoting Rhizobacteria and Plant Growth Regulators on Wheat Physiology Grown in Sandy Soil. Arch. Microbiol. 2019, 201, 769–785. [Google Scholar] [CrossRef] [PubMed]
- Julkowska, M.M.; Testerink, C. Tuning Plant Signaling and Growth to Survive Salt. Trends Plant Sci. 2015, 20, 586–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Annunziata, M.G.; Ciarmiello, L.F.; Woodrow, P.; Maximova, E.; Fuggi, A.; Carillo, P. Durum Wheat Roots Adapt to Salinity Remodeling the Cellular Content of Nitrogen Metabolites and Sucrose. Front. Plant Sci. 2017, 7, 2035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, F.; Kamal, A.; Singh, A.; Ashfaque, F.; Alamri, S.; Siddiqui, M.H. Salicylic Acid Modulates Antioxidant System, Defense Metabolites, and Expression of Salt Transporter Genes in Pisum sativum under Salinity Stress. J. Plant Growth Regul. 2020, 1–14. [Google Scholar] [CrossRef]
- Reddy, I.N.B.L.; Kim, S.-M.; Kim, B.-K.; Yoon, I.-S.; Kwon, T.-R. Identification of Rice Accessions Associated with K+/Na+ Ratio and Salt Tolerance Based on Physiological and Molecular Responses. Rice Sci. 2017, 24, 360–364. [Google Scholar] [CrossRef]
- Mohamed, A.K.S.; Qayyum, M.F.; Abdel-Hadi, A.M.; Rehman, R.A.; Ali, S.; Rizwan, M. Interactive Effect of Salinity and Silver Nanoparticles on Photosynthetic and Biochemical Parameters of Wheat. Arch. Agron. Soil Sci. 2017, 63, 1736–1747. [Google Scholar] [CrossRef]
- Gratão, P.L.; Polle, A.; Lea, P.J.; Azevedo, R.A. Making the Life of Heavy Metal-Stressed Plants a Little Easier. Funct. Plant Biol. 2005, 32, 481–494. [Google Scholar] [CrossRef] [PubMed]
- Abouzari, A.; Fakheri, B.A. Reactive Oxygen Species: Generation, Oxidative Damage, and Signal Transduction. Int. J. Life Sci. 2015, 9, 3–17. [Google Scholar] [CrossRef] [Green Version]
- Waszczak, C.; Carmody, M.; Kangasjärvi, J. Reactive Oxygen Species in Plant Signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bela, K.; Horváth, E.; Gallé, Á.; Szabados, L.; Tari, I.; Csiszár, J. Plant Glutathione Peroxidases: Emerging Role of the Antioxidant Enzymes in Plant Development and Stress Responses. J. Plant Physiol. 2015, 176, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, P.; Jaleel, C.A.; Salem, M.A.; Nabi, G.; Sharma, S. Roles of Enzymatic and Nonenzymatic Antioxidants in Plants during Abiotic Stress. Crit. Rev. Biotechnol. 2010, 30, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Selmar, D.; Kleinwächter, M. Stress Enhances the Synthesis of Secondary Plant Products: The Impact of Stress-Related over-Reduction on the Accumulation of Natural Products. Plant Cell Physiol. 2013, 54, 817–826. [Google Scholar] [CrossRef] [PubMed]
- Jamshidi, M.; Ghanati, F. Taxanes Content and Cytotoxicity of Hazel Cells Extract after Elicitation with Silver Nanoparticles. Plant Physiol. Biochem. 2017, 110, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Ascorbate and Glutathione: The Heart of the Redox Hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasanuzzaman, M.; Bhuyan, M.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive Oxygen Species and Antioxidant Defense in Plants under Abiotic Stress: Revisiting the Crucial Role of a Universal Defense Regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef] [PubMed]
- Dorion, S.; Ouellet, J.C.; Rivoal, J. Glutathione Metabolism in Plants under Stress: Beyond Reactive Oxygen Species Detoxification. Metabolites 2021, 11, 641. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, L.; Chen, Y.; Shen, H.; Gong, Y.; Limera, C.; Liu, L. Transcriptome Profiling of Radish (Raphanus Sativus, L.) Root and Identification of Genes Involved in Response to Lead (Pb) Stress with next Generation Sequencing. PLoS ONE 2013, 8, e66539. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Chauhan, N.; Singh, S. Understanding the Cross-Talk of Redox Metabolism and Fe-s Cluster Biogenesis in Leishmania through Systems Biology Approach. Front. Cell. Infect. Microbiol. 2019, 9, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanou, G.; Fotopoulos, V.; Molassiotis, A. Priming against Environmental Challenges and Proteomics in Plants: Update and Agricultural Perspectives. Front. Plant Sci. 2012, 3, 216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, R.; Puthur, J.T. Seed Priming as a Cost Effective Technique for Developing Plants with Cross Tolerance to Salinity Stress. Plant Physiol. Biochem. 2021, 162, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Méndez, A.A.; Pena, L.B.; Benavides, M.P.; Gallego, S.M. Priming with NO Controls Redox State and Prevents Cadmium-Induced General up-Regulation of Methionine Sulfoxide Reductase Gene Family in Arabidopsis. Biochimie 2016, 131, 128–136. [Google Scholar] [CrossRef]
- Wiszniewska, A.; Muszyńska, E.; Kołton, A.; Kamińska, I.; Hanus-Fajerska, E. In Vitro Acclimation to Prolonged Metallic Stress Is Associated with Modulation of Antioxidant Responses in a Woody Shrub Daphne Jasminea. Plant Cell Tissue Organ Cult. 2019, 139, 339–357. [Google Scholar] [CrossRef] [Green Version]
- Paparella, S.; Araújo, S.; Rossi, G.; Wijayasinghe, M.; Carbonera, D.; Balestrazzi, A. Seed Priming: State of the Art and New Perspectives. Plant Cell Rep. 2015, 34, 1281–1293. [Google Scholar] [CrossRef]
- Filippou, P.; Tanou, G.; Molassiotis, A.; Fotopoulos, V. Plant Acclimation to Environmental Stress Using Priming Agents. In Plant Acclimation to Environmental Stress; Springer: New York, NY, USA, 2013; pp. 1–27. [Google Scholar]
- Sani, E.; Herzyk, P.; Perrella, G.; Colot, V.; Amtmann, A. Hyperosmotic Priming of Arabidopsis Seedlings Establishes a Long-Term Somatic Memory Accompanied by Specific Changes of the Epigenome. Genome Biol. 2013, 14, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Saha, P.; Chatterjee, P.; Biswas, A.K. NaCl Pretreatment Alleviates Salt Stress by Enhancement of Antioxidant Defense System and Osmolyte Accumulation in Mungbean (Vigna Radiata, L. Wilczek). Indian J. Exp. Biol. 2010, 48, 593–600. [Google Scholar]
- Thomas, D.T.; Puthur, J.T. Amplification of Abiotic Stress Tolerance Potential in Rice Seedlings with a Low Dose of UV-B Seed Priming. Funct. Plant Biol. 2019, 46, 455–466. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive Oxygen Species and Antioxidant Machinery in Abiotic Stress Tolerance in Crop Plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Alnusairi, G.S.; Mazrou, Y.S.; Qari, S.H.; Elkelish, A.A.; Soliman, M.H.; Eweis, M.; Abdelaal, K.; El-Samad, G.A.; Ibrahim, M.F.; ElNahhas, N. Exogenous Nitric Oxide Reinforces Photosynthetic Efficiency, Osmolyte, Mineral Uptake, Antioxidant, Expression of Stress-Responsive Genes and Ameliorates the Effects of Salinity Stress in Wheat. Plants 2021, 10, 1693. [Google Scholar] [CrossRef] [PubMed]
- Nasrallah, A.K.; Kheder, A.A.; Kord, M.A.; Fouad, A.S.; El-Mogy, M.M.; Atia, M.A. Mitigation of Salinity Stress Effects on Broad Bean Productivity Using Calcium Phosphate Nanoparticles Application. Horticulturae 2022, 8, 75. [Google Scholar] [CrossRef]
- Frukh, A.; Siddiqi, T.O.; Khan, M.I.R.; Ahmad, A. Modulation in Growth, Biochemical Attributes and Proteome Profile of Rice Cultivars under Salt Stress. Plant Physiol. Biochem. 2020, 146, 55–70. [Google Scholar] [CrossRef] [PubMed]
- Adly, W.M.; Mazrou, Y.S.; EL-Denary, M.E.; Mohamed, M.A.; El-Salam, A.; El-Sayed, T.; Fouad, A.S. Boosting Polyamines to Enhance Shoot Regeneration in Potato (Solanum tuberosum, L.) Using AgNO3. Horticulturae 2022, 8, 113. [Google Scholar] [CrossRef]
- Fouad, A.; Hegazy, A.E.; Azab, E.; Khojah, E.; Kapiel, T. Boosting of Antioxidants and Alkaloids in Catharanthus Roseus Suspension Cultures Using Silver Nanoparticles with Expression of CrMPK3 and STR Genes. Plants 2021, 10, 2202. [Google Scholar] [CrossRef]
- Hafez, R.; Fouad, A. Mitigation of Genotoxic and Cytotoxic Effects of Silver Nanoparticles on Onion Root Tips Using Some Antioxidant Scavengers. Egypt. J. Bot. 2020, 60, 133–145. [Google Scholar] [CrossRef]
- Taffouo, V.; Wamba, O.; Youmbi, E.; Nono, G.; Akoa, A. Growth, Yield, Water Status and Ionic Distribution Response of Three Bambara Groundnut (Vigna subterranea (L.) Verdc.) Landraces Grown under Saline Conditions. Int. J. Bot. 2010, 6, 53–58. [Google Scholar] [CrossRef] [Green Version]
- Parida, A.K.; Das, A.B. Salt Tolerance and Salinity Effects on Plants: A Review. Ecotoxicol. Environ. Saf. 2005, 60, 324–349. [Google Scholar] [CrossRef]
- Soussi, M.; Ocana, A.; Lluch, C. Effects of Salt Stress on Growth, Photosynthesis and Nitrogen Fixation in Chick-Pea (Cicer arietinum L.). J. Exp. Bot. 1998, 49, 1329–1337. [Google Scholar] [CrossRef]
- Rady, M.M. Effect of 24-Epibrassinolide on Growth, Yield, Antioxidant System and Cadmium Content of Bean (Phaseolus vulgaris L.) Plants under Salinity and Cadmium Stress. Sci. Hortic. 2011, 129, 232–237. [Google Scholar] [CrossRef]
- Mohammadi, F.; Kavousi, H.R.; Mansouri, M. Effects of Salt Stress on Physio-Biochemical Characters and Gene Expressions in Halophyte Grass Leptochloa fusca (L.) Kunth. Acta Physiol. Plant. 2019, 41, 1–10. [Google Scholar] [CrossRef]
- Wahid, A.; Ghazanfar, A. Possible Involvement of Some Secondary Metabolites in Salt Tolerance of Sugarcane. J. Plant Physiol. 2006, 163, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Stefanov, M.A.; Rashkov, G.D.; Yotsova, E.K.; Borisova, P.B.; Dobrikova, A.G.; Apostolova, E.L. Different Sensitivity Levels of the Photosynthetic Apparatus in Zea mays, L. and Sorghum bicolor, L. under Salt Stress. Plants 2021, 10, 1469. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Xu, H.; Cao, W.; Chen, X. Salt Priming Improved Salt Tolerance in Sweet Sorghum by Enhancing Osmotic Resistance and Reducing Root Na+ Uptake. Acta Physiol. Plant. 2015, 37, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Maswada, H.F.; Djanaguiraman, M.; Prasad, P. Response of Photosynthetic Performance, Water Relations and Osmotic Adjustment to Salinity Acclimation in Two Wheat Cultivars. Acta Physiol. Plant. 2018, 40, 1–15. [Google Scholar] [CrossRef]
- He, J.; Ng, O.W.J.; Qin, L. Salinity and Salt-Priming Impact on Growth, Photosynthetic Performance, and Nutritional Quality of Edible Mesembryanthemum crystallinum L. Plants 2022, 11, 332. [Google Scholar] [CrossRef]
- Liebthal, M.; Maynard, D.; Dietz, K.-J. Peroxiredoxins and Redox Signaling in Plants. Antioxid. Redox Signal. 2018, 28, 609–624. [Google Scholar] [CrossRef] [Green Version]
- Hasanuzzaman, M.; Nahar, K.; Anee, T.I.; Fujita, M. Glutathione in Plants: Biosynthesis and Physiological Role in Environmental Stress Tolerance. Physiol. Mol. Biol. Plants 2017, 23, 249–268. [Google Scholar] [CrossRef]
- Khan, I.; Raza, M.A.; Awan, S.A.; Shah, G.A.; Rizwan, M.; Ali, B.; Tariq, R.; Hassan, M.J.; Alyemeni, M.N.; Brestic, M.; et al. Amelioration of Salt Induced Toxicity in Pearl Millet by Seed Priming with Silver Nanoparticles (AgNPs): The Oxidative Damage, Antioxidant Enzymes and Ions Uptake Are Major Determinants of Salt Tolerant Capacity. Plant Physiol. Biochem. 2020, 156, 221–232. [Google Scholar] [CrossRef]
- Ahmad, Z.; Waraich, E.A.; Akhtar, S.; Anjum, S.; Ahmad, T.; Mahboob, W.; Hafeez, O.B.A.; Tapera, T.; Labuschagne, M.; Rizwan, M. Physiological Responses of Wheat to Drought Stress and Its Mitigation Approaches. Acta Physiol. Plant. 2018, 40, 1–13. [Google Scholar] [CrossRef]
- ElSayed, A.I.; Rafudeen, M.S.; Gomaa, A.M.; Hasanuzzaman, M. Exogenous Melatonin Enhances the Reactive Oxygen Species Metabolism, Antioxidant Defense-Related Gene Expression, and Photosynthetic Capacity of Phaseolus vulgaris L. to Confer Salt Stress Tolerance. Physiol. Plant. 2021, 173, 1369–1381. [Google Scholar] [CrossRef] [PubMed]
- El-Mogy, M.M.; Atia, M.A.; Dhawi, F.; Fouad, A.S.; Bendary, E.S.; Khojah, E.; Samra, B.N.; Abdelgawad, K.F.; Ibrahim, M.F.; Abdeldaym, E.A. Towards Better Grafting: SCoT and CDDP Analyses for Prediction of the Tomato Rootstocks Performance under Drought Stress. Agronomy 2022, 12, 153. [Google Scholar] [CrossRef]
- Chojak-Koźniewska, J.; Linkiewicz, A.; Sowa, S.; Radzioch, M.; Kuźniak, E. Interactive Effects of Salt Stress and Pseudomonas Syringae Pv. Lachrymans Infection in Cucumber: Involvement of Antioxidant Enzymes, Abscisic Acid and Salicylic Acid. Environ. Exp. Bot. 2017, 136, 9–20. [Google Scholar] [CrossRef]
- Imran, M.; Boelt, B.; Mühling, K.-H. Zinc Seed Priming Improves Salt Resistance in Maize. J. Agron. Crop Sci. 2018, 204, 390–399. [Google Scholar] [CrossRef]
- Abdelhamid, M.T.; El-Masry, R.R.; Darwish, D.S.; Abdalla, M.M.; Oba, S.; Ragab, R.; EL Sabagh, A.; El Kholy, M.H.; Omer, E. Mechanisms of Seed Priming Involved in Salt Stress Amelioration. In Priming and Pretreatment of Seeds and Seedlings; Springer: Singapore, 2019; pp. 219–251. [Google Scholar]
- Guo, X.; Zhi, W.; Feng, Y.; Zhou, G.; Zhu, G. Seed Priming Improved Salt-Stressed Sorghum Growth by Enhancing Antioxidative Defense. PLoS ONE 2022, 17, e0263036. [Google Scholar] [CrossRef]
- Castañares, J.; Bouzo, C.A. Seed Priming Induces Biochemical Changes in Melon Plants and Increase Salt Tolerance. RIA Rev. Investig. Agropecu. 2020, 46, 208–217. [Google Scholar]
- Gondim, F.A.; Gomes-Filho, E.; Lacerda, C.F.; Prisco, J.T.; Azevedo Neto, A.D.; Marques, E.C. Pretreatment with H2O2 in Maize Seeds: Effects on Germination and Seedling Acclimation to Salt Stress. Braz. J. Plant Physiol. 2010, 22, 103–112. [Google Scholar] [CrossRef] [Green Version]
- Milić, B.L.; Djilas, S.M.; Čanadanović-Brunet, J.M. Antioxidative Activity of Phenolic Compounds on the Metal-Ion Breakdown of Lipid Peroxidation System. Food Chem. 1998, 61, 443–447. [Google Scholar] [CrossRef]
- Verstraeten, S.V.; Keen, C.L.; Schmitz, H.H.; Fraga, C.G.; Oteiza, P.I. Flavan-3-Ols and Procyanidins Protect Liposomes against Lipid Oxidation and Disruption of the Bilayer Structure. Free Radic. Biol. Med. 2003, 34, 84–92. [Google Scholar] [CrossRef]
- Arora, A.; Byrem, T.M.; Nair, M.G.; Strasburg, G.M. Modulation of Liposomal Membrane Fluidity by Flavonoids and Isoflavonoids. Arch. Biochem. Biophys. 2000, 373, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.M.; Madany, M. Coumarin Pretreatment Alleviates Salinity Stress in Wheat Seedlings. Plant Physiol. Biochem. 2015, 88, 27–35. [Google Scholar] [CrossRef] [PubMed]
- El-Badri, A.M.; Batool, M.; Mohamed, I.A.A.; Wang, Z.; Khatab, A.; Sherif, A.; Ahmad, H.; Khan, M.N.; Hassan, H.M.; Elrewainy, I.M.; et al. Antioxidative and Metabolic Contribution to Salinity Stress Responses in Two Rapeseed Cultivars during the Early Seedling Stage. Antioxidants 2021, 10, 1227. [Google Scholar] [CrossRef] [PubMed]
- Falcinelli, B.; Sileoni, V.; Marconi, O.; Perretti, G.; Quinet, M.; Lutts, S.; Benincasa, P. Germination under Moderate Salinity Increases Phenolic Content and Antioxidant Activity in Rapeseed (Brassica napus var oleifera Del.) Sprouts. Molecules 2017, 22, 1377. [Google Scholar] [CrossRef] [Green Version]
- Nouman, W.; Aziz, U. Seed Priming Improves Salinity Tolerance in Calotropis Procera (Aiton) by Increasing Photosynthetic Pigments, Antioxidant Activities, and Phenolic Acids. Biologia 2022, 77, 1–18. [Google Scholar] [CrossRef]
- Ti, H.; Zhang, R.; Zhang, M.; Li, Q.; Wei, Z.; Zhang, Y.; Tang, X.; Deng, Y.; Liu, L.; Ma, Y. Dynamic Changes in the Free and Bound Phenolic Compounds and Antioxidant Activity of Brown Rice at Different Germination Stages. Food Chem. 2014, 161, 337–344. [Google Scholar] [CrossRef]
- Amini, F.; Ehsanpour, A. Expression Pattern Analysis of TomPRO2 and LaPA1 Genes in Tomato under in Vitro Salt Stress by Semi-Quantitative RT-PCR. Int. J. Plant Prod. 2009, 3, 69–76. [Google Scholar]
- Zhang, L.; Yan, S.; Zhang, S.; Yan, P.; Wang, J.; Zhang, H. Glutathione, Carbohydrate and Other Metabolites of Larix olgensis A. Henry Reponse to Polyethylene Glycol-Simulated Drought Stress. PLoS ONE 2021, 16, e0253780. [Google Scholar] [CrossRef]
- Antoniou, C.; Zarza, X.; Gohari, G.; Panahirad, S.; Filippou, P.; Tiburcio, A.F.; Fotopoulos, V. Involvement of Polyamine Metabolism in the Response of Medicago Truncatula Genotypes to Salt Stress. Plants 2021, 10, 269. [Google Scholar] [CrossRef]
- Paul, S.; Roychoudhury, A. Effect of Seed Priming with Spermine/Spermidine on Transcriptional Regulation of Stress-Responsive Genes in Salt-Stressed Seedlings of an Aromatic Rice Cultivar. Plant Gene 2017, 11, 133–142. [Google Scholar] [CrossRef]
- Tanou, G.; Ziogas, V.; Belghazi, M.; Christou, A.; Filippou, P.; Job, D.; Fotopoulos, V.; Molassiotis, A. Polyamines Reprogram Oxidative and Nitrosative Status and the Proteome of Citrus Plants Exposed to Salinity Stress. Plant Cell Environ. 2014, 37, 864–885. [Google Scholar] [CrossRef] [PubMed]
- Souza, M.O.; Pelacani, C.R.; Willems, L.A.; Castro, R.D.; Hilhorst, H.W.; Ligterink, W. Effect of Osmopriming on Germination and Initial Growth of Physalis angulata L. under Salt Stress and on Expression of Associated Genes. An. Acad. Bras. Ciências 2016, 88, 503–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubala, S.; Wojtyla, Ł.; Quinet, M.; Lechowska, K.; Lutts, S.; Garnczarska, M. Enhanced Expression of the Proline Synthesis Gene P5CSA in Relation to Seed Osmopriming Improvement of Brassica Napus Germination under Salinity Stress. J. Plant Physiol. 2015, 183, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, D.; Wang, W.; Gao, T.; Fang, X.; Gao, X.; Li, J.; Bu, H.; Mu, J. Calcium Alleviates Decreases in Photosynthesis under Salt Stress by Enhancing Antioxidant Metabolism and Adjusting Solute Accumulation in Calligonum Mongolicum. Conserv. Physiol. 2017, 5, cox060. [Google Scholar] [CrossRef] [Green Version]
- Atia, M.A.; El-Khateeb, E.A.; El-Maksoud, A.; Reem, M.; Abou-Zeid, M.A.; Salah, A.; Abdel-Hamid, A.M. Mining of leaf rust resistance genes content in Egyptian bread wheat collection. Plants 2021, 10, 1378. [Google Scholar] [CrossRef] [PubMed]
- Sagervanshi, A.; Naeem, A.; Geilfus, C.-M.; Kaiser, H.; Mühling, K.H. One-Time Abscisic Acid Priming Induces Long-Term Salinity Resistance in Vicia faba: Changes in Key Transcripts, Metabolites, and Ionic Relations. Physiol. Plant. 2021, 172, 146–161. [Google Scholar] [CrossRef]
- Qados, A.M.A. Mechanism of Nanosilicon-Mediated Alleviation of Salinity Stress in Faba Bean (Vicia faba L.) Plants. Am. J. Exp. Agric. 2015, 7, 78–95. [Google Scholar] [CrossRef]
- Hassanein, R.; Hashem, H.; Khalil, R. Stigmasterol Treatment Increases Salt Stress Tolerance of Faba Bean Plants by Enhancing Antioxidant Systems. Plant Omics 2012, 5, 476–485. [Google Scholar]
- Fadeel, A. Location and Properties of Chloroplasts and Pigment Determination in Roots. Physiol. Plant. 1962, 15, 130–146. [Google Scholar] [CrossRef]
- Sestak, Z.; Catsky, J.; Jarvis, P. (Eds.) Determination of Chlorophylls a and b. In Plant Photosynthetic Production; Dr. W. Junk Publishers: The Hague, The Netherlands, 1971; pp. 672–701. [Google Scholar]
- Junglee, S.; Urban, L.; Sallanon, H.; Lopez-Lauri, F. Optimized Assay for Hydrogen Peroxide Determination in Plant Tissue Using Potassium Iodide. Am. J. Anal. Chem. 2014, 5, 730. [Google Scholar] [CrossRef] [Green Version]
- Heath, R.L.; Packer, L. Photoperoxidation in Isolated Chloroplasts: I. Kinetics and Stoichiometry of Fatty Acid Peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]
- Sauvesty, A.; Page, F.; Huot, J. A Simple Method for Extracting Plant Phenolic Compounds. Can. J. For. Res. 1992, 22, 654–659. [Google Scholar] [CrossRef]
- Lowe, L. Total and Labile Polysaccharide Analysis of Soils. In Soil Samples and Methods of Analysis; Lewis Publishers: Boca Raton, FL, USA, 1993; pp. 373–376. [Google Scholar]
- Grace, S.C.; Logan, B.A. Acclimation of Foliar Antioxidant Systems to Growth Irradiance in Three Broad-Leaved Evergreen Species. Plant Physiol. 1996, 112, 1631–1640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagrimini, L. Plant Peroxidases: Under-and over-Expression in Transgenic Plants and Physiological Consequences. Plant Peroxidases 1980, 1990, 59–69. [Google Scholar]
- Aebi, H. [13] Catalase in Vitro. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1984; Volume 105, pp. 121–126. [Google Scholar]
- Dhindsa, R.S.; Plumb-Dhindsa, P.; Thorpe, T.A. Leaf Senescence: Correlated with Increased Levels of Membrane Permeability and Lipid Peroxidation, and Decreased Levels of Superoxide Dismutase and Catalase. J. Exp. Bot. 1981, 32, 93–101. [Google Scholar] [CrossRef]
- Nakano, Y.; Asada, K. Hydrogen Peroxide Is Scavenged by Ascorbate-Specific Peroxidase in Spinach Chloroplasts. Plant Cell Physiol. 1981, 22, 867–880. [Google Scholar]
- Carlberg, I.; Mannervik, B. Purification and Characterization of the Flavoenzyme Glutathione Reductase from Rat Liver. J. Biol. Chem. 1975, 250, 5475–5480. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein Measurement with the Folin Phenol Reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Maness, N. Extraction and Analysis of Soluble Carbohydrates. In Plant Stress Tolerance; Springer: Berlin/Heidelberg, Germany, 2010; pp. 341–370. [Google Scholar]
- Yemm, E.; Willis, A. The Estimation of Carbohydrates in Plant Extracts by Anthrone. Biochem. J. 1954, 57, 508. [Google Scholar] [CrossRef] [Green Version]
- Abdallah, I.S.; Atia, M.A.; Nasrallah, A.K.; El-Beltagi, H.S.; Kabil, F.F.; El-Mogy, M.M.; Abdeldaym, E.A. Effect of New Pre-Emergence Herbicides on Quality and Yield of Potato and Its Associated Weeds. Sustainability 2021, 13, 9796. [Google Scholar] [CrossRef]
- Shabnam, N.; Tripathi, I.; Sharmila, P.; Pardha-Saradhi, P. A Rapid, Ideal, and Eco-Friendlier Protocol for Quantifying Proline. Protoplasma 2016, 253, 1577–1582. [Google Scholar] [CrossRef] [PubMed]
- Abid, G.; Muhovski, Y.; Mingeot, D.; Saidi, M.N.; Aouida, M.; Aroua, I.; M’hamdi, M.; Barhoumi, F.; Rezgui, S.; Jebara, M. Identification and characterization of two faba bean (Vicia faba L.) WRKY transcription factors and their expression analysis during salt and drought stress. J. Agric. Sci. 2017, 155, 791–803. [Google Scholar] [CrossRef]
- Mokhtar, M.M.; Hussein, E.H.; El-Assal, S.E.-D.S.; Atia, M.A. Vf ODB: A Comprehensive Database of ESTs, EST-SSRs, MtSSRs, MicroRNA-Target Markers and Genetic Maps in Vicia faba. AoB Plants 2020, 12, plaa064. [Google Scholar] [CrossRef] [PubMed]
- Abid, G.; Ouertani, R.N.; Jebara, S.H.; Boubakri, H.; Muhovski, Y.; Ghouili, E.; Abdelkarim, S.; Chaieb, O.; Hidri, Y.; Kadri, S.; et al. Alleviation of drought stress in faba bean (Vicia faba L.) by exogenous application of β-aminobutyric acid (BABA). Physiol. Mol. Biol. Plants 2020, 26, 1173–1186. [Google Scholar] [CrossRef]




| Gene | Primer Sequence (5′–3′) | References | |
|---|---|---|---|
| Forward | Reverse | ||
| VfEFα (Housekeeping gene) | GACAACATGATTGAGAGGTCCACC | GGCTCCTTCTCAATCTCCTTACC | [108] |
| L-APX gene | ACCGTATTGTGCAGTGCTCA | TTAGCCCTCTCCTGCTGCTA | [109,110] |
| SpS gene | TTGGTAGGCACATTCGTCCC | AATGTGGCCTGGAGAAGCTC | |
| L-AP gene | CCTCAAATCGCCGTCCCTAG | TCTCCGGTGACAGTTCTTGC | |
| AP-N gene | CCGGTCACCTCCTACTGGTA | GGAAGCCTTCTGCCTCACAT | |
| RDR-M gene | ATCAGCATGCCATGGTTCCA | CCAATGCTGCGTGTGTTCAA | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nasrallah, A.K.; Atia, M.A.M.; Abd El-Maksoud, R.M.; Kord, M.A.; Fouad, A.S. Salt Priming as a Smart Approach to Mitigate Salt Stress in Faba Bean (Vicia faba L.). Plants 2022, 11, 1610. https://doi.org/10.3390/plants11121610
Nasrallah AK, Atia MAM, Abd El-Maksoud RM, Kord MA, Fouad AS. Salt Priming as a Smart Approach to Mitigate Salt Stress in Faba Bean (Vicia faba L.). Plants. 2022; 11(12):1610. https://doi.org/10.3390/plants11121610
Chicago/Turabian StyleNasrallah, Amira K., Mohamed A. M. Atia, Reem M. Abd El-Maksoud, Maimona A. Kord, and Ahmed S. Fouad. 2022. "Salt Priming as a Smart Approach to Mitigate Salt Stress in Faba Bean (Vicia faba L.)" Plants 11, no. 12: 1610. https://doi.org/10.3390/plants11121610










