Abstract
Ammanniaauriculata is a troublesome broadleaf weed, widely distributed in the paddy fields of southern China. In this study, 10 biotypes of A. auriculata were sampled from Yangzhou City, China, where the paddy fields were seriously infested with A. auriculata, and their resistance levels to acetolactate synthase (ALS) inhibitor bensulfuron-methyl were determined. The whole-plant response assays showed that nine A. auriculata biotypes were highly resistant (from 16.4- to 183.1-fold) to bensulfuron-methyl in comparison with a susceptible YZ-S biotype, and only one YZ-6 biotype was susceptible. ALS gene sequencing revealed that three ALS gene copies existed in A. auriculata, and four different amino acid substitutions (Pro197-Leu, -Ala, -Ser, and -His) at site 197 in the AaALS1 or 2 genes were found in eight resistant biotypes. In addition, no amino acid mutations in three ALS genes were found in the YZ-3 biotype. These results suggested that target-site mutations or non-target-site resistance mechanisms were involved in tested resistant A. auriculata biotypes. Finally, a cleaved amplified polymorphic sequence (CAPS) marker was identified to rapidly detect the Pro197 mutations in A. auriculata.
1. Introduction
Modern herbicides are the primary ways of controlling weeds due to their high-efficiency and labor-saving characteristics, and they have made great contributions to food safety production in the world. However, strong selection pressures have made weeds obtain a powerful weapon in their battle with herbicides—herbicide resistance. To date, there are 267 weed species from 96 crops in 71 countries, which are evolving resistance to herbicides across most sites of action [1]. Target-site resistance due to amino acid substitution in the herbicide target-site genes is one of the most common mechanisms facilitating resistance evolution [2,3,4].
Sulfonylurea herbicides, targeting the acetolactate synthase (ALS) enzyme, are the most important group of ALS inhibitors, and more than 50 active ingredients have been commercialized [5]. ALS inhibitors lead to the death of plants by inhibiting the synthesis of branched-chain amino acids [6,7]. Those herbicides have been widely used in a variety of crop fields worldwide due to their super high effectivity and low toxicity to mammalian species [8]. However, ALS inhibitors have a high resistance risk, and the number of weeds that have developed resistance to ALS inhibitors is the highest among all the different types of herbicides [1]. Resistance to ALS inhibitors can be endowed by several different mechanisms. In most cases, target-site mutations in ALS genes are known to cause the high resistance to herbicides in many field-evolved resistant-weed populations. Nine conserved amino acid substitutions (Ala122 and 205, Pro197, Phe206, Asp376, Arg377, Trp574, Ser653, and Gly654) in ALS genes have so far been identified to be closely related to ALS inhibitor resistance [9,10,11]. In addition to ALS mutations, metabolic resistance is also an important mechanism of resistance. Metabolic resistance can evolve independently in resistant weeds, and has been reported in Sinapis arvensis L. [12], Bromus rigidus Roth [13], Amaranthus tuberculatus [14], Sagittaria trifolia L. [15], Ipomoea purpurea (L.) Roth. [16], and Bromus japonicus Thunb. [17]. More often, metabolic resistance and ALS mutations co-occur in the same biotypes, such as in Papaver rhoeas L. [18,19], Amaranthus palmeri [20], and Descurainia sophia L. [21]. Furthermore, a few studies have found that ALS gene overexpression can play an important role in resistance to ALS inhibitors in Cyperus compressus L. [22] and Bromus sterilis L. [23].
Ammanniaauriculata is one of the most serious weeds in the rice fields of southern China. In recent years, this species has spread rapidly in Jiangsu, Anhui, and Zhejiang provinces, and poses a serious threat to rice production. Control of A. auriculata in China relies heavily on the ALS inhibitor bensulfuron-methyl. Many species of weeds in rice fields, including A. auriculata, have developed resistance to this herbicide [24,25,26,27]. In California, A. auriculata first evolved resistance to bensulfuron-methyl in 1997 [1]. In addition, Wang et al. [28] reported that 85 out of 88 A. auriculata biotypes from Zhejiang Province tested by the agar method were resistant to bensulfuron-methyl. However, there is no report about the target-site resistance mechanisms for ALS inhibitors in A. auriculata. The aims of this study were to: (1) determine the resistance levels to bensulfuron-methyl in 11 A. auriculata biotypes from Yangzhou City in Jiangsu Province; (2) identify the ALS mutations in underlying resistant biotypes; and (3) develop a cleaved amplified polymorphic sequence (CAPS) marker to rapidly detect resistant A. auriculata plants with the ALS mutations.
2. Results
2.1. Bensulfuron-Methyl Dose Response
The whole-plant response bioassays indicated that nine A. auriculata biotypes showed the high resistance to bensulfuron-methyl based on the classification of resistance grades (RF ≥ 10) [29]. The GR50 values of two susceptible biotypes, YZ-S and -6, were 0.18 and 0.26 g ai ha−1, respectively. By contrast, the GR50 values of the nine resistant biotypes (YZ-1 to 5 and YZ-7 to 10) to bensulfuron-methyl were between 2.95 and 32.96 g ai ha−1, and were from 16.4- to 183.1-fold greater than that of the susceptible YZ-S biotype (Table 1, Figure 1). These results revealed that the bensulfuron-methyl-resistant A. auriculata biotypes were prevalent in the paddy fields.

Table 1.
Parameter values of log-logistic equation for whole-plant response assays of 11 A. auriculata populations to bensulfuron-methyl.
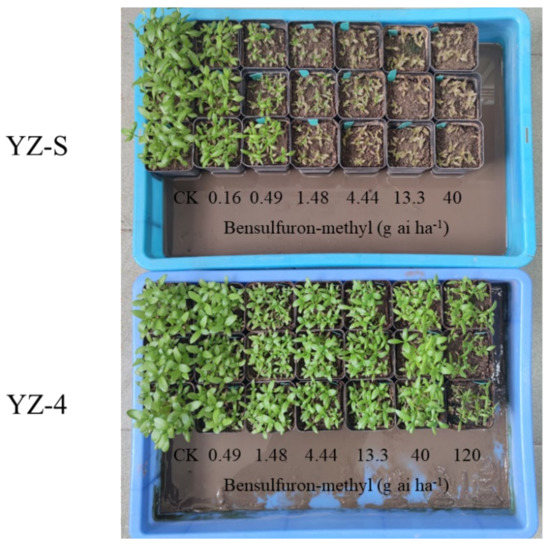
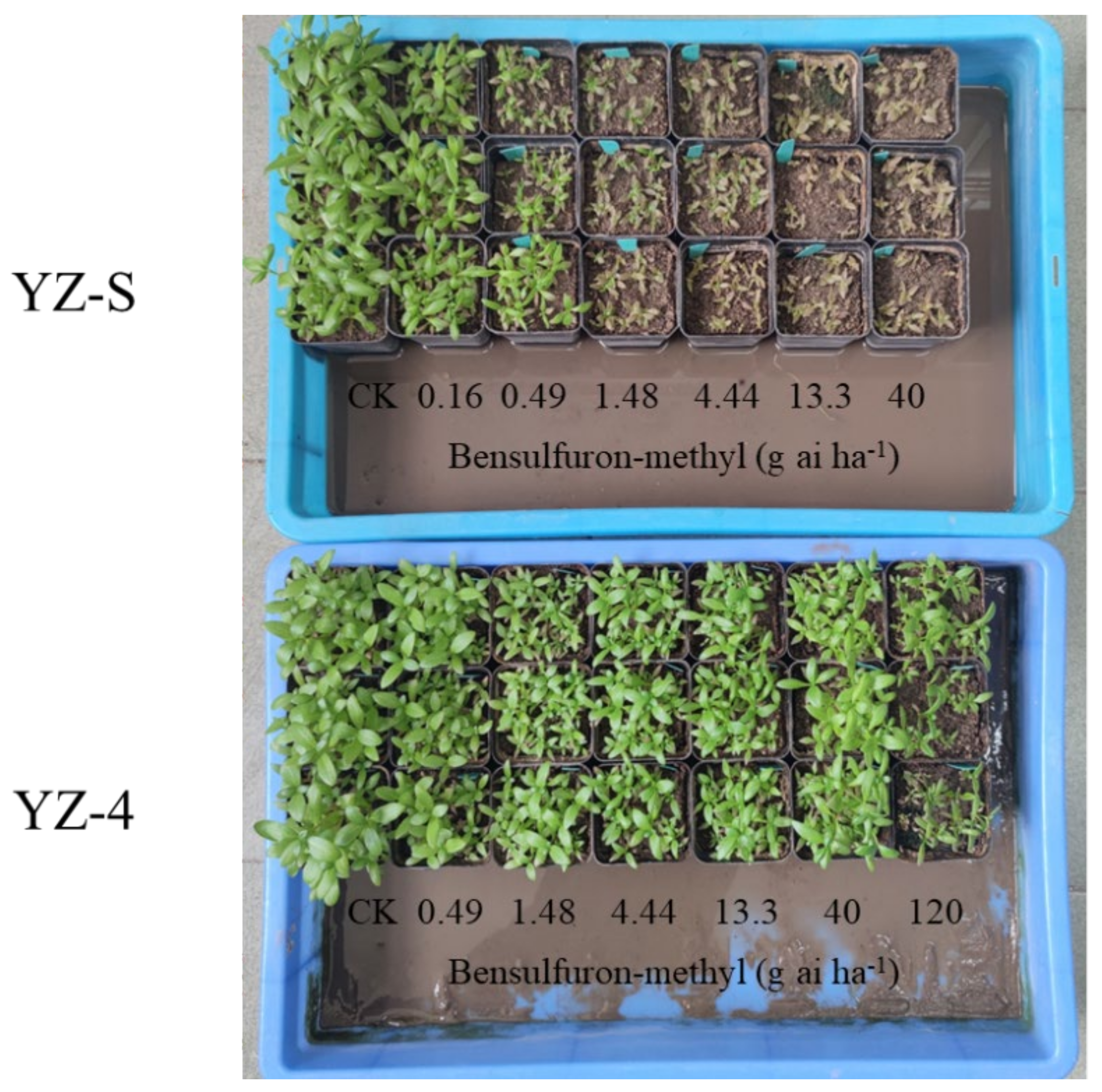
Figure 1.
Growth status of YZ-S and -4 A. auriculata plants 21 days after bensulfuron-methyl treatment.
2.2. Identification of ALS Gene Mutations
A direct sequencing of PCR products showed multiple peaks in certain bases of the chromatogram, indicating the existence of multiple ALS gene copies. Three different sequences (AaALS1, 2, and 3) were separated by cloning sequencing. The ALS gene sequences of nine resistant A. auriculata biotypes were compared with those of the susceptible biotypes. Four different ALS mutations (Pro197-Ser, -Leu, -Ala, and -His) in ALS genes were identified in eight resistant biotypes, and no ALS mutations known to endow resistance to ALS inhibitors were observed in the resistant YZ-3 biotype (Table 2, Figure 2). Interestingly, the frequency of resistance-endowing ALS mutations in AaALS1 was highest in nine tested biotypes, followed by 2, and no mutations in 3 were observed. These findings indicated that the target-site resistance was the main resistance mechanisms to bensulfuron-methyl, and the Pro197 mutations were very common in A. auriculata.

Table 2.
Codon and encoded amino acids at site of Pro197 in ALS genes in 11 A. auriculata biotypes.
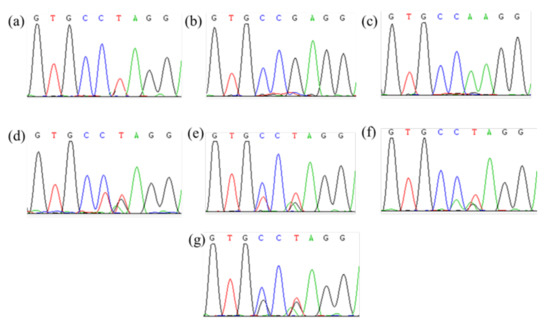
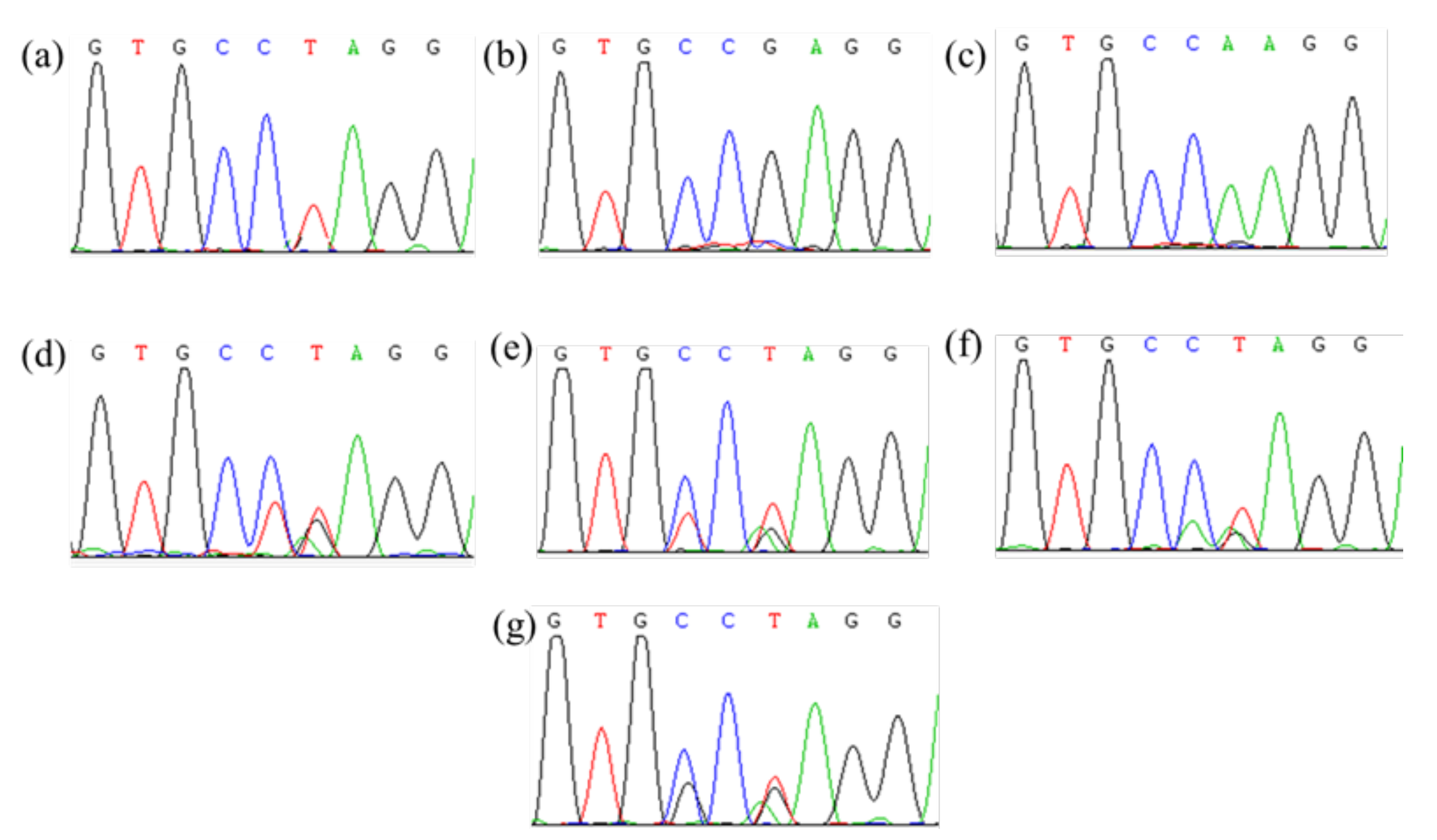
Figure 2.
Partial peak diagram of ALS sequencing containing Pro197 site. (a–c) Isolated three ALS genes; (d–g) four different amino acid mutations at Pro197.
2.3. CAPS Method for the Pro197 Mutations
The BanI restriction sequences were: 5′G↓GYRCC3′, and the two cytosine bases were the first two bases of codon 197. Thus, the A. auriculata plants with or without the Pro197 mutation can be distinguished by product bands after enzyme digestion. The wild-type ALS alleles were cut into two fragments of 125 and 200 bp, and the mutant alleles showed three bands of 125, 200, and 325 bp. The YZ-S and -3 biotypes with the wild-type ALS genes, and the YZ-2, -4, -8, and -9 biotypes with the mutant ALS genes, were selected for CAPS analysis. As expected, the products from YZ-S and -3 plants showed two bands, and products from plants with different Pro197 mutations showed three bands (Figure 3). The results of the CAPS analysis were consistent with the ALS gene sequencing results, indicating that the CAPS method can be used to accurately detect the Pro197 mutations in A. auriculata.
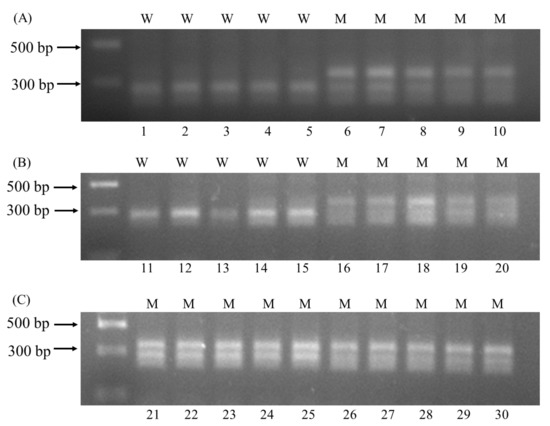
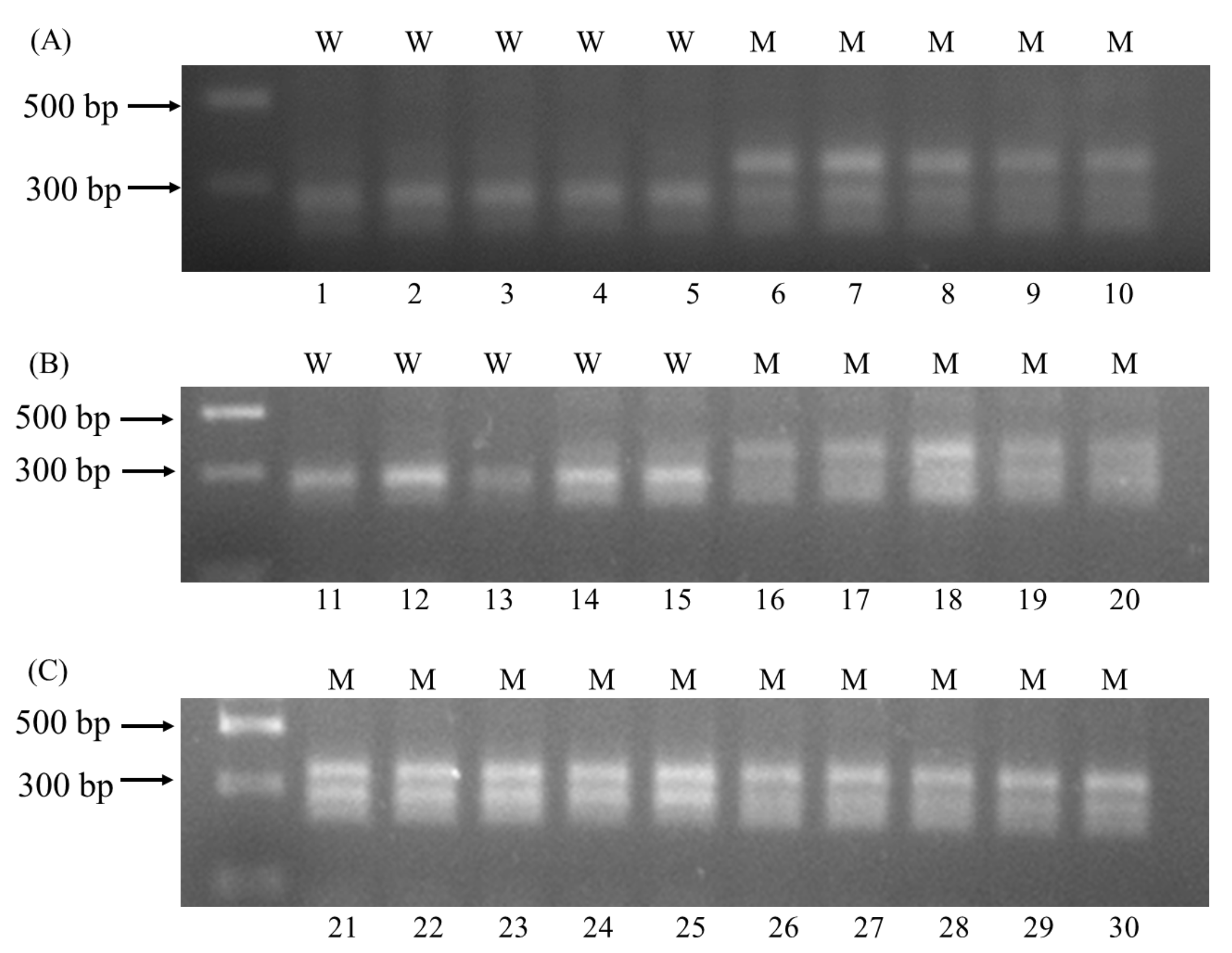
Figure 3.
CAPS analysis of 30 A. auriculata plants carrying wild-type ALS alleles (W) or mutant ALS alleles (M). (A) Lane 1–5 were samples of YZ-S, lane 6–10 were samples of YZ-2; (B) lane 11–15 were samples of YZ-3, lane 16–20 were samples of YZ-4; (C) lane 21–25 were samples of YZ-8, lane 26–30 were samples of YZ-9.
3. Discussion
The bensulfuron-methyl resistance levels of 10 A. auriculata biotypes from rice fields in Yangzhou City were investigated, and the results showed that 90% of the biotypes evolved resistance to this herbicide. This indicated that bensulfuron-methyl-resistant A. auriculata biotypes are widespread in rice-growing areas in China. Indeed, Wang et al. [28] reported that 96.6% of A. auriculata biotypes from the Ningshao plain in China were resistant to bensulfuron-methyl. Zhang et al. [30] found that four A. auriculata biotypes collected from Zhejiang, Jiangsu, and Anhui provinces exhibited medium to high resistance to bensulfuron-methyl. Bensulfuron-methyl has been used as a pre-emergence or post-emergence herbicide to control broadleaf weeds, including A. auriculata, for about 30 years in rice fields in China. The continuous herbicide application may be the driving force of rapid and widespread developments of resistance to bensulfuron-methyl in A. auriculata. Thus, herbicides with different sites of action, such as fluroxypyr or bentazon, are recommended as alternative options to control the resistant A. auriculata.
In this study, three ALS gene copies were identified in A. auriculata. Multiple ALS genes have been reported in many weed species. For example, Monochoria korsakowii and Monochoria vaginalis have three and five ALS gene copies, respectively [31]. Decurainia sophia possesses four ALS gene copies [32]. It is easy to miss the target-site mutations in a gene for weed species with multiple ALS genes. Thus, it is necessary to detect the sequences of all copies.
It is widely accepted that target-site mutations are key factors in most cases of ALS resistance [1]. In the present study, four different types of Pro197 mutations were determined in eight out of nine resistant A. auriculata biotypes. The results of ALS mutation identifications are strongly associated with those of resistance tests, which demonstrated that the Pro197 mutations in ALS play a major role in bensulfuron-methyl resistance in A. auriculata. In addition, the Pro197 mutations occurred in either AaALS1 or 2, but not in 3. Similar results were found in other resistant weeds. Xu et al. [32] reported that target-site mutations had a preference for ALS1 or 2 in four ALS gene copies in D. sophia. Tanigali et al. [33] found that the resistant mutations exclusively occurred in ALS1 or 3 in M. vaginalis. In our previous study, amino acid mutations in ALS1 or 2 independently, or in both, were found in Ammannia multiflora [34]. The preference of resistant mutations may be related to gene expression levels, herbicide use, and weed species. Interestingly, we found that a A. auriculata biotype (YZ-3) showed significant resistance to bensulfuron-methyl, but no resistance-conferring mutations were detected in ALS genes, suggesting that other resistance mechanisms may be involved in this case. A further study will investigate the potential resistance mechanisms, such as metabolic resistance.
Several molecular techniques, including PCR-RFLP, CAPS, dCAPS, and LAMP, have been developed for the diagnosis of target-site gene mutations [35,36,37,38,39,40]. Compared with the time-consuming whole-plant response bioassays, these molecular detection methods are rapid, cheap, and reliable, and thus suitable for resistance detections of hundreds of samples. In our study, a CAPS method was established to screen the Pro197 mutations in ALS genes in A. auriculata. The Pro197-Ser, -Leu, -His, and -Ala mutations in AaALS1 or 2 can be detected without DNA sequencing. The downside was that plants with non-target-site resistance cannot be detected by this method.
4. Materials and Methods
4.1. Plant Materials
We collected seed material of A. auriculata from rice fields in Yangzhou City in October 2019. Each sampling location was more than 5 kilometers apart. We collected the known susceptible A. auriculata biotype (YZ-S) from experimental fields with no history of herbicide use. We harvested the 10 suspected resistant A. auriculata biotypes from rice fields in which bensulfuron-methyl was used for over 15 years (Table 3).

Table 3.
Geographical origin of 11 A. auriculata populations collected from paddy fields.
4.2. Whole-Plant Response to Bensulfuron-Methyl
We carried whole-plant response assays out to determine the GR50 reaction (herbicide dose causing 50% plant-growth inhibition) of 11 A. auriculata biotypes with bensulfuron-methyl. We cultivated A. auriculata plants in a glasshouse during the growing seasons (May–July; 35/25℃). We used seedlings at four-leaf stage for dose response assays. We applied bensulfuron-methyl (10% wettable powder; Jiangsu Dongbao agrochemical Co., Ltd., Yangzhou, China) at various rates of 0, 0.055, 0.16, 0.49, 1.48, 4.44, 13.3, 40, 120, and 360 g ai ha−1 using a cabinet sprayer with a fan nozzle delivering 450 L ha−1 at 0.2 MPa. We returned plants to the glasshouse after herbicide treatment. We measured the fresh weight of above-ground part 21 d after treatment. We conducted each treatment in a random design with three replications, and each replication contained at least 12 A. auriculata plants. We repeated the experiment twice.
4.3. ALS Gene Sequencing
We isolated the total RNA from the leaf samples of A. auriculata biotypes using the RNAprep Plant Kit (Tiangen, Beijing, China), and we reverse transcribed into cDNA using the Fast RT Kit (Tiangen, Beijing, China). We designed two primer pairs (F1: 5′-GGACATCCTCGTGGAGGC-3′/R1: 5′-TTGTTCTTTCCAATTTCAGC-3′ and F2: 5′-TGTTCAGGAGTTAGCCACGATCAGG-3′/R2: 5′-TTAGTAAGATGTCCGCCCGTCACCC-3′) to amplify the fragment of ALS genes. We conducted the PCR reaction in a volume of 50 µL that contained 25 µL of 2×Taq Plus PCR Mix, 1 µL of each primer, 2 µL of cDNA, and 21 µL of ddH2O. We ran the PCR using the following cycles: 94℃ for 5 min, 35 cycles of 94 ℃ for 30 s, 58 ℃ (F1/R1) or 62 ℃ (F2/R2) for 30 s, and 72 ℃ for 40 to 60 s, followed by 72 ℃ elongation for 10 min. We purified and sequenced the PCR products from both ends with above amplification primers by commercial services (Sangon Biotech, China). Because there were three ALS gene copies in A. auriculata, for samples with ALS mutations detected by directly sequencing, we cloned the amplified fragments into the pGM-T vector to distinguish which ALS genes were mutated. We randomly selected a total of 10 plants from each A. auriculata biotype for identification of ALS mutations.
4.4. CAPS Detection for Pro197 Mutations
We used a total of 30 plants from 6 A. auriculata biotypes (YZ-S, -2, -3, -4, -8, and -9) for CAPS genotyping. We designed primers (F: 5′-GGTCAGCGGCCTCGCC-3′/R: 5′-TGGCTTGGGCAACCTGGA-3′) to amplify the ALS gene fragments (325 bp) containing the Pro197 site. We performed the procedures in the same PCR conditions described above. Then, we used the amplified products for enzyme digestion. We carried the digestion reaction out in a 50 µL volume containing 1 µL of restriction enzyme BanI (NEB, USA), 5 µL of 10×NEBuffer, 10 µL of PCR product, and 34 µL of ddH2O. We ran the reactions at 37 ℃ for 60 min, and inactivated them at 65 ℃ for 10 min. After that, we visualized the digestion products by electrophoresis on 2% agarose gels at 110 V for 50 min.
4.5. Statistical Analysis
In the whole-plant response experiments, we converted the data of fresh weight into a percentage of the nontreated control. We evaluated the GR50 values using the equation: y = C + (D - C) / [1 + (x / GR50) b] [41]. The resistance factor (RF) was expressed by the GR50 of the resistant A. auriculata biotypes divided by the GR50 of the susceptible biotypes.
5. Conclusions
In conclusion, 9 out of 10 A. auriculata biotypes showed a high level of resistance to bensulfuron-methyl. Herbicides with different sites of action should be used to control the resistant A. auriculata. A. auriculata plants possess three ALS gene copies, and resistance-conferring Pro197 mutations were found in AaALS1 and 2. Moreover, we established the CAPS method to detect the Pro197-Ser, -Leu, -Ala, and -His mutations in A. auriculata.
Author Contributions
Conceptualization, W.D. and S.Y.; methodology, W.D.; software, Y.L.; validation, W.D. and S.Y.; formal analysis, Z.D.; investigation, L.L.; resources, P.W.; data curation, L.L.; writing—original draft preparation, W.D.; writing—review and editing, S.Y.; visualization, C.P.; supervision, S.Y.; project administration, W.D.; funding acquisition, W.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Agricultural Science and Technology Innovation Fund (CX (20) 3131), the National Natural Science Foundation of China (32001925), and the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (19KJB210005).
Institutional Review Board Statement
Not appliable.
Informed Consent Statement
Not appliable.
Data Availability Statement
The data are available on request from the corresponding author.
Acknowledgments
We thank Qian Yang from the Institute of Agricultural Sciences for Lixiahe Region in Jiangsu for assistance with the sample.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Heap, I. The International Herbicide-Resistant Weed Database. Available online: http://www.weedscience.org (accessed on 30 June 2022).
- Powles, S.B.; Yu, Q. Evolution in action: Plants resistant to herbicides. Annu. Rev. Plant Biol. 2010, 61, 317–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Délye, C.; Jasieniuk, M.; Le Corre, V. Deciphering the evolution of herbicide resistance in weeds. Trends Genet. 2013, 29, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Murphy, B.P.; Tranel, P.J. Target-site mutations conferring herbicide resistance. Plants 2019, 8, 382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.; Wu, Y.; Liu, H. Research progress of sulfonylurea herbicides. Mod. Agrochem. 2022, 21, 14–21. [Google Scholar]
- Ray, T.B. Site of action of chlorsulfuron: Inhibition of valine and isoleucine biosynthesis in plants. Plant Physiol. 1984, 75, 827–831. [Google Scholar] [CrossRef] [Green Version]
- Duggleby, R.G.; McCourt, J.A.; Guddat, L.W. Structure and mechanism of inhibition of plant acetohydroxyacid synthase. Plant Physiol. Biochem. 2008, 46, 309–324. [Google Scholar] [CrossRef]
- Mazur, B.J.; Falco, S.C. The development of herbicide resistant crops. Ann. Rev. Plant Biol. 1989, 40, 441–470. [Google Scholar] [CrossRef]
- Tranel, P.J.; Wright, T.R. Resistance of weeds to ALS-inhibiting herbicides: What have we learned? Weed Sci. 2002, 50, 700–712. [Google Scholar] [CrossRef]
- Fang, J.; Yang, D.; Zhao, Z.; Chen, J.; Dong, L. A novel Phe-206-Leu mutation in acetolactate synthase confers resistance to penoxsulam in barnyardgrass (Echinochloa crusgalli (L.) P. Beauv). Pest Manag. Sci. 2022, 78, 2560–2570. [Google Scholar] [CrossRef]
- Tranel, P.J.; Wright, T.R.; Heap, I.M. Mutations in Herbicide-Resistant Weeds to ALS Inhibitors. Available online: https://www.weedscience.org/mutations/mutationdisplayall.aspx (accessed on 30 June 2022).
- Veldhuis, L.J.; Hall, L.M.; O’Donovan, J.T.; Dyer, W.; Hall, J.C. Metabolism-based resistance of a wild mustard (Sinapis arvensis L.) biotype to ethametsulfuron-methyl. J. Agri. Food Chem. 2000, 48, 2986–2990. [Google Scholar] [CrossRef]
- Owen, M.J.; Goggin, D.E.; Powles, S.B. Non-target-site-based resistance to ALS-inhibiting herbicides in six Bromus rigidus populations from Western Australian cropping fields. Pest Manag. Sci. 2012, 68, 1077–1082. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Riggins, C.W.; Hausman, N.E.; Hager, A.G.; Riechers, D.E.; Davis, A.S.; Tranel, P.J. Nontarget-site resistance to ALS inhibitors in waterhemp (Amaranthus tuberculatus). Weed Sci. 2015, 63, 399–407. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.; Fu, D.; Yu, Y.; Huang, C.; Yan, K.; Li, P.; Shafi, J.; Zhu, H.; Wei, S.; Ji, M. Non-target-site resistance to ALS-inhibiting herbicides in a Sagittaria trifolia L. population. Pestic. Biochem. Physiol. 2017, 140, 79–84. [Google Scholar] [CrossRef]
- Cao, S.; Zhao, B.; Zou, Y.; Sun, Z.; Zhang, H.; Wei, S.; Ji, M. P450s mediated enhanced herbicide metabolism involved in the thifensulfuron-methyl resistance in Ipomoea purpurea L. Pestic. Biochem. Physiol. 2022, 184, 105111. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.; Zhou, X.; Lin, S.; Cao, Y.; Wei, S.; Huang, H.; Li, Y.; Huang, Z. Pro-197-Ser mutation and cytochrome P450-mediated metabolism conferring resistance to flucarbazone-sodium in Bromus japonicus. Plants 2022, 11, 1641. [Google Scholar] [CrossRef]
- Délye, C.; Pernin, F.; Scarabel, L. Evolution and diversity of the mechanisms endowing resistance to herbicides inhibiting acetolactate-synthase (ALS) in corn poppy (Papaver rhoeas L.). Plant Sci. 2011, 180, 333–342. [Google Scholar] [CrossRef]
- Rey-Caballero, J.; Menéndez, J.; Osuna, M.D.; Salas, M.; Torra, J. Target-site and non-target-site resistance mechanisms to ALS inhibiting herbicides in Papaver rhoeas. Pestic. Biochem. Physiol. 2017, 138, 57–65. [Google Scholar] [CrossRef] [Green Version]
- Nakka, S.; Thompson, C.R.; Peterson, D.E.; Jugulam, M. Target site–based and non–target site based resistance to ALS inhibitors in palmer amaranth (Amaranthus palmeri). Weed Sci. 2017, 65, 681–689. [Google Scholar] [CrossRef]
- Yang, Q.; Deng, W.; Li, X.; Yu, Q.; Bai, L.; Zheng, M. Target-site and non-target-site based resistance to the herbicide tribenuron-methyl in flixweed (Descurainia sophia L.). BMC Genom. 2016, 17, 551–563. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.L.; McCullough, P.K.; McElroy, J.S.; Jespersen, D.; Shilling, D.G. Gene expression and target-site mutations are associated with resistance to ALS inhibitors in annual sedge (Cyperus compressus) biotypes from Georgia. Weed Sci. 2020, 68, 460–466. [Google Scholar] [CrossRef]
- Sen, M.K.; Hamouzová, K.; Mikulka, J.; Bharati, R.; Košnarová, P.; Hamouz, P.; Royc, A.; Soukupa, J. Enhanced metabolism and target gene overexpression confer resistance against acetolactate synthase-inhibiting herbicides in Bromus sterilis. Pest Manag. Sci. 2021, 77, 2122–2128. [Google Scholar] [CrossRef]
- Lu, Z.; Zhang, C.; Fu, J.; Li, M.; Li, G. Molecular basis of resistance to bensulfuron-methyl in Monochoria korsakowii. Sci. Agric. Sin. 2009, 42, 3516–3521. [Google Scholar]
- Li, D.; Li, X.J.; Yu, H.L.; Wang, J.J.; Cui, H.L. Cross-resistance of eclipta (Eclipta prostrata) in China to ALS inhibitors due to a Pro-197-Ser point mutation. Weed Sci. 2017, 65, 547–556. [Google Scholar] [CrossRef]
- Deng, W.; Yang, M.; Duan, Z.; Peng, C.; Xia, Z.; Yuan, S. Molecular basis of resistance to bensulfuron-methyl and cross-resistance patterns to ALS-inhibiting herbicides in Ludwigia prostrata. Weed Technol. 2021, 35, 656–661. [Google Scholar] [CrossRef]
- Li, Z.; Li, X.; Chen, J.; Peng, L.; Wang, J.; Cui, H. Variation in mutations providing resistance to acetohydroxyacid synthase inhibitors in Cyperus difformis in China. Pestic. Biochem. Physiol. 2020, 166, 104571. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, Q.; Zhu, J.; Liu, R.; Wang, S.; Liu, Y.; Lu, Q.; Wang, G. Resistance comparison of Ammannia arenaria to bensulfuron-methyl in different paddy rice growing regions of Zhejiang province. Chin. J. Pestic. Sci. 2013, 15, 52–58. [Google Scholar]
- Beckie, H.J.; Tardif, F.J. Herbicide cross resistance in weeds. Crop Prot. 2012, 35, 15–28. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, B.; Cai, X.; Zhou, W.; Wang, H.; Lu, Q.; Zhou, G.; Liu, Y.; Liang, W.; Wang, S.; et al. Resistance and its resistant molecular mechanism of Ammannia arenaria to ALS inhibiting herbicides. Chin. J. Pestic. Sci. 2020, 22, 60–67. [Google Scholar]
- Iwakami, S.; Tanigaki, S.; Uchino, A.; Ozawa, Y.; Tominaga, T.; Wang, G.X. Characterization of the acetolactate synthase gene family in sensitive and resistant biotypes of two tetraploid Monochoria weeds, M. vaginalis and M. korsakowii. Pestic. Biochem. Physiol. 2020, 165, 104506. [Google Scholar] [CrossRef]
- Xu, Y.; Xu, L.; Li, X.; Zheng, M. Investigation of resistant level to tribenuron-methyl, diversity and regional difference of the resistant mutations on acetolactate synthase (ALS) isozymes in Descurainia sophia L. from China. Pestic. Biochem. Physiol. 2020, 169, 104653. [Google Scholar] [CrossRef]
- Tanigaki, S.; Uchino, A.; Okawa, S.; Miura, C.; Hamamura, K.; Matsuo, M.; Yoshino, N.; Ueno, N.; Toyama, Y.; Fukumi, N.; et al. Gene expression shapes the patterns of parallel evolution of herbicide resistance in the agricultural weed Monochoria vaginalis. New Phytol. 2021, 232, 928–940. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Duan, Z.; Li, Y.; Cui, H.; Peng, C.; Yuan, S. Characterization of target-site resistance to ALS-inhibiting herbicides in Ammannia multiflora populations. Weed Sci. 2022, 70, 292–297. [Google Scholar] [CrossRef]
- Yu, Q.; Han, H.; Powles, S.B. Mutations of the ALS gene endowing resistance to ALS-inhibiting herbicides in Lolium rigidum populations. Pestic. Sci. 2008, 64, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Kaloumenos, N.S.; Dordas, C.A.; Diamantidis, G.C.; Eleftherohorinos, I.G. Multiple Pro197 substitutions in the acetolactate synthase of corn poppy (Papaver rhoeas) confer resistance to tribenuron. Weed Sci. 2009, 57, 362–368. [Google Scholar] [CrossRef]
- Deng, W.; Liu, M.J.; Yang, Q.; Mei, Y.; Li, X.F.; Zheng, M.Q. Tribenuron-methyl resistance and mutation diversity of Pro197 in flixweed (Descurainia sophia L.) accessions from China. Pestic. Biochem. Physiol. 2015, 117, 68–74. [Google Scholar] [CrossRef]
- Pan, L.; Li, J.; Xia, W.; Zhang, D.; Dong, L. An effective method, composed of LAMP and dCAPS, to detect different mutations in fenoxaprop-P-ethyl-resistant American sloughgrass (Beckmannia syzigachne Steud.) populations. Pestic. Biochem. Physiol. 2015, 117, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Li, J.; Zhang, W.N.; Dong, L. Detection of the I1781L mutation in fenoxaprop-p-ethyl-resistant American sloughgrass (Beckmannia syzigachne Steud.), based on the loop-mediated isothermal amplification method. Pest Manag. Sci. 2015, 71, 123–130. [Google Scholar] [CrossRef]
- Peng, Y.; Pan, L.; Liu, D.; Cheng, X.; Ma, G.; Li, S.; Liu, X.; Wang, L.; Bai, L. Confirmation and characterization of cyhalofop-butyl–resistant Chinese sprangletop (Leptochloa chinensis) populations from China. Weed Sci. 2020, 68, 253–259. [Google Scholar] [CrossRef]
- Seefeldt, S.S.; Jensen, J.E.; Fuerst, E.P. Log-logistic analysis of herbicide dose response relationships. Weed Technol. 1995, 9, 218–227. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).