Abstract
Hemp (Cannabis sativa L., 2n = 20) is a valuable crop that is successfully used as a food, technical and medicinal crop. It is a dioecious plant with an XX\XY sex determination system. Some chromosomes of C. sativa have almost the same lengths and centromeric indexes. Cytogenetic markers help to distinguish similar plant chromosomes, including sex chromosomes, which is important for the breeding process. Two repeats (CS-1 and CS-237) were used to develop labeled oligo-probes for rapid and low-cost oligo-FISH. These oligos can be recommended for use as cytological markers to distinguish sex chromosomes (X and Y) and somatic chromosome pairs 3, 6, and 8 by rapid oligo-FISH in a short time.
1. Introduction
Hemp (Cannabis sativa L.) is an important agricultural plant, that has been cultivated for more than 5000 years [1]. Furthermore, it is one of the oldest plant sources to serve as a food and technical crop [2,3,4,5,6]. C. sativa has long been used for medical purposes dating back to ancient times. The plant has medicinal usage in the treatment of burns, pain, glaucoma, nausea, cardiovascular and bronchopulmonary diseases, depression, neuralgia, anemia, and bone fragility, among others [7,8,9,10,11,12,13,14,15].
Plant sex chromosomes are very rare; they have been reported in about 40 species [16,17,18]. C. sativa is a dioecious plant with an XX\XY sex determination system [19,20,21,22]; the chromosome number is 2n = 20 [23]. The haploid nuclear genome size for males and females is calculated to be 843 mega-base pairs (Mbp) and 818 Mbp, respectively [24,25]. Sex determination is important for cannabis production and depends on the specific application. For instance, the unfertilized female flowers contain higher levels of cannabinoids and seeded flowers are undesirable for medicinal applications [24]. C. sativa can be used for better understanding of the evolution of sex chromosomes [23,26,27]. A high level of polymorphism has been noted in C. sativa [28,29]. Furthermore, the chromosomal polymorphism has been detected by FISH on the inter- and intra-cultivar levels of C. sativa [23]. The high level of polymorphism determines the importance of C. sativa cytological studies for cultivar identification and characterization.
Cytogenetic markers help to distinguish similar plant chromosomes, including sex chromosomes, which is important for the breeding process [19,30]. Cytogenetic markers also help us to associate genomic assemblies with physical chromosomes [31,32]. DNA repeats for FISH analysis usually need to be amplified in a bacterial plasmid, extracted, and then labeled by nick translation [33,34]. In addition, FISH probes can be generated by PCR, but DNA isolation is previously required [35,36,37]. Thus, the procedure for preparing the probes is time consuming. Synthetic oligonucleotides with a fluorescent label can also be used as a probe for FISH [38,39] or ND-FISH analysis [40,41,42,43,44,45,46,47,48]. Such a probe is convenient to use since fluorochrome labeled oligonucleotides can be purchased directly from commercial sources. ND-FISH have a short hybridization time and no denaturation process, saving the structure of chromosomes, reagents and other resources [40,46,47,49]. Based on these advantages, the method is widely used to identify plant chromosomes [39,40,42,47,48]. Moreover, ND-FISH technology can be used to identify crop varieties [50]. ND-FISH is simple and convenient, but less reproducible in a series of experiments [46].
Oligos are being developed for easy application, which allows researchers to obtain results by fast oligo-FISH or ND-FISH in a short time. Oligos have been developed and successfully used for cereals [40,41,42,43,51,52] and other crops [53,54,55]. In this article, we report the development of oligo-labeled probes, based on CS-1 [56] and CS-237 [31] repeats for rapid oligo-FISH. The possibility of their use as cytological markers for C. sativa sex and some somatic chromosomes is discussed.
2. Results and Discussion
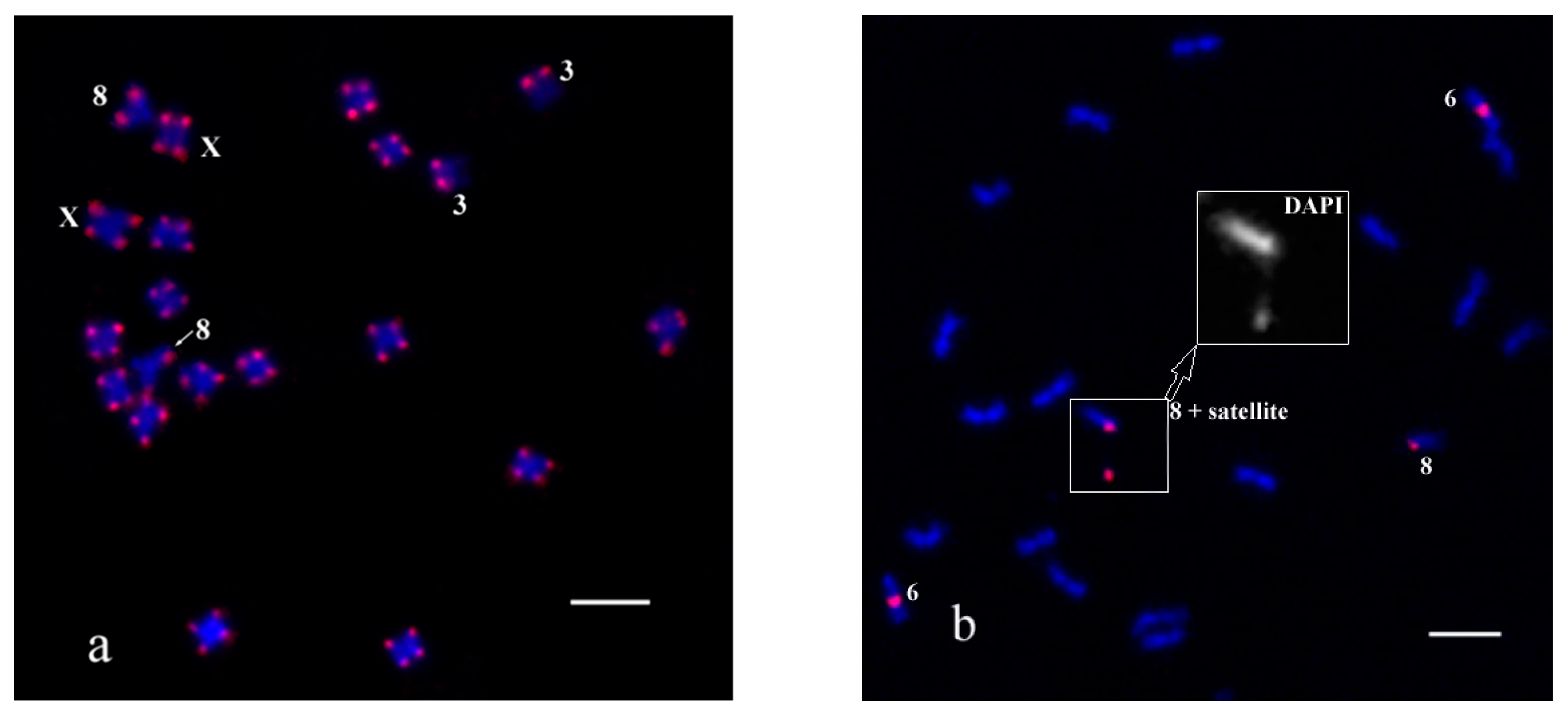
To compare the efficiency between conventional FISH and oligo-FISH we used them in turn. The first experiment was carried out on C. sativa chromosomes with PCR labelled probes. As a result, hybridization signals of CS-1 (Figure 1a) and CS-237 (Figure 1b) were detected as described by Divashuk et al. [56] and Alexandrov et al. [31], respectively. Satellites are often separated from the chromosome 8, as shown in Figure 1b.

Figure 1.
The results of FISH experiments with the PCR labelled probes CS-1 and CS-237 on C. sativa chromosomes: (a) pink—CS-1 probe; (b) pink—CS-237 probe. DAPI layer of chromosome 8 with satellite is enlarged and shown in gray. Distinguished chromosomes are indicated according to Alexandrov et al. [31]. Bar equals 5 μm.
Labeled oligos were designed and synthesized for the repeats CS-1 and CS-237 by Primer3 software [57] for more simple use of them as cytogenetic markers. For each repeat we designed two different sequences of oligos to increase the chances of successful hybridization. Labeling was carried out by using various fluorophores (FAM, TAMRA, Cy5) (Table 1) for more convenient use in combination with other labeled probes.

Table 1.
Labeled oligo-probes designed for the repeats CS-1 and CS-237.
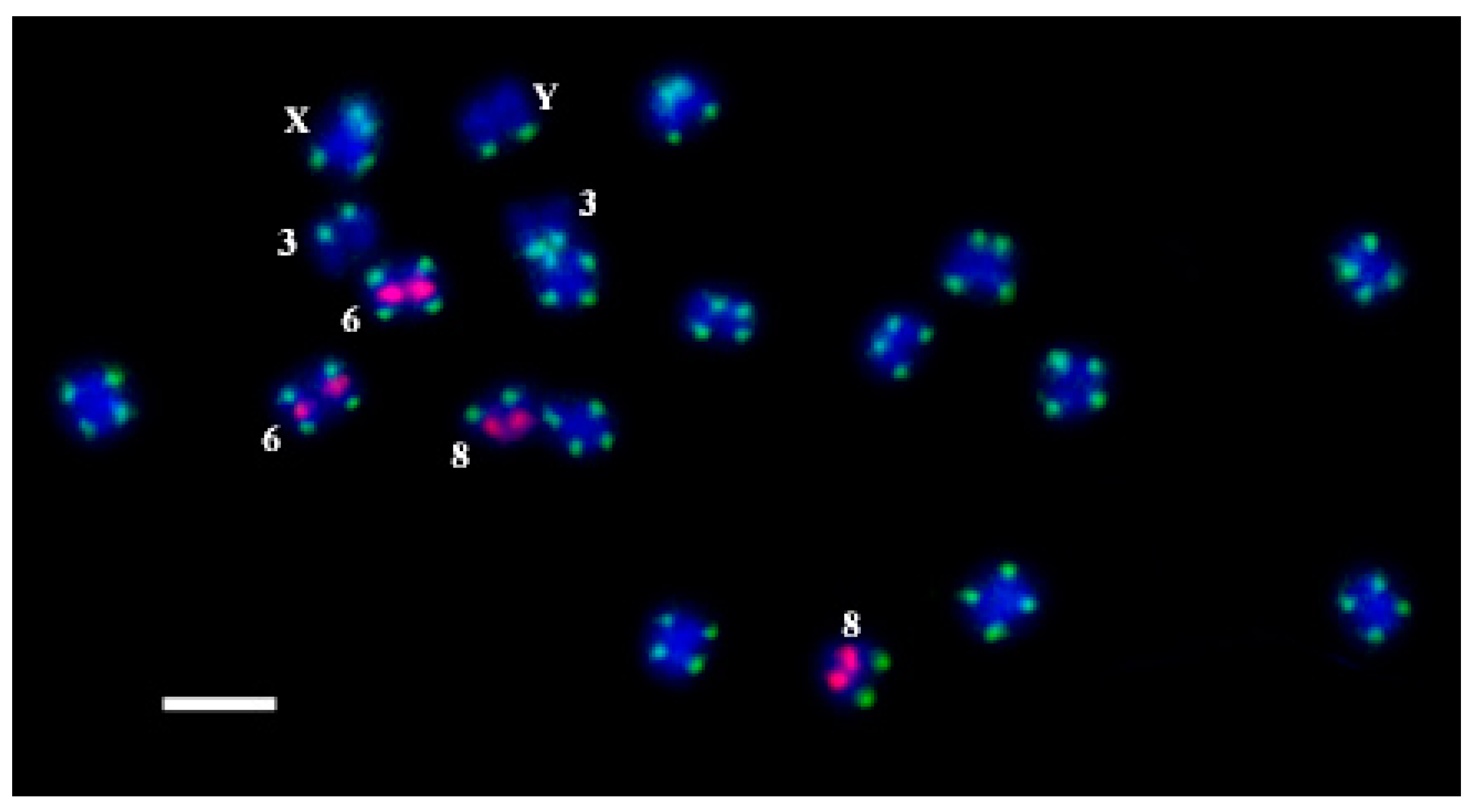
All the oligos were hybridized on C. sativa chromosomes by rapid oligo-FISH (Figure 2). Hybridization signals were observed for all the developed oligos. The results of the oligo-FISH probe detection with all the oligos on C. sativa chromosomes were similar as for FISH with the PCR labelled probes. There were no non-specific signals interfering with the work with the metaphase plates.
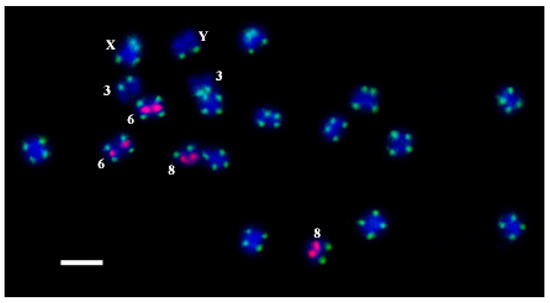
Figure 2.
The results of oligo-FISH experiments with the oligo-probes CS-1-FAM (green) and CS-237-TAMRA (pink) on C. sativa chromosomes. Distinguished chromosomes are indicated according to Alexandrov et al. [31]. Bar equals 5 μm.
We found that the oligo-probes oligoCS-1-FAM, oligoCS-1-Cy5, and oligoCS-1-TAMRA are located in the subtelomeric region on both arms of all C. sativa chromosomes, except for the long arm of chromosome 3, the short arm of chromosome 8, and the long arm of the Y chromosome at the same positions. The oligo-probes oligoCS-237-FAM, oligoCS-237-Cy5, and oligoCS-237-TAMRA are located in the proximal part of the short arm of chromosome 6 and in the distal part of the short arm of chromosome 8 at the same positions. Thus, we can use any oligo with any dye from Table 1 suitable for a particular experiment. The positions of the oligos completely coincide with the positions of the corresponding repeats on C. sativa chromosomes [31,56]. We noted a different intensity of signals in different plants of C. sativa, which is quite consistent with Razumova et al. [23], where subtelomere hybridization sites disappear or new hybridization sites appear. This interesting phenomenon for the cross-pollinated plant is relatively expected and requires further study.
Some chromosomes of C. sativa are difficult to distinguish from each other without the use of cytological markers (chromosomes 5 and 6, X and Y) since they have almost the same lengths and centromeric indexes. Differences in the position of the oligo-probes for CS-1 and CS-237 repeats on C. sativa chromosomes make it possible to distinguish chromosomes 5 and 6, X and Y and easily identify chromosomes 3 and 8. Razumova et al. [23] showed different variations of FISH signals for the CS-1 repeat on chromosome 8 and on chromosome X. Nevertheless, the combined use of the oligo-probes for CS-1 and CS-237 repeats can identify these chromosomes. Thus, the probes can be recommended as cytogenetic markers for better identification of C. sativa chromosomes 3, 6, 8, X and Y by rapid oligo-FISH.
3. Conclusions
In conclusion, the oligonucleotide probes oligoCS-1-FAM, oligoCS-1-Cy5, oligoCS-1-TAMRA, oligoCS-237-FAM, oligoCS-237-Cy5, and oligoCS-237-TAMRA have been developed in the present study. They can be used to conveniently identify sex and somatic chromosomes and their polymorphism in hemp using rapid oligo-FISH.
4. Materials and Methods
4.1. Plant Material
For the study of mitosis metaphase chromosomes, cv ‘‘Zenitsa’’ (dioecious) seeds of C. sativa were used (originated by P.P. Lukyanenko Krasnodar Research and the Development Institute of Agriculture, Krasnodar, Russia). The seeds were harvested in 2020 and then stored at +4 °C.
4.2. Chromosome Preparation
C. sativa seeds were germinated on moist filter paper in Petri dishes at 24 °C in the dark for 72 h. The seedlings (2–3 cm long) were used to prepare the slides of mitotic metaphase chromosomes as described by Romanov et al. [58].
4.3. DNA Probes and Labeling
Young seedlings of C. sativa were used for DNA isolation by the CTAB method [59]. Repeat sequences CS-1 and CS-237, previously described by Divashuk et al. [56] and Alexandrov et al. [31], respectively, were used to design primers and oligos by using the Primer3 software v.4.1.0 [57]. DNA labeling was performed by PCR with biotin-16-dUTP according to the manufacturer’s instruction (Boehringer, Germany). The oligos were synthesized and labeled by FAM, TAMRA or Cy5 (Evrogen JSC, Moscow, Russia).
4.4. Fluorescence In Situ Hybridization (FISH) and Oligo-FISH
The FISH experiments were performed as described by Karlov et al. [60]. Post-hybridization washing was performed in 50% (v/v) formamide in 2× SSC for 15 min at 42 °C, while the theoretical washing stringency was about 80%. The chromosomes were counterstained with 1 mg/mL DAPI and mounted in Vectashield (Vector laboratories, Burlingame, CA, USA). Oligo-FISH experiments were performed as FISH, but without the detection step, as described by Kuznetsova et al. [46]. The hybridization procedure for FISH and oligo-FISH lasted for 1 h and 16 h, respectively, at 37 °C.
4.5. Microscopy and Image Analysis
Chromosome preparations were viewed by using a THUNDER 3D Tissue microscope with a filter set DFT51111, a fluorescence light source LED3, and a digital camera DFC9000 GTC (Leica Microsystems, Wetzlar, Germany). Multichannel fluorescence recording, image processing for brightness/contrast and color settings were performed using LasX software (Leica Microsystems, Wetzlar, Germany). The karyotype for C. sativa metaphase chromosomes was developed by Divashuk et al. [56] and improved by Alexandrov et al. [31]. In this article, chromosome numbering is in accordance with Alexandrov et al. [31]. Chromosomes 3, 5, 6, 8, X, and Y according to Alexandrov et al. [31] correspond to chromosomes 4, 6, 3, 9, X, and Y, respectively, according to Divashuk et al. [56].
Author Contributions
Project administration, investigation, discussion, and writing the article, D.V.R.; methodology and discussion, G.I.K.; conceptualization, methodology and discussion, M.G.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Russian Science Foundation, grant number 20-76-00036.
Data Availability Statement
Not applicable.
Acknowledgments
The authors are grateful to P.P. Lukyanenko Krasnodar Research and the Development Institute of Agriculture (Krasnodar, Russia) for the seeds of C. sativa drug-free varieties.
Conflicts of Interest
The authors declare no conflict of interest.
References
- ElSohly, M.A.; Radwan, M.M.; Gul, W.; Chandra, S.; Galal, A. Phytochemistry of Cannabis sativa L. Prog. Chem. Org. Nat. Prod. 2017, 103, 1–36. [Google Scholar] [CrossRef]
- Cherney, J.H.; Small, E. Industrial Hemp in North America: Production, Politics and Potential. Agronomy 2016, 6, 58. [Google Scholar] [CrossRef] [Green Version]
- Crini, G.; Lichtfouse, E.; Chanet, G.; Morin-Crini, N. Applications of hemp in textiles, paper industry, insulation and building materials, horticulture, animal nutrition, food and beverages, nutraceuticals, cosmetics and hygiene, medicine, agrochemistry, energy production and environment: A review. Environ. Chem. Lett. 2020, 18, 1451–1476. [Google Scholar] [CrossRef]
- Jiang, H.E.; Li, X.; Zhao, Y.X.; Ferguson, D.K.; Hueber, F.; Bera, S.; Wang, Y.F.; Zhao, L.C.; Liu, C.J.; Li, C.S. A new insight into Cannabis sativa (Cannabaceae) utilization from 2500-year-old Yanghai Tombs, Xinjiang, China. J. Ethnopharmacol. 2006, 108, 414. [Google Scholar] [CrossRef]
- Kriese, U.; Schumann, E.; Weber, W.E.; Beyer, M.; Brühl, L.; Matthus, B. Oil content, tocopherol composition and fatty acid patterns of the seeds of 51 C. sativa L. genotypes. Euphytica 2004, 137, 339. [Google Scholar] [CrossRef]
- Werf, H.V.D.; Mathussen, E.W.J.M.; Haverkort, A.J. The potential of hemp (Cannabis sativa L.) for sustainable fibre production: A crop physiological appraisal. Ann. Appl. Biol. 1996, 129, 109–123. [Google Scholar] [CrossRef]
- Abrams, D.I.; Jay, C.A.; Shade, S.B.; Vizoso, H.; Reda, H.; Press, S.; Kelly, M.E.; Rowbotham, M.C.; Petersen, K.L. Cannabis in painful HIV-associated sensory neuropathy: A randomized placebo-controlled trial. Neurology 2007, 68, 515. [Google Scholar] [CrossRef]
- Andre, C.M.; Hausman, J.F.; Guerriero, G. Cannabis sativa: The plant of the thousand and one molecules. Front. Plant Sci. 2016, 7, 19. [Google Scholar] [CrossRef] [Green Version]
- Bonini, S.A.; Premoli, M.; Tambaro, S.; Kumar, A.; Maccarinelli, G.; Memo, M.; Mastinu, A. Cannabis sativa: A comprehensive ethnopharmacological review of a medicinal plant with a long history. J. Ethnopharmacol. 2018, 227, 300–315. [Google Scholar] [CrossRef]
- Guindon, J.; Hohmann, A.G. The endocannabinoid system and pain. Curr. Drug Targets CNS Neurol. Disord. 2009, 8, 403. [Google Scholar] [CrossRef]
- Merlin, M.D. Archaeological evidence for the tradition of psychoactive plant use in the old world. Econ. Bot. 2003, 57, 295–323. [Google Scholar] [CrossRef]
- Potter, D.J. A review of the cultivation and processing of cannabis (Cannabis sativa L.) for production of prescription medicines in the UK. Drug Test. Anal. 2014, 6, 31–38. [Google Scholar] [CrossRef]
- Russo, E. Hemp for headache: An in-depth historical and scientific review of cannabis in migraine treatment. J. Cannabis Ther. 2001, 1, 21–92. [Google Scholar] [CrossRef]
- Slatkin, N.E. Cannabinoids in the treatment of chemotherapy-induced nausea and vomiting: Beyond prevention of acute emesis. J. Support. Oncol. 2007, 5, 1–9. [Google Scholar]
- Zuardi, A.W. History of Cannabis as a medicine: A review. Braz. J. Psychiat. 2006, 28, 153. [Google Scholar] [CrossRef] [Green Version]
- Renner, S.S. The relative and absolute frequencies of angiosperm sexual systems: Dioecy, monoecy, gynodioecy, and an updated online database. Am. J. Bot. 2014, 101, 1588–1596. [Google Scholar] [CrossRef] [Green Version]
- Muyle, A.; Shearn, R.; Marais, G.A. The evolution of sex chromosomes and dosage compensation in plants. Genome Biol. Evol. 2017, 9, 627–645. [Google Scholar] [CrossRef] [Green Version]
- Fruchard, C.; Marais, G.A. Evolutionary Developmental Biology: A Reference Guide; Nuno de la Rosa, L., Müller, G., Eds.; Springer: Cham, Switzerland, 2021; pp. 683–696. [Google Scholar] [CrossRef] [Green Version]
- Divashuk, M.G.; Alexandrov, O.S.; Kroupin, P.Y.; Karlov, G.I. Molecular cytogenetic mapping of Humulus lupulus sex chromosomes. Cytogenet. Genome Res. 2011, 134, 213–219. [Google Scholar] [CrossRef]
- Grabowska-Joachimiak, A.; Mosiolek, M.; Lech, A.; Góralski, G. C-Banding/DAPI and in situ hybridization reflect karyotype structure and sex chromosome differentiation in Humulus japonicus Siebold & Zucc. Cytogenet. Genome Res. 2011, 132, 203–211. [Google Scholar] [CrossRef]
- Matsunaga, S.; Kawano, S. Sex determination by sex chromosomes in dioecious plants. Plant Biol. 2001, 3, 481–488. [Google Scholar] [CrossRef]
- Ming, R.; Bendahmane, A.; Renner, S.S. Sex chromosomes in land plants. Annu. Rev. Plant Biol. 2011, 62, 485–514. [Google Scholar] [CrossRef] [Green Version]
- Razumova, O.V.; Alexandrov, O.S.; Divashuk, M.G.; Sukhorada, T.I.; Karlov, G.I. Molecular cytogenetic analysis of monoecious hemp (Cannabis sativa L.) cultivars reveals its karyotype variations and sex chromosomes constitution. Protoplasma 2016, 253, 895–901. [Google Scholar] [CrossRef]
- Hesami, M.; Pepe, M.; Alizadeh, M.; Rakei, A.; Baiton, A.; Jones, A.M.P. Recent advances in cannabis biotechnology. Ind. Crops Prod. 2020, 158, 113026. [Google Scholar] [CrossRef]
- Hurgobin, B.; Tamiru-Oli, M.; Welling, M.T.; Doblin, M.S.; Bacic, A.; Whelan, J.; Lewsey, M.G. Recent advances in Cannabis sativa genomics research. New Phytol. 2021, 230, 73–89. [Google Scholar] [CrossRef]
- Prentout, D.; Razumova, O.; Rhoné, B.; Badouin, H.; Henri, H.; Feng, C.; Käfer, J.; Karlov, G.; Marais, G.A. A high-throughput segregation analysis identifies the sex chromosomes of Cannabis sativa. bioRxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Razumova, O.V.; Divashuk, M.G.; Alexandrov, O.S.; Karlov, G.I. GISH painting of the Y chromosomes suggests advanced phases of sex chromosome evolution in three dioecious Cannabaceae species (Humulus lupulus, H. japonicus, and Cannabis sativa). Protoplasma 2022, 1774, 1–8. [Google Scholar] [CrossRef]
- Faeti, V.; Mandolino, G.; Ranalli, P. Genetic diversity of Cannabis sativa germplasm based on RAPD markers. Plant Breed. 1996, 115, 367–370. [Google Scholar] [CrossRef]
- Forapani, S.; Carboni, A.; Paoletti, C.; Moliterni, V.C.; Ranalli, P.; Mandolino, G. Comparison of hemp varieties using random amplified polymorphic DNA markers. Crop Sci. 2001, 41, 1682–1689. [Google Scholar] [CrossRef] [Green Version]
- Kroupin, P.Y.; Kuznetsova, V.M.; Nikitina, E.A.; Martirosyan, Y.T.; Karlov, G.I.; Divashuk, M.G. Development of new cytogenetic markers for Thinopyrum ponticum (Podp.) Z.-W. Liu & R.-C. Wang. Comp. Cytogenet. 2019, 13, 231–243. [Google Scholar] [CrossRef]
- Alexandrov, O.S.; Romanov, D.V.; Divshuk, M.G.; Razumova, O.V.; Ulyanov, D.S.; Karlov, G.I. Studying and physical mapping of the 45S IGS linked CS-237 DNA repeat in Cannabis sativa L. Plants 2022, 11, 1396. [Google Scholar] [CrossRef]
- Chao, Y.T.; Chen, W.C.; Chen, C.Y.; Ho, H.Y.; Yeh, C.H.; Kuo, Y.T.; Su, C.L.; Yen, S.H.; Hsueh, H.Y.; Yeh, J.H.; et al. Chromosome-level assembly, genetic and physical mapping of Phalaenopsis aphrodite genome provides new insights into species adaptation and resources for orchid breeding. Plant Biotechnol. J. 2018, 16, 2027–2041. [Google Scholar] [CrossRef] [Green Version]
- Bray-Ward, P. FISH Probes and Labelling Techniques. FISH. A Practical Approach; Oxford University Press: Oxford, UK, 2002; pp. 5–28. [Google Scholar]
- Giorgetti, L.; Piolot, T.; Heard, E. High-resolution 3D DNA FISH using plasmid probes and computational correction of optical aberrations to study chromatin structure at the sub-megabase scale. In Nuclear Bodies and Noncoding RNAs; Humana Press: New York, NY, USA, 2015; Volume 1262, pp. 37–53. [Google Scholar] [CrossRef]
- Huber, D.; von Voithenberg, L.V.; Kaigala, G.V. Fluorescence in situ hybridization (FISH): History, limitations and what to expect from micro-scale FISH? Micro Nano Eng. 2018, 1, 15–24. [Google Scholar] [CrossRef]
- Jiang, J. Fluorescence in situ hybridization in plants: Recent developments and future applications. Chromosome Res. 2019, 27, 153–165. [Google Scholar] [CrossRef]
- Kroupin, P.; Kuznetsova, V.; Romanov, D.; Kocheshkova, A.; Karlov, G.; Dang, T.X.; Khuat, T.M.L.; Kirov, I.; Alexandrov, O.; Polkhovskiy, A.; et al. Pipeline for the rapid development of cytogenetic markers using genomic data of related species. Genes 2019, 10, 113. [Google Scholar] [CrossRef] [Green Version]
- Lang, T.; Li, G.; Wang, H.; Yu, Z.; Chen, Q.; Yang, E.; Fu, S.; Tang, Z.; Yang, Z. Physical location of tandem repeats in the wheat genome and application for chromosome identification. Planta 2019, 249, 663–675. [Google Scholar] [CrossRef]
- Tang, Z.; Yang, Z.; Fu, S. Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119. 2, pTa-535, pTa71, CCs1, and pAWRC. 1 for FISH analysis. J. Appl. Genet. 2014, 55, 313–318. [Google Scholar] [CrossRef]
- Fu, S.; Chen, L.; Wang, Y.; Li, M.; Yang, Z.; Qiu, L.; Yan, B.; Ren, Z.; Tang, Z. Oligonucleotide probes for ND-FISH analysis to identify rye and wheat chromosomes. Sci. Rep. 2015, 5, 10552. [Google Scholar] [CrossRef] [Green Version]
- Tang, S.; Qiu, L.; Xiao, Z.; Fu, S.; Tang, Z. New oligonucleotide probes for ND-FISH analysis to identify barley chromosomes and to investigate polymorphisms of wheat chromosomes. Genes 2016, 7, 118. [Google Scholar] [CrossRef] [Green Version]
- Tang, S.; Tang, Z.; Qiu, L.; Yang, Z.; Li, G.; Lang, T.; Zhu, W.; Zhang, J.; Fu, S. Developing new oligo probes to distinguish specific chromosomal segments and the A, B, D genomes of wheat (Triticum aestivum L.) using ND-FISH. Front. Plant Sci. 2018, 9, 1104. [Google Scholar] [CrossRef]
- Xi, W.; Tang, S.; Du, H.; Luo, J.; Tang, Z.; Fu, S. ND-FISH-positive oligonucleotide probes for detecting specific segments of rye (Secale cereale L.) chromosomes and new tandem repeats in rye. Crop J. 2020, 8, 171–181. [Google Scholar] [CrossRef]
- Cuadrado, Á.; Golczyk, H.; Jouve, N. A novel, simple and rapid nondenaturing FISH (ND-FISH) technique for the detection of plant telomeres. Potential used and possible target structures detected. Chromosome Res. 2009, 17, 755–762. [Google Scholar] [CrossRef]
- Cuadrado, Á.; Jouve, N. Chromosomal detection of simple sequence repeats (SSRs) using nondenaturing FISH (ND-FISH). Chromosoma 2010, 119, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, V.M.; Razumova, O.V.; Karlov, G.I.; Dang, T.X.; Kroupin, P.Y.; Divashuk, M.G. Some peculiarities in application of denaturating and non-denaturating in situ hybridization on chromosomes of cereals. Mosc. Univ. Biol. Sci. Bull. 2019, 74, 75–80. [Google Scholar] [CrossRef]
- Li, Z.; Ren, Z.; Tan, F.; Tang, Z.; Fu, S.; Yan, B.; Ren, T. Molecular cytogenetic characterization of new wheat-rye 1R (1B) substitution and translocation lines from a Chinese Secale cereal L. Aigan with resistance to stripe rust. PLoS ONE 2016, 11, e0163642. [Google Scholar]
- Xiao, Z.; Tang, S.; Qiu, L.; Tang, Z.; Fu, S. Oligonucleotides and ND-FISH displaying different arrangements of tandem repeats and identification of Dasypyrum villosum chromosomes in wheat backgrounds. Molecules 2017, 22, 973. [Google Scholar] [CrossRef] [Green Version]
- Ren, T.; He, M.; Sun, Z.; Tan, F.; Luo, P.; Tang, Z.; Fu, S.; Yan, B.; Ren, Z.; Li, Z. The polymorphisms of oligonucleotide probes in wheat cultivars determined by ND-FISH. Molecules 2019, 24, 1126. [Google Scholar] [CrossRef] [Green Version]
- Jiang, M.; Xaio, Z.Q.; Fu, S.L.; Tang, Z.X. FISH karyotype of 85 common wheat cultivars/lines displayed by ND-FISH using oligonucleotide probes. Cereal Res. Commun. 2017, 45, 549–563. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Zhang, T.; Yu, Z.; Wang, H.; Yang, E.; Yang, Z. An efficient Oligo-FISH painting system for revealing chromosome rearrangements and polyploidization in Triticeae. Plant J. 2021, 105, 978–993. [Google Scholar] [CrossRef]
- Liu, X.; Sun, S.; Wu, Y.; Zhou, Y.; Gu, S.; Yu, H.; Yi, C.; Gu, M.; Jiang, J.; Liu, B.; et al. Dual-color oligo-FISH can reveal chromosomal variations and evolution in Oryza species. Plant J. 2020, 101, 112–121. [Google Scholar] [CrossRef]
- Fu, L.; Wang, Q.; Li, L.; Lang, T.; Guo, J.; Wang, S.; Sun, Z.; Han, S.; Huang, B.; Dong, W.; et al. Physical mapping of repetitive oligonucleotides facilitates the establishment of a genome map-based karyotype to identify chromosomal variations in peanut. BMC Plant Biol. 2021, 21, 107. [Google Scholar] [CrossRef]
- Luo, X.; He, Z. Distribution of FISH oligo-5S rDNA and oligo-(AGGGTTT) 3 in Hibiscus mutabilis L. Genome 2021, 64, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Liu, J.; He, Z. Oligo-FISH Can Identify Chromosomes and Distinguish Hippophaë rhamnoides L. Taxa. Genes 2022, 13, 195. [Google Scholar] [CrossRef] [PubMed]
- Divashuk, M.G.; Alexandrov, O.S.; Razumova, O.V.; Kirov, I.V.; Karlov, G.I. Molecular cytogenetic characterization of the dioecious Cannabis sativa with an XY chromosome sex determination system. PLoS ONE 2014, 9, e85118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [Green Version]
- Romanov, D.V.; Shirnin, S.Y.; Karlov, G.I.; Divashuk, M.G. Cytogenetic Study of Aegopodium podagraria (Umbelliferae) for Use in Breeding. Mosc. Univ. Biol. Sci. Bull. 2020, 75, 65–70. [Google Scholar] [CrossRef]
- Rogers, S.O.; Bendich, A.J. Extraction of DNA from plant tissues. In Plant Molecular Biology Manual; Springer: Dordrecht, The Netherlands, 1989; Volume 1, pp. 73–83. [Google Scholar] [CrossRef]
- Karlov, G.I.; Danilova, T.V.; Horlemann, C.; Weber, G. Molecular cytogenetics in hop (Humulus lupulus L.) and identification of sex chromosomes by DAPI-banding. Euphytica 2003, 132, 185–190. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).