A New Method for Rapid Subcellular Localization and Gene Function Analysis in Cotton Based on Barley Stripe Mosaic Virus
Abstract
:1. Introduction
2. Results
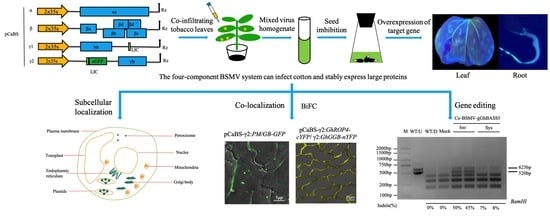
2.1. The Four-Component BSMV System Can Infect Cotton and Stably Express Large Proteins
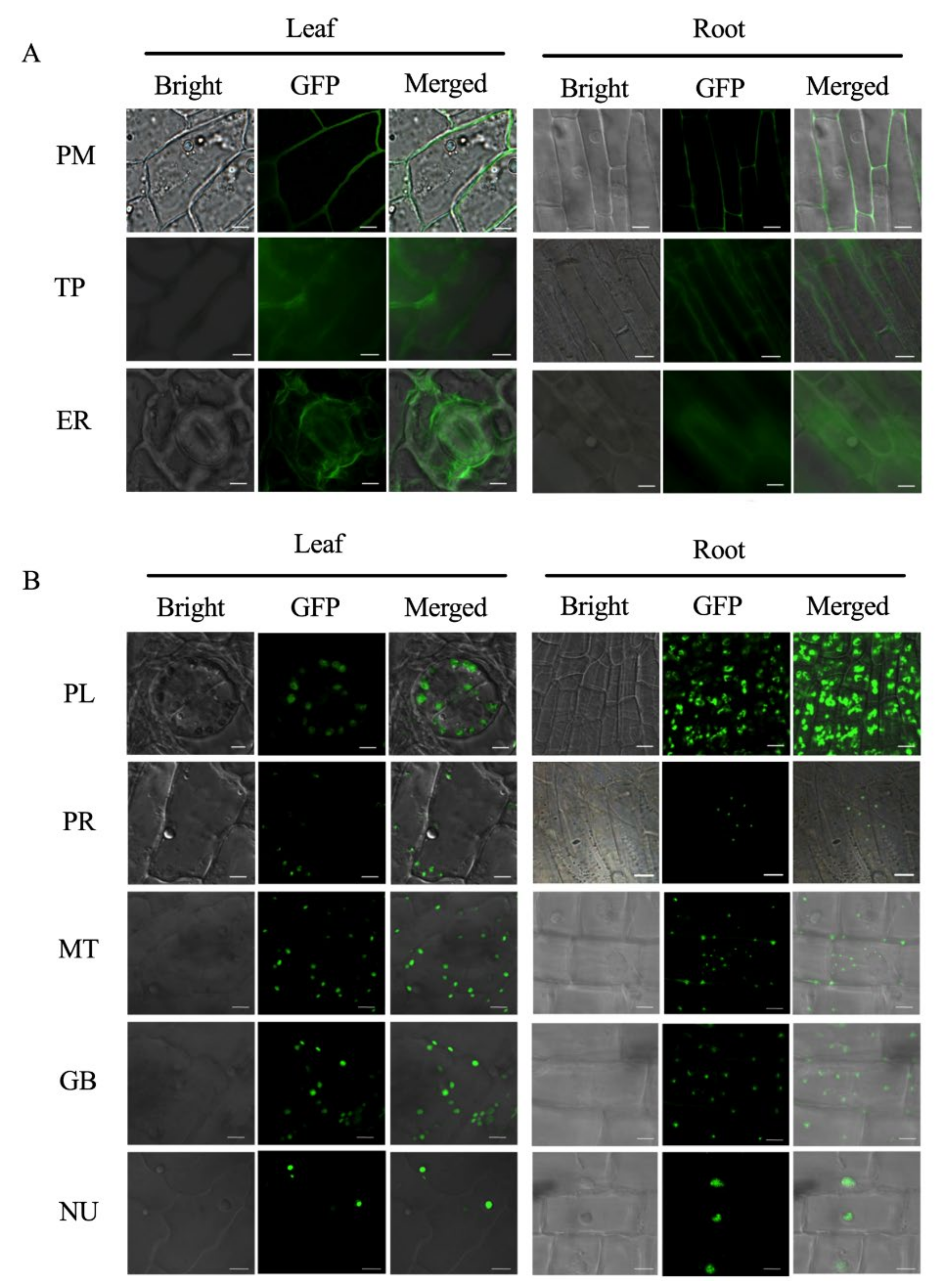
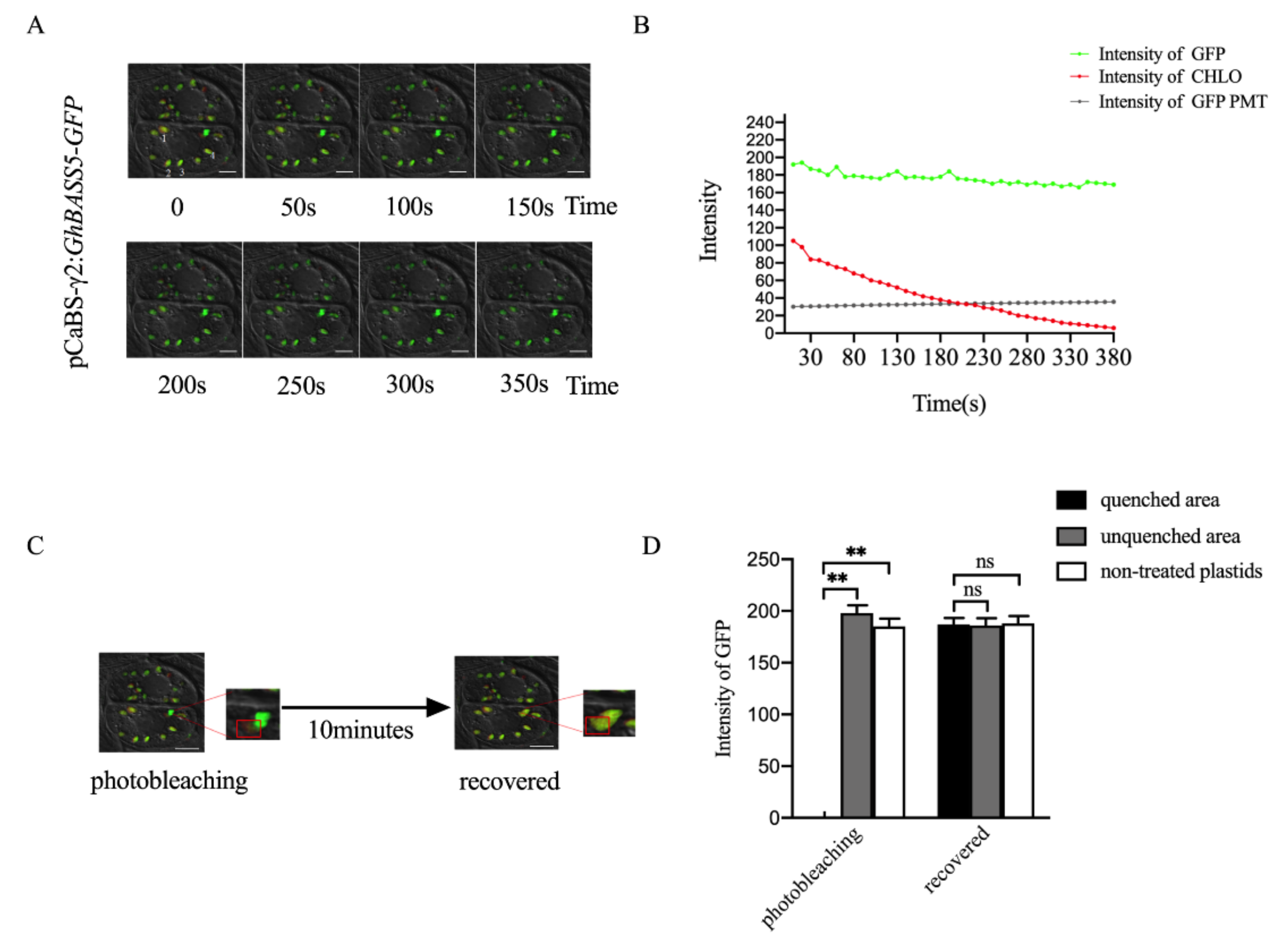
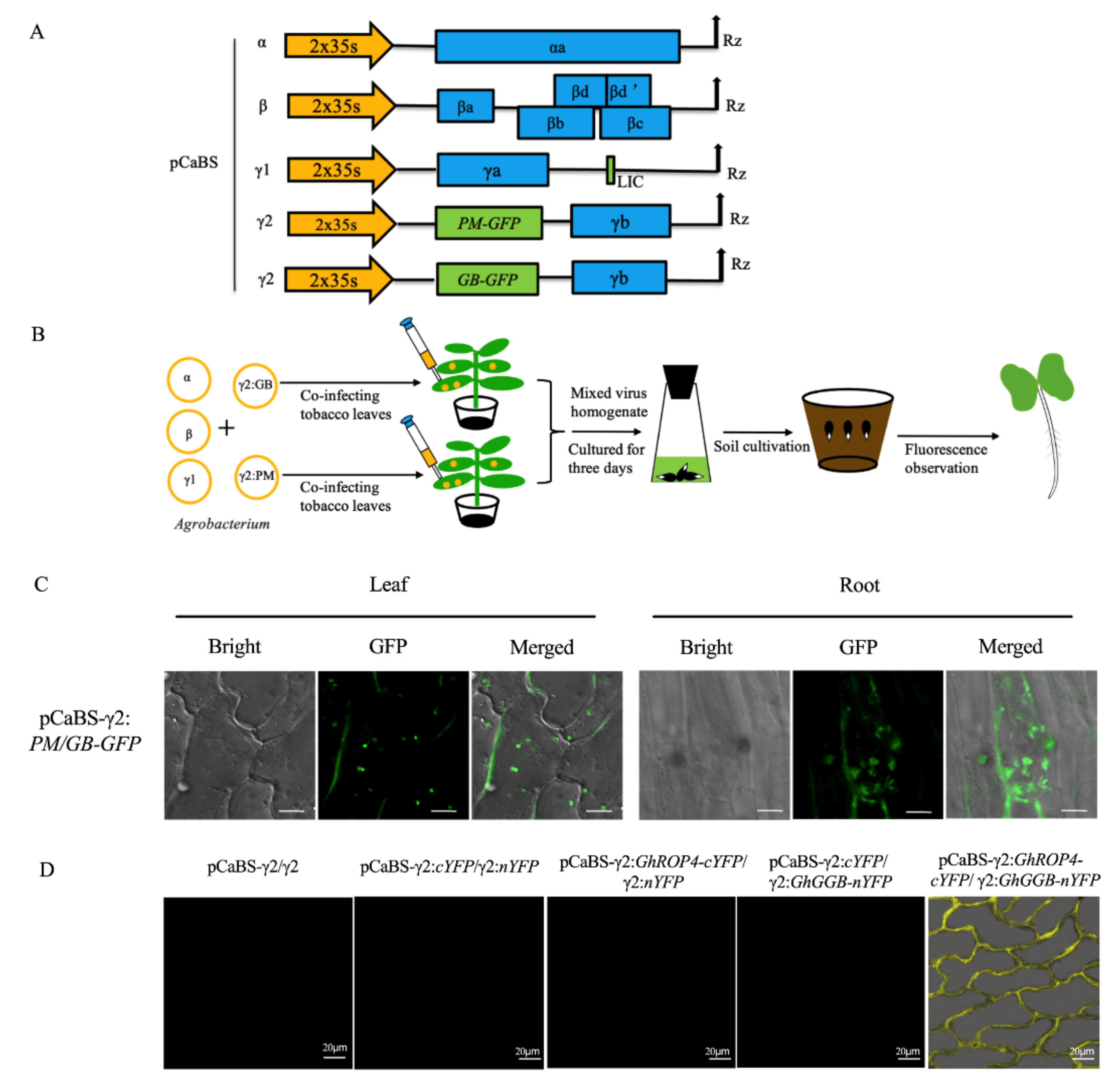
2.2. Establishing BSMV-Mediated Organelle Marker Lines in Cotton
2.3. BSMV Allows Stable Gene Expression in Cotton
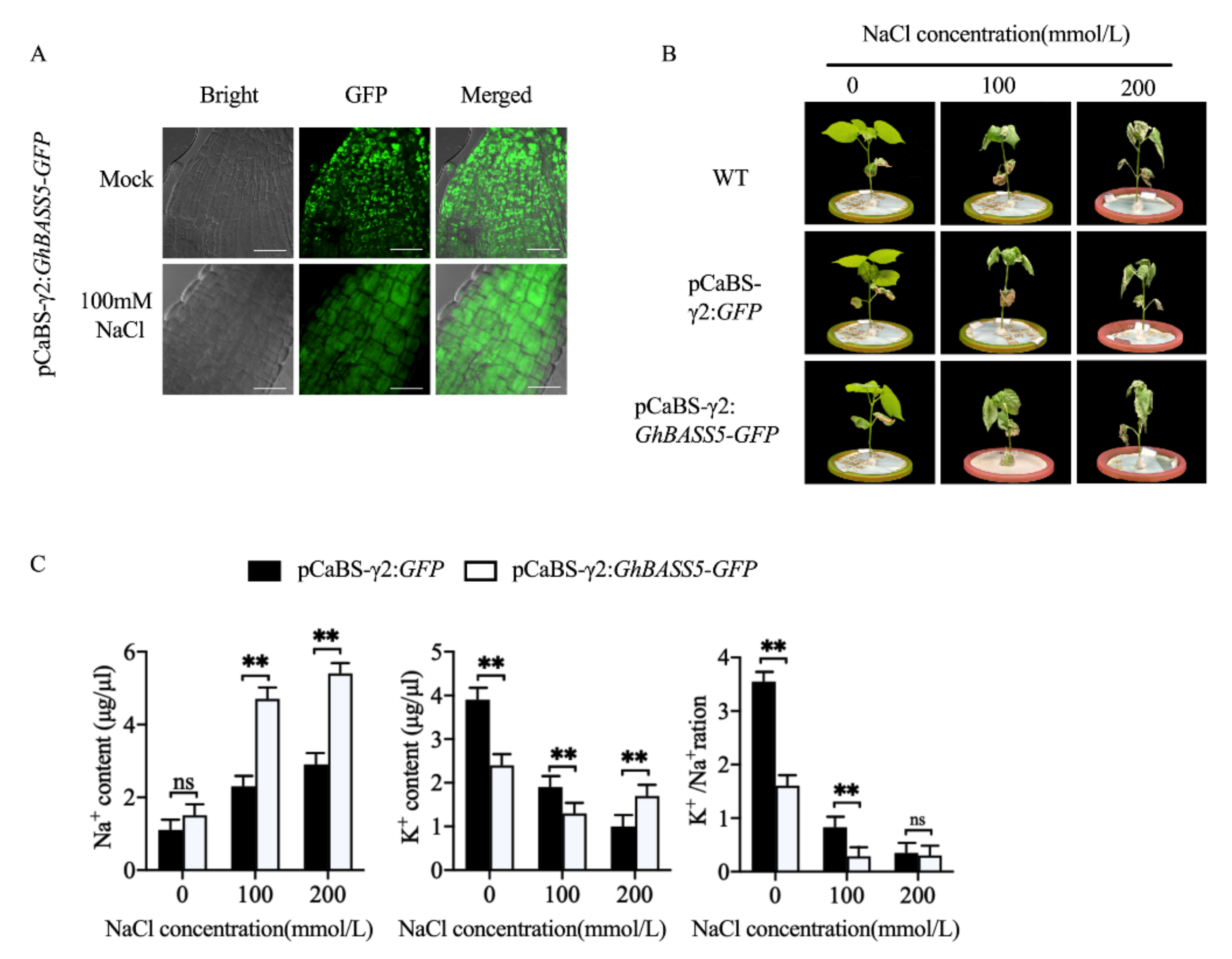
2.4. BSMV Allowed for Rapid Gene Function Analysis in Cotton
2.5. BSMV-Mediated Co-Expression of Two Proteins for Co-Localization and Interactions Studies in Cotton
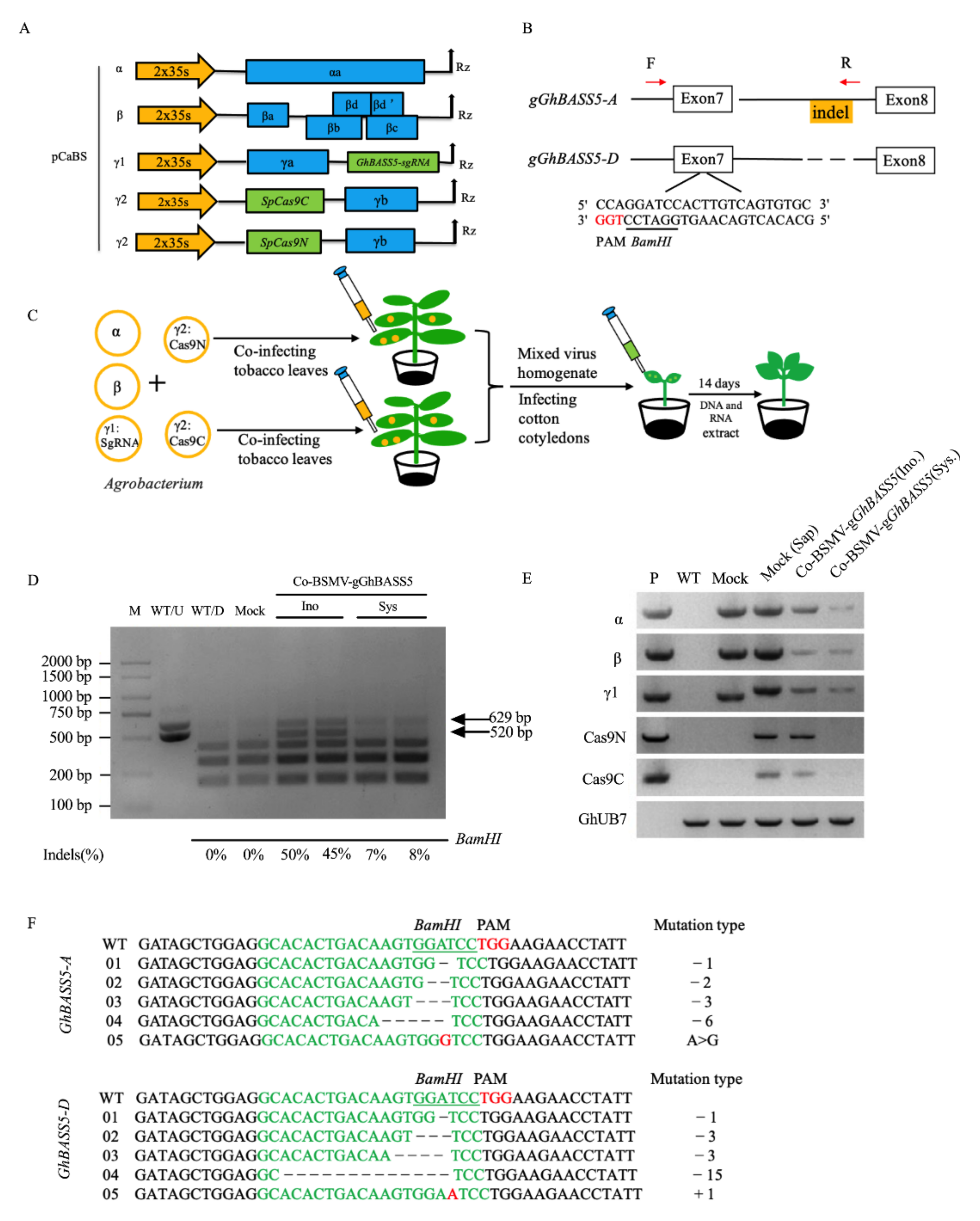
2.6. BSMV Allows the Delivery of CRISPR/Cas9 Reagents for DNA-Free Gene Editing in Cotton
3. Discussion
3.1. BSMV System Makes It Possible to Carry Out Rapid and Simple Gene Overexpression in Cotton
3.2. Advantages of Establishing Subcellular Localization Marker Lines in Cotton
3.3. The BSMV System Can Be Successfully Used for Gene Function Studies
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Plasmid Construction
4.3. Isolation and Transformation of Cotton Protoplasts
4.4. Agroinfiltration of N. benthamiana and Viral Inoculation
4.5. Quantitative RT-PCR Analysis
4.6. Fluorescence Imaging
4.7. Measurement of Na+ and K+ Content
4.8. Detection of Gene Editing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sattar, M.N.; Kvarnheden, A.; Saeed, M.; Briddon, R.W. Cotton leaf curl disease—An emerging threat to cotton production worldwide. J. Gen. Virol. 2013, 94, 695–710. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanz, S.K.; Castleden, I.; Small, I.D.; Millar, A.H. Fluorescent protein tagging as a tool to define the subcellular distribution of proteins in plants. Front. Plant Sci. 2013, 4, 214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B. Transgenic cotton: From biotransformation methods to agricultural application. Methods Mol. Biol. 2013, 958, 3–15. [Google Scholar] [PubMed]
- Sun, H.J.; Sun, X.W.; Wang, H.; Ma, X.L. Advances in salt tolerance molecular mechanism in tobacco plants. Hereditas 2020, 157, 5. [Google Scholar] [CrossRef]
- Hefferon, K. Plant virus expression vector development: New perspectives. Biomed. Res. Int. 2014, 2014, 785382. [Google Scholar] [CrossRef]
- Chapman, S.; Kavanagh, T.; Baulcombe, D. Potato virus-X as a vector for Gene-Expression in Plants. Plant J. 1992, 2, 549–557. [Google Scholar]
- Killiny, N.; Jones, S.E.; Gonzalez-Blanco, P. Silencing of delta-aminolevulinic acid dehydratase via virus induced gene silencing promotes callose deposition in plant phloem. Plant Signal Behav. 2022, 1, 2024733. [Google Scholar] [CrossRef]
- Ellison, E.E.; Nagalakshmi, U.; Gamo, M.E.; Huang, P.J.; Dinesh-Kumar, S.; Voytas, D.F. Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs. Nat. Plants 2020, 6, 620–624. [Google Scholar] [CrossRef]
- Lawrence, D.M.; Jackson, A.O. Requirements for cell-to-cell movement of Barley stripe mosaic virus in monocot and dicot hosts. Mol. Plant Pathol. 2001, 2, 65–75. [Google Scholar] [CrossRef]
- Bruun-Rasmussen, M.; Madsen, C.T.; Jessing, S.; Albrechtsen, M. Stability of Barley stripe mosaic virus—Induced gene silencing in barley. Mol. Plant-Microbe Interact. 2007, 20, 1323–1331. [Google Scholar] [CrossRef] [Green Version]
- Cheuk, A.; Houde, M. A rapid and efficient method for uniform gene expression using the barley stripe mosaic virus. Plant Methods 2017, 13, 24. [Google Scholar] [CrossRef]
- Holzberg, S.; Brosio, P.; Gross, C.; Pogue, G.P. Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 2002, 30, 315–327. [Google Scholar] [CrossRef]
- Yuan, C.; Li, C.; Yan, L.J.; Jackson, A.O.; Liu, Z.Y.; Han, C.G.; Yu, J.L.; Li, D.W. A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS ONE 2011, 6, e26468. [Google Scholar] [CrossRef]
- Lee, W.S.; Hammond-Kosack, K.E.; Kanyuka, K. Barley stripe mosaic virus-mediated tools for investigating gene function in cereal plants and their pathogens: Virus-induced gene silencing, host-mediated gene silencing, and virus-Mediated overexpression of heterologous protein. Plant Physiol. 2012, 160, 582–590. [Google Scholar] [CrossRef] [Green Version]
- Cheuk, A.; Houde, M. A new barley stripe mosaic virus allows large protein overexpression for rapid function analysis. Plant Physiol. 2018, 176, 1919–1931. [Google Scholar] [CrossRef] [Green Version]
- Li, X.J.; Wang, X.H.; Yang, Y.; Li, R.L.; He, Q.H.; Fang, X.H.; Luu, D.T.; Maurel, C.; Lin, J.X. Single-Molecule Analysis of PIP2;1 Dynamics and partitioning reveals multiple modes of Arabidopsis plasma membrane aquaporin regulation. Plant Cell 2011, 23, 3780–3797. [Google Scholar] [CrossRef] [Green Version]
- Li, D.D.; Tai, F.J.; Zhang, Z.T.; Li, Y.; Zheng, Y.; Wu, Y.F.; Li, X.B. A cotton gene encodes a tonoplast aquaporin that is involved in cell tolerance to cold stress. Genes 2009, 438, 26–32. [Google Scholar] [CrossRef]
- Tamura, T.; Kuroda, M.; Oikawa, T.; Kyozuka, J.; Terauchi, K.; Ishimaru, Y.; Abe, K.; Asakura, T. Signal peptide peptidases are expressed in the shoot apex of rice, localized to the endoplasmic reticulum. Plant Cell Rep. 2009, 28, 1615–1621. [Google Scholar] [CrossRef]
- Dangol, S.; Singh, R.; Chen, Y.F.; Jwa, N.S. Visualization of multicolored in vivo organelle markers for co-localization studies in Oryza sativa. Mol. Cells 2017, 40, 828–836. [Google Scholar]
- Teixeira, F.K.; Menezes-Benavente, L.; Galvao, V.C.; Margis, R.; Margis-Pinheiro, M. Rice ascorbate peroxidase gene family encodes functionally diverse isoforms localized in different subcellular compartments. Planta 2006, 224, 300–314. [Google Scholar] [CrossRef]
- Nakazono, M.; Tsuji, H.; Li, Y.H.; Saisho, D.; Arimura, S.; Tsutsumi, N.; Hirai, A. Expression of a gene encoding mitochondrial aldehyde dehydrogenase in rice increases under submerged conditions. Plant Physiol. 2000, 124, 587–598. [Google Scholar] [CrossRef] [Green Version]
- Saint-Jore-Dupas, C.; Nebenfuhr, A.; Boulaflous, A.; Follet-Gueye, M.L.; Plasson, C.; Hawes, C.; Driouich, A.; Faye, L.; Gomord, V. Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell 2006, 18, 3182–3200. [Google Scholar] [CrossRef] [Green Version]
- Myo, T.; Wei, F.; Zhang, H.H.; Hao, J.F.; Zhang, B.; Liu, Z.X.; Cao, G.Q.; Tian, B.M.; Shi, G.Y. Genome-wide identification of the BASS gene family in four Gossypium species and functional characterization of GhBASSs against salt stress. Sci. Rep. 2021, 11, 11342. [Google Scholar] [CrossRef]
- Myo, T.; Tian, B.; Zhang, Q.; Niu, S.; Shi, G. Ectopic overexpression of a cotton plastidial Na+ transporter GhBASS5 impairs salt tolerance in Arabidopsis via increasing Na+ loading and accumulation. Planta 2020, 252, 41. [Google Scholar] [CrossRef]
- Li, W.; Wugalihan, A.; Dai, P.; Liu, C.; Yao, Z.; Guo, W.; Liu, X. Interaction between cotton small GTP binding protein GhROP4 and protein geranyl acyltransferase GhGGB. Mol. Plant Breed. 2020, 18, 8126–8130. (In Chinese) [Google Scholar]
- Zetsche, B.; Volz, S.E.; Zhang, F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 2015, 33, 139–142. [Google Scholar] [CrossRef] [Green Version]
- Lin, Z.F.; Hong, Y.G.; Yin, M.G.; Li, C.Y.; Zhang, K.; Grierson, D. A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J. 2008, 55, 301–310. [Google Scholar] [CrossRef] [Green Version]
- Quadrana, L.; Rodriguez, M.C.; Lopez, M.; Bermudez, L.; Nunes-Nesi, A.; Fernie, A.R.; Descalzo, A.; Asis, R.; Rossi, M.; Asurmendi, S.; et al. Coupling virus-induced gene silencing to exogenous Green Fluorescence Protein expression provides a highly efficient system for functional genomics in Arabidopsis and across all stages of tomato fruit development. Plant Physiol. 2011, 156, 1278–1291. [Google Scholar] [CrossRef] [Green Version]
- Smith, O.; Clapham, A.; Rose, P.; Liu, Y.; Wang, J.; Allaby, R.G. A complete ancient RNA genome: Identification, reconstruction and evolutionary history of archaeological Barley Stripe Mosaic Virus. Sci. Rep. 2014, 4, 4003. [Google Scholar] [CrossRef] [Green Version]
- Bennypaul, H.S.; Mutti, J.S.; Rustgi, S.; Kumar, N.; Okubara, P.A.; Gill, K.S. Virus-induced gene silencing (VIGS) of genes expressed in root, leaf, and meiotic tissues of wheat. Funct. Integr. Genom. 2012, 12, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.Y.; Hao, M.Y.; Tian, B.M.; Cao, G.Q.; Wei, F.; Xie, Z.Q. A methodological advance of tobacco rattle virus-induced gene silencing for functional genomics in plants. Front. Plant Sci. 2021, 12, 671091. [Google Scholar] [CrossRef] [PubMed]
- Lunn, J.E. Compartmentation in plant metabolism. J. Exp. Bot. 2007, 58, 35–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heazlewood, J.L.; Tonti-Filippini, J.S.; Gout, A.M.; Day, D.A.; Whelan, J.; Millar, A.H. Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 2004, 16, 241–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.J.; Ehrhardt, D.W.; Rhee, S.Y. Systematic analysis of Arabidopsis organelles and a protein localization database for facilitating fluorescent tagging of full-length Arabidopsis proteins. Plant Physiol. 2006, 141, 527–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katta, S.; Talakayala, A.; Reddy, M.K.; Addepally, U.; Garladinne, M. Development of transgenic cotton (Narasimha) using triple gene Cry2Ab-Cry1F-Cry1Ac construct conferring resistance to lepidopteran pest. J. Biosci. 2020, 45, 31. [Google Scholar] [CrossRef] [PubMed]
- Dhanoa, P.K.; Sinclair, A.M.; Mullen, R.T.; Mathur, J. Illuminating subcellular structures and dynamics in plants: A fluorescent protein toolbox. Botany 2006, 84, 515–522. [Google Scholar]
- Luo, B.; Nakata, P.A. A set of GFP organelle marker lines for intracellular localization studies in Medicago truncatula. Plant Sci. 2012, 188, 19–24. [Google Scholar] [CrossRef]
- Wu, T.M.; Lin, K.C.; Liau, W.S.; Chao, Y.Y.; Yang, L.H.; Chen, S.Y.; Lu, C.A.; Hong, C.Y. A set of GFP-based organelle marker lines combined with DsRed-based gateway vectors for subcellular localization study in rice (Oryza sativa L.). Plant Mol. Biol. 2016, 90, 107–115. [Google Scholar] [CrossRef]
- Nelson, B.K.; Cai, X.; Nebenfuhr, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef]
- Gong, J.; Tian, Z.; Qu, X.; Meng, Q.; Guan, Y.; Liu, P.; Chen, C.; Deng, X.; Guo, W.; Cheng, Y.; et al. Illuminating the cells: Transient transformation of citrus to study gene functions and organelle activities related to fruit quality. Hortic. Res. 2021, 8, 175. [Google Scholar] [CrossRef]
- Nagalakshmi, U.; Meier, N.; Liu, J.Y.; Voytas, D.F.; Dinesh-Kumar, S.P. High-efficiency multiplex biallelic heritable editing in Arabidopsis using an RNA virus. Plant Physiol. 2022, 4, 159. [Google Scholar] [CrossRef]
- Mohanraju, P.; Makarova, K.S.; Zetsche, B.; Zhang, F.; Koonin, E.V.; Van der Oost, J. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 2016, 353, 6299. [Google Scholar] [CrossRef] [Green Version]
- Oh, Y.; Kim, H.; Kim, S.G. Virus-induced plant genome editing. Curr. Opin. Plant Biol. 2021, 60, 101992. [Google Scholar] [CrossRef]
- Wright, A.V.; Sternberg, S.H.; Taylor, D.W.; Staahl, B.T.; Bardales, J.A.; Kornfeld, J.E.; Doudna, J.A. Rational design of a split-Cas9 enzyme complex. Proc. Natl. Acad. Sci. USA 2015, 112, 2984–2989. [Google Scholar] [CrossRef] [Green Version]
- Kaya, H.; Ishibashi, K.; Toki, S. A Split Staphylococcus aureus Cas9 as a compact genome-editing tool in plants. Plant Cell Physiol. 2017, 58, 643–649. [Google Scholar] [CrossRef] [Green Version]
- Shi, G.Y.; Hao, M.Y.; Tian, B.M.; Cao, G.Q.; Chen, W.W.; Zhang, Q.; Zhang, Y.S.; Ling, H.; Li, J.; Xie, Z.Q.; et al. A newly established virus-induced gene silencing method via seed imbibition for functional genomics at early germination stages in cotton. Ind. Crop. Prod. 2021, 172, 114040. [Google Scholar] [CrossRef]
- Ni-Na, L.I.; Ding, L.Y.; Zhang, Z.Y.; Guo, W.Z. Isolation of mesophyll protoplast and establishment of gene transient expression system in cotton. Acta Agron. Sin. 2014, 40, 231–239. [Google Scholar]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [Green Version]

| Organelle | Gene Name | Binary Plasmids | Virus Vectors | Gene Description | Reference |
|---|---|---|---|---|---|
| Plasma membrane | GhPIP2 | pCambia1300-eGFP-PM * | BSMV-γ2: PM-GFP | Transporter activity; aquaporin PIP | Li et al., 2011 [17] |
| Tonoplast | GhTIP2 | pCambia1300-eGFP-TP | BSMV-γ2: TP-GFP | Transporter activity; aquaporin TIP | Li et al., 2009 [18] |
| Endoplasmic reticulum | GhSPP | pCambia1300-eGFP-ER | BSMV-γ2: ER-GFP | Aspartic-type endopeptidase activity; integral component of membrane | Tamura et al., 2009 [19] |
| Plastids | GhClpD | pCambia1300-eGFP-PL | BSMV-γ2: PL-GFP | Chaperone protein ClpD, chloroplastic | Dangol et al., 2017 [20] |
| Peroxisome | GhAPX3 | pCambia1300-eGFP-PR | BSMV-γ2: PR-GFP | Response to oxidative stress; peroxidase activity | Teixeira et al., 2006 [21] |
| Mitochondria | GhALDH2 | pCambia1300-eGFP-MT | BSMV-γ2: MT-GFP | Oxidoreductase activity; metabolic process | Nakazono et al., 2000 [22] |
| Golgi body | GhMNS1 | pCambia1300-eGFP-GB | BSMV-γ2: GB-GFP | Mannosyl-oligosaccharide 1,2-alpha-mannosidase activity | Saint-Jore-Dupas et al., 2006 [23] |
| Nucleus | GhTAF2 | pCambia1300-eGFP-NU | BSMV-γ2: NU-GFP | DNA-dependent transcription, initiation | Dangol et al., 2017 [20] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, W.; Huang, C.; Luo, C.; Zhang, Y.; Zhang, B.; Xie, Z.; Hao, M.; Ling, H.; Cao, G.; Tian, B.; et al. A New Method for Rapid Subcellular Localization and Gene Function Analysis in Cotton Based on Barley Stripe Mosaic Virus. Plants 2022, 11, 1765. https://doi.org/10.3390/plants11131765
Chen W, Huang C, Luo C, Zhang Y, Zhang B, Xie Z, Hao M, Ling H, Cao G, Tian B, et al. A New Method for Rapid Subcellular Localization and Gene Function Analysis in Cotton Based on Barley Stripe Mosaic Virus. Plants. 2022; 11(13):1765. https://doi.org/10.3390/plants11131765
Chicago/Turabian StyleChen, Weiwei, Chaolin Huang, Chenmeng Luo, Yongshan Zhang, Bin Zhang, Zhengqing Xie, Mengyuan Hao, Hua Ling, Gangqiang Cao, Baoming Tian, and et al. 2022. "A New Method for Rapid Subcellular Localization and Gene Function Analysis in Cotton Based on Barley Stripe Mosaic Virus" Plants 11, no. 13: 1765. https://doi.org/10.3390/plants11131765
APA StyleChen, W., Huang, C., Luo, C., Zhang, Y., Zhang, B., Xie, Z., Hao, M., Ling, H., Cao, G., Tian, B., Wei, F., & Shi, G. (2022). A New Method for Rapid Subcellular Localization and Gene Function Analysis in Cotton Based on Barley Stripe Mosaic Virus. Plants, 11(13), 1765. https://doi.org/10.3390/plants11131765