Distribution and Climatic Adaptation of Wild Tomato (Solanum lycopersicum L.) Populations in Mexico
Abstract
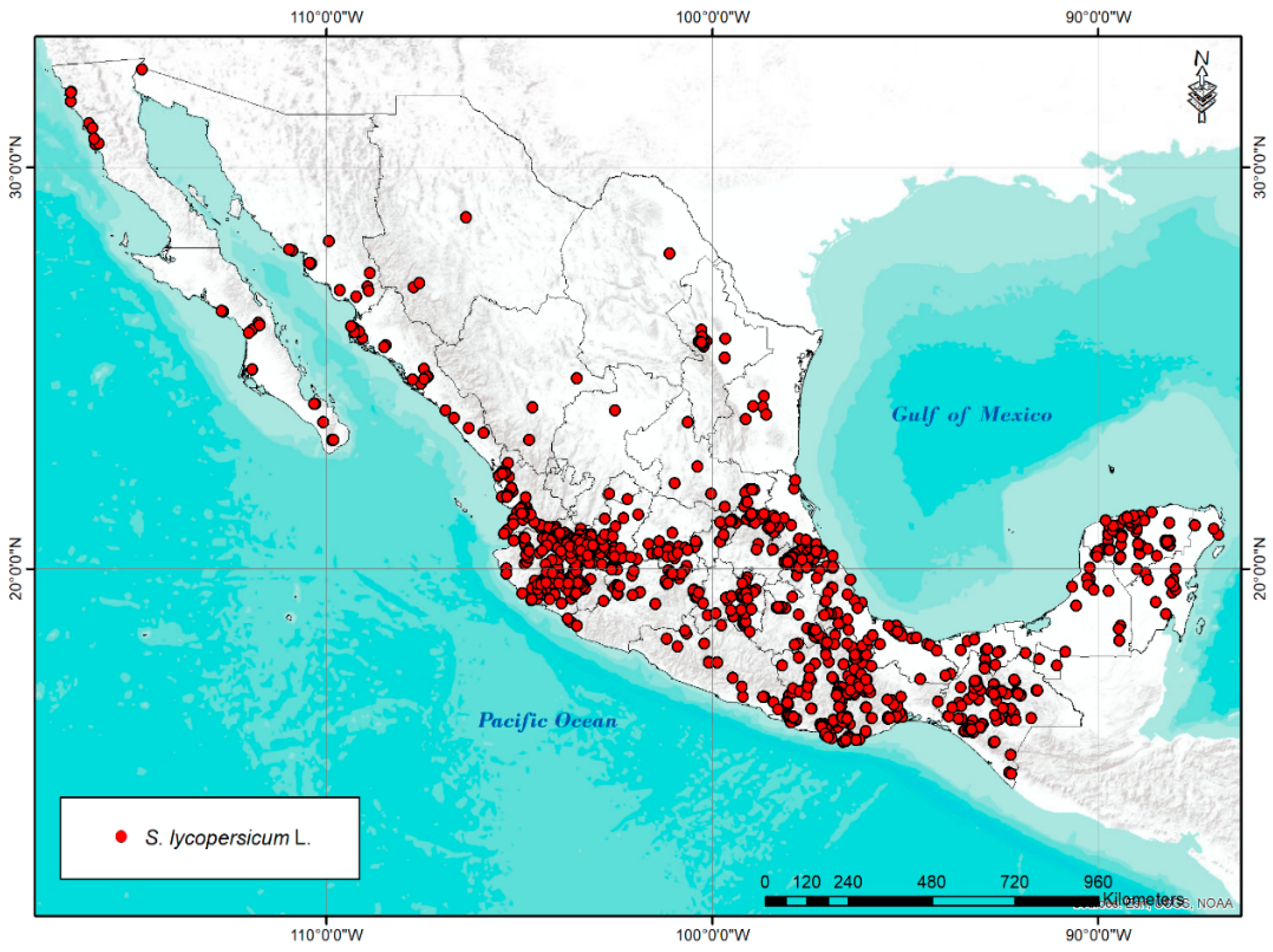
1. Introduction
2. Results
2.1. Statistical Analyses and Ecological Descriptors
2.2. Climatic Diversity and Hotspot Analysis
2.3. Potential Distribution
3. Discussion
4. Materials and Methods
4.1. Database
4.2. Environmental Information
4.3. Statistical Analysis and Ecological Descriptors
4.4. Climate Diversity and Hotspot Analysis
4.5. Potential Distribution
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peralta, I.E.; Spooner, D.M.; Knapp, S. Taxonomy of wild tomatoes and their relatives (Solanum sect. Lycopersicoides, sect. Juglandifolia, sect. Lycopersicon; Solanaceae). Syst. Bot. Monogr. 2008, 84, 1–186. [Google Scholar]
- Chávez-Servia, J.L.; Vera-Guzmán, A.M.; Carrillo-Rodríguez, J.C.; Pachecho-Triste, I.A.; Crisanto-Juárez, A.U. Variación morfológica y de licopeno en frutos de tomate semidomesticado y cultivado en Oaxaca, México. J. Int. Am. Soc. Trop. Agric. Hortic. 2011, 54, 151–153. [Google Scholar]
- Rick, C.M.; Fobes, J.F. Allozyme variation in the cultivated tomato and closely related species. Bull. Torrey Bot. Club 1975, 102, 376–384. [Google Scholar] [CrossRef]
- Peralta, I.E.; Spooner, D.M. Morphological characterization and relationships of wild tomatoes (Solanum L. section Lycopersicon [Mill.]). In A Festschrift for William, G. D’Arcy: The Legacy of a Taxonomist; Keating, R.C., Hollowell, V.A., Croat, T.B., Eds.; Missouri Botanical Garden Press: St. Louis, MO, USA, 2005; pp. 227–257. [Google Scholar]
- Blanca, J.; Cañizares, J.; Cordero, L.; Pascual, L.; Diez, M.J.; Nuez, F. Variation revealed by SNP genotyping and morphology provides insight into the origin of the tomato. PLoS ONE 2012, 7, e48198. [Google Scholar] [CrossRef]
- Blanca, J.; Montero-Pau, J.; Sauvage, C.; Bauchet, G.; Illa, E.; Díez, M.J.; Francis, D.; Causse, M.; van der Knaap, E.; Cañizares, J. Genomic variation in tomato, from wild ancestors to contemporary breeding accessions. BMC Genom. 2015, 16, 257. [Google Scholar] [CrossRef]
- Razifard, H.; Ramos, A.; Valle, A.L.D.; Bodary, C.; Goetz, E.; Manser, E.J.; Li, X.; Zhang, L.; Visa, S.; Tieman, D.; et al. Evidence for complex domestication history of the cultivated tomato in Latin America. Mol. Biol. Evol. 2020, 37, 1118–1132. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Guzmán, E.; Vargas-Canela, D.; de Sánchez-González, J.J.; Lépiz-Idelfonso, R.; Rodríguez-Contreras, A.; Ruiz-Corral, J.A.; Puente-Ovalle, P.; Miranda-Medrano, R. Etnobotánica de Solanum var cerasiforme en el occidente de México. Nat. Desarro. 2009, 7, 45–57. [Google Scholar]
- Muller, C.H. A revision of the genus Lycopersicon. USDA Misc. Publ. 1940, 328, 29. [Google Scholar]
- Moyle, C.L. Ecological and evolutionary genomics in the wild tomatoes (Solanum sect. Lycopersicon). Evolution 2008, 62, 2995–3013. [Google Scholar] [CrossRef] [PubMed]
- Pacheco-Triste, I.A.; Chávez-Servia, J.L.; Carrillo-Rodríguez, J.C. Relación entre variación ecológica- orográfica y variabilidad morfológica de tomate (Solanum lycopersicum L.) en Oaxaca. Rev. Mex. Agroecosistemas 2014, 1, 28–39. [Google Scholar]
- Délices, G.; Leyva, O.; Mota-Vargas, C.; Núñez, P.R.; Gámez, P.R.; Andrés, M.P.; Serna-Lagunes, R. Biogeografía del tomate Solanum lycopersicum var, cerasiforme (Solanaceae) en su centro de origen (sur de América) y de domesticación (México). Rev. Biol. Trop. 2019, 67, 1023–1036. [Google Scholar] [CrossRef]
- Ladizinsky, G. Plant Evolution under Domestication; Kluwer Academic: Dordrecht, The Netherlands, 1998; p. 262. [Google Scholar]
- Flores-Hernández, L.A.; Lobato-Ortíz, R.; Sangerman-Jarquín, D.M.; García-Zavala, J.J.; Molina-Galán, J.D.; Velasco-Alvarado, M.J.; Marín-Montes, I.M. Genetic diversity within wild species of Solanum. Rev. Chapingo Ser. Hortic. 2018, 24, 85–96. [Google Scholar] [CrossRef]
- Zamir, D. Improving plant breeding with exotic genetic libraries. Nat. Rev. Genet. 2001, 2, 983–989. [Google Scholar] [CrossRef] [PubMed]
- Bao, B.; Ke, L.; Jiang, J.; Ying, T. Fruit quality of transgenic tomatoes with suppressed expression of LeETR1 and LeETR2 genes. Asia Pac. J. Clin. Nutr. 2007, 16, 122–126. [Google Scholar]
- Flores-Hernández, L.A.; Lobato-Ortiz, R.; García-Zavala, J.J.; Molina-Galán, J.D.; Sangerman-Jarquín, D.M.; Velazco-Alvarado, M.J. Parientes silvestres del tomate como fuente de germoplasma para el mejoramiento genético de la especie. Rev. Fitotec. Mex. 2017, 40, 83–91. [Google Scholar] [CrossRef]
- Perrino, E.V.; Wagensommer, R.P. Crop wild relatives (CWRs) threatened and endemic to Italy: Urgent actions for protection and use. Biology 2022, 11, 193. [Google Scholar] [CrossRef]
- Marín-Montes, I.M.; Lobato-Ortiz, R.; Carrillo-Castañeda, G.; Rodríguez-Pérez, J.E.; García-Zavala, J.J.; Velasco-García, A.M. Alellic richness of native tomato populations (Solanum lycopersicum L.) for plant breeding. Agrociencia 2019, 53, 355–370. [Google Scholar]
- Cervantes-Moreno, R.; Rodríguez-Pérez, J.E.; Carrillo-Fonseca, C.; Sahagún-Castellanos, J.; Rodríguez-Guzmán, E. Tolerancia de 26 colectas de tomates nativos de México al nemátodo Meloidogyne incognita (Kofoid y White) Chitwood. Rev. Chapingo Ser. Hortic. 2014, 20, 5–18. [Google Scholar] [CrossRef]
- Vela-Hinojosa, C.; Escalona-Buendía, H.B.; Mendoza-Espinoza, J.A.; Villa-Hernández, J.M.; Lobato-Ortiz, R.; Rodríguez-Pérez, J.E.; Pérez-Flores, L.J. Antioxidant balance and regulation in tomato genotypes of different color. J. Am. Soc. Hortic. Sci. 2019, 144, 45–54. [Google Scholar] [CrossRef]
- Juárez-López, P.; Castro-Brindis, R.; Colinas-León, T.; Ramírez-Vallejo, P.; Sandoval-Villa, M.; Reed, D.W.; Cisneros-Zevallos, L.; King, S. Evaluación de calidad de frutos de siete genotipos nativos de tomate (Lycopersicon esculentum var. cerasiforme). Rev. Chapingo Ser. Hortic. 2009, 15, 5–9. [Google Scholar]
- Crisanto-Juárez, A.; Vera-Guzmán, A.M.; Chávez-Servia, J.L.; Carrillo-Rodríguez, J.C. Calidad de frutos de tomates silvestres (Lycopersicon esculentum var. cerasiforme Dunal) de Oaxaca. México. Rev. Fitotec. Mex. 2010, 33, 7–13. [Google Scholar]
- Vela-Hinojosa, C.; Barbosa-Martínez, C.; Escalona-Buendía, H.B.; Mendoza-Espinoza, J.A.; Lobato-Ortiz, R.; Rodríguez-Pérez, J.E.; Villa-Hernández, J.M.; Pérez-Flores, L.J. Architectural diversity of the cuticle and epidermis of native and hybrid tomato fruit genotypes and the relation to polygalacturonase expression. Not. Bot. Horti Agrobot. 2018, 46, 45–51. [Google Scholar] [CrossRef]
- Magallanes-López, A.M.; Martínez-Damián, M.T.; Sahagún-Castellanos, J.; Pérez-Flores, L.J.; Marín-Montes, I.M.; Rodríguez-Pérez, J.E. Post-harvest quality of 40 collections of tomato (Solanum lycopersicum L.) native of Mexico. Agrociencia 2020, 54, 779–795. [Google Scholar] [CrossRef]
- Velázquez, A.; Bocco, G.; Torres, A. Turning scientific approaches into practical conservation actions: The case of comunidad indigena de Nuevo San Juan Parangaricutiro, Mexico. Environ. Manag. 2001, 27, 655–665. [Google Scholar] [CrossRef]
- Rosete, F.; Bocco, G. Los sistemas de información geográfica y la percepción remota. Herramientas integradas para los planes de manejo en comunidades forestales. Gac. Ecológica 2003, 68, 43–54. [Google Scholar]
- Sánchez-González, J.; Ruiz-Corral, J.A.; García-Medina, G.; Ramírez-Ojeda, G.; Larios, L.D.L.C.; Holland, J.B. Ecogeography of teosinte. PLoS ONE 2018, 13, e0192676. [Google Scholar] [CrossRef] [PubMed]
- Beck, H.E.; Zimmermann, N.E.; Mcvicar, T.R.; Vergopolan, N.; Berg, A.; Wood, E.F. Present and future Köppen-Geiger climate classification maps at 1-km resolution. Sci. Data 2018, 5, 180214. [Google Scholar] [CrossRef] [PubMed]
- Phillips, S.J.; Dudík, M.; Schapire, R.E. Maxent Software for Modeling Species Niches and Distributions (Version 3.4.1). Available online: http://biodiversityinformatics.amnh.org/open_source/maxent/ (accessed on 24 February 2022).
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Nee, M. Solanaceae I (III). In Flora de Veracruz. 1986 Fascículo 49; Sosa, V., Ed.; Instituto de Ecología: Xalapa, Veracruz, México, 1986. [Google Scholar]
- Perrino, E.V.; Wagensommer, R.P. Crop Wild Relatives (CWR) Priority in Italy: Distribution, Ecology, In Situ and Ex Situ Conservation and Expected Actions. Sustainability 2021, 13, 1682. [Google Scholar] [CrossRef]
- Intergovernmental Panel on Climate Change (IPCC). Climate Change 2007: Mitigation of Climate Change. Contribution of Working Group III to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change; Metz, B., Davidson, O., Bosch, P., Dave, R., Meyer, L., Eds.; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2007; p. 851. [Google Scholar]
- Ramírez-Ojeda, G.; Ruiz-Corral, J.A.; Pérez-Mendoza, C.; Villavicencio-García, R.; Mena-Munguía, S.; Durán-Puga, N. Impactos del cambio climático en la distribución geográfica de Gossypium hirsutum L. en México. Rev. Mex. Cienc. Agríc. 2014, 5, 1885–1895. [Google Scholar]
- Organization of the United Nations Food and Agriculture Organization (FAO). Adaptation to Climate Change in Agriculture, Forestry and Fisheries: Perspective, Framework and Priorities. FAO Interdepartmental Working Group on Climate Change; FAO Electronic Publication: Rome, Italy, 2007; p. 24. [Google Scholar]
- Roland, T.; Zumbo, B.D.; Kwan, E.; Schweitzer, L. On Johnson’s (2000) relative weights method for assessing variable importance: A reanalysis. Multivar. Behav. Res. 2014, 49, 329–338. [Google Scholar] [CrossRef]
- Ramírez-Ojeda, G.; Peralta, I.E.; Rodríguez-Guzmán, E.; Chávez-Servia, J.L.; Sahagún-Castellanos, J.; Rodríguez-Pérez, J.E. Climatic diversity and ecological descriptors of wild tomato species (Solanum sect. Lycopersicon) and close related species (Solanum sect. Juglandifolia y sect. Lycopersicoides) in Latin America. Plants 2021, 10, 855. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Ojeda, G.; Peralta, I.E.; Rodríguez-Guzmán, E.; Sahagún-Castellanos, J.; Chávez-Servia, J.L.; Medina-Hinostroza, T.C.; Rijalba-Vela, J.R.; Vásquez-Núñez, L.P.; Rodríguez-Pérez, J.E. Edaphoclimatic descriptors of wild tomato species (Solanum sect. Lycopersicon) and closely related species (Solanum sect. Juglandifolia and Sect. Lycopersicoides) in South America. Front. Genet. 2021, 12, 748979. [Google Scholar] [CrossRef] [PubMed]
- Maldonado-Peralta, R.; Ramírez-Vallejo, P.; González-Hernández, V.A.; Castillo-González, F.; Sandoval-Villa, M.; Livera-Muñoz, M.; Cruz-Huerta, N. Riqueza agronómica en colectas mexicanas de tomates (Solanum lycopersicum L.) nativos. Agroproductividad 2016, 9, 68–75. [Google Scholar]
- Velasco-Alvarado, M.J.; Lobato-Ortiz, R.; García-Zavala, J.J.; Castro-Brindis, R.; Cruz-Izquierdo, S.; Corona-Torres, T.; Moedano-Mariano, M.K. Mexican native tomatoes as rootstocks to increase fruit yield. Chilean. J. Agric. Res. 2017, 77, 187–193. [Google Scholar] [CrossRef][Green Version]
- Arellano-Rodríguez, L.J.; Rodríguez-Guzmán, E.; Ron-Parra, J.; Martínez-Ramírez, J.L.; Lozoya-Saldaña, H.; Sánchez-Martínez, J.; Lépiz-Idelfonso, R. Evaluación de resistencia a Phytophthora en poblaciones silvestres de Solanum lycopersicum var. cerasiforme. Rev. Mex. Cien. Agríc. 2013, 4, 753–766. [Google Scholar]
- Bonilla-Barrientos, O.; Lobato-Ortíz, R.; García-Zavala, J.J.; Cruz-Izquierdo, S.; Reyes-López, D.; Hernández-Leal, E.; Hernández-Bautista, A. Diversidad agronómica y morfológica de tomates arriñonados y tipo pimiento de uso local en Puebla y Oaxaca, Mexico. Rev. Fitotec. Mex. 2014, 37, 129–139. [Google Scholar] [CrossRef]
- Elith, J.; Phillips, S.J.; Hastie, T.; Dudík, M.; Chee, Y.E.; Yates, C.J. A statistical explanation of MaxEnt for ecologists. Divers. Distrib. 2011, 17, 43–57. [Google Scholar] [CrossRef]
- Fourcade, Y.; Engler, J.O.; Rödder, D.; Secondi, J. Mapping species distributions with MaxEnt using a geographically biased sample of presence data: A performance assessment of methods for correcting sampling bias. PLoS ONE 2014, 9, e97122. [Google Scholar] [CrossRef] [PubMed]
- Gollin, D. Conserving genetic resources for agriculture: Economic implications of emerging science. Food Secur. 2020, 12, 919–927. [Google Scholar] [CrossRef]
- REMIB (Red Mundial de Información sobre Biodiversidad). Comisión Nacional para el Conocimiento y Uso de la Biodiversidad. México. Available online: http://www.conabio.gob.mx/remib/ (accessed on 19 December 2020).
- TGRC (Tomato Genetics Resource Center). Available online: https://tgrc.ucdavis.edu/ (accessed on 19 December 2020).
- GBIF (Global Biodiversity Information Facility). Available online: https://www.gbif.org/ (accessed on 19 December 2020).
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Trabucco, A.; Zomer, R.J. Global high-resolution soil-water balance. Figshare Fileset 2019, 10, m9. [Google Scholar] [CrossRef]
- Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (CONABIO). Provincias Biogeográficas de Mexico; Escala 1:4 000 000; Comisión Nacional para el Conocimiento y Uso de la Biodiversidad: Ciudad de México, Mexico, 2016. [Google Scholar]
- SAS Institute. SAS/STAT User’s Guide; Software Version 9.4; Statistical Analysis System Institute: Cary, NC, USA, 2013. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Kassambara, A. Practical Guide to Principal Component Methods in R, 1st ed.; CreateSpace Independent Publishing Platform: Scotts Valley, Ca, USA, 2017; p. 29. Available online: https://payhip.com/b/shrk (accessed on 24 February 2022).
- Yilan, L.; Rutong, Z. Clustertend: Check the Clustering Tendency, R Package Version 1.4. 2015. Available online: https://CRAN.R-project.org/package=clustertend (accessed on 24 February 2022).
- Brock, G.; Pihur, V.; Datta, S.; Datta, S. clValid: An R Package for Cluster Validation. J. Stat. Softw. 2008, 25, 1–22. [Google Scholar] [CrossRef]
- Charrad, M.; Ghazzali, N.; Boiteau, V.; Niknafs, A. NbClust: An R package for determining the relevant number of clusters in a data set. J. Stat. Softw. 2014, 61, 1–36. [Google Scholar] [CrossRef]
- Steiner, J.J.; Greene, S.L. Proposed ecological descriptors and their utility for plant germplasm collections. Crop Sci. 1996, 36, 439–451. [Google Scholar] [CrossRef]
- Ruiz-Corral, J.A.; Durán-Puga, N.; Sánchez-González, J.J.; Ron-Parra, J.; González-Eguiarte, D.R.; Holland, J.B.; Medina-García, G. Climatic adaptation and ecological descriptors of 42 Mexican Maize (Zea mays L.) races. Crop Sci. 2008, 48, 1502–1512. [Google Scholar] [CrossRef]
- Cerda-Hurtado, I.M.; Mayek-Pérez, N.; Hernández-Delgado, S.; Muruaga-Martínez, J.S.; Reyes-Lara, M.A.; Reyes-Valdés, M.H.; González-Prieto, J.M. Climatic adaptation and ecological descriptors of wild beans from Mexico. Ecol. Evol. 2018, 8, 6492–6504. [Google Scholar] [CrossRef] [PubMed]
- ArcGis, version 10.3; Redlands, Ca. Environmental Systems Research Institute, Inc.: Redlands, CA, USA, 2010.
- Hijmans, J.R.; Spooner, D.M. Geographic distribution of wild potato species. Am. J. Bot. 2001, 88, 2101–2112. [Google Scholar] [CrossRef] [PubMed]
- Spooner, D.M.; Gavrilenko, T.; Jansky, S.H.; Ovchinnikova, A.; Krylova, E.; Knapp, S. Ecogeography of ploidy variation in cultivated potato (Solanum sect. Petota). Am. J. Bot. 2010, 97, 2049–2060. [Google Scholar] [CrossRef] [PubMed]
- Getis, A.; Ord, J.K. The analysis of spatial association by use of distance statistics. Geogr. Anal. 1992, 24, 189–206. [Google Scholar] [CrossRef]
- Avila, C.A.; Villavicencio, G.R.; Ruiz Corral, J.A. Distribución potencial de Pinus herrerae Martínez en el Occidente del estado de Jalisco. Rev. Mex. Cien. For. 2014, 5, 92–108. [Google Scholar]
- Hanley, J.A.; McNeil, B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982, 143, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Soberón, J.; Peterson, T. Ecological niche shifts and environmental space anisotropy: A cautionary note. Rev. Mex. Biodivers. 2011, 82, 1348–1355. [Google Scholar]

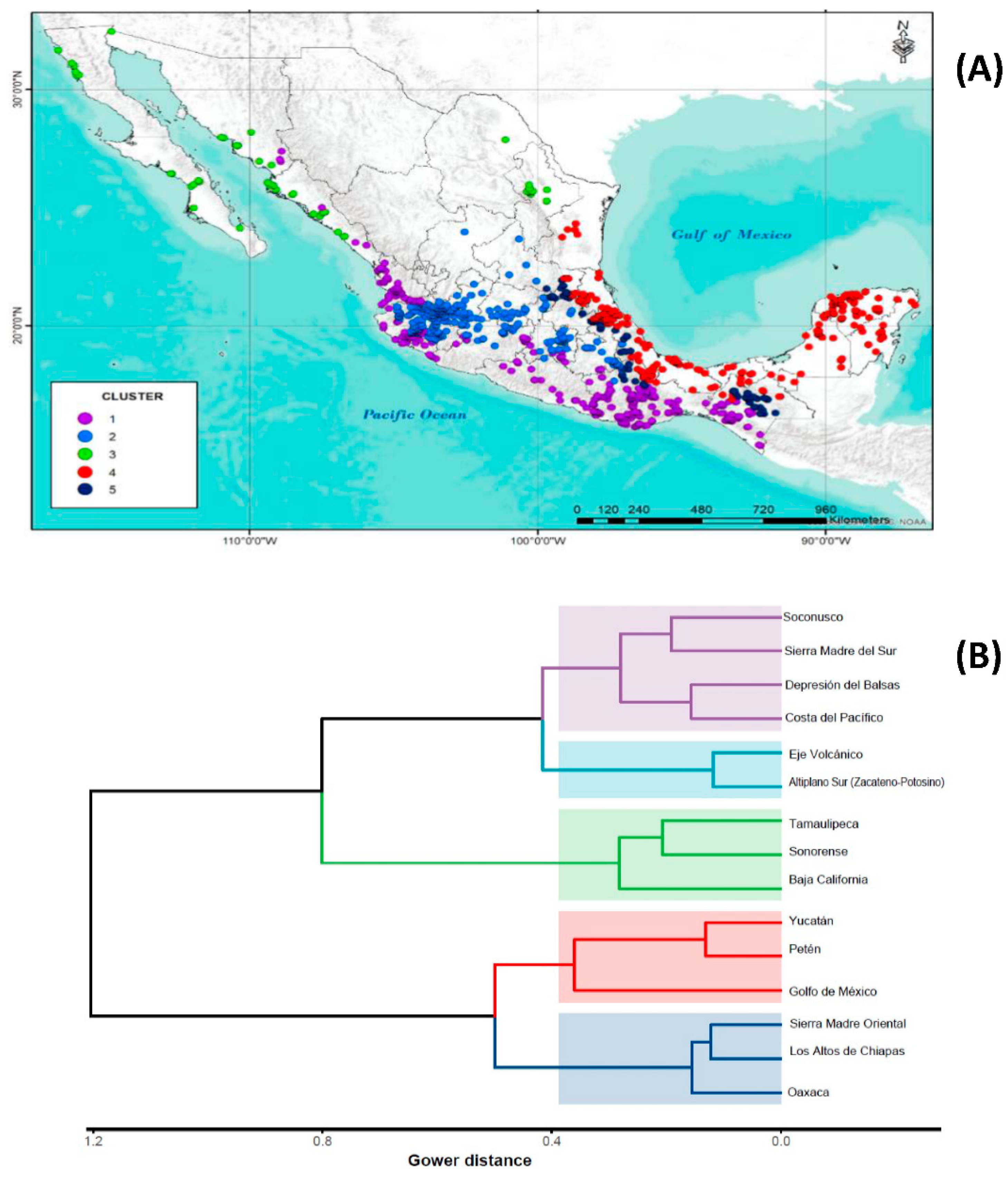
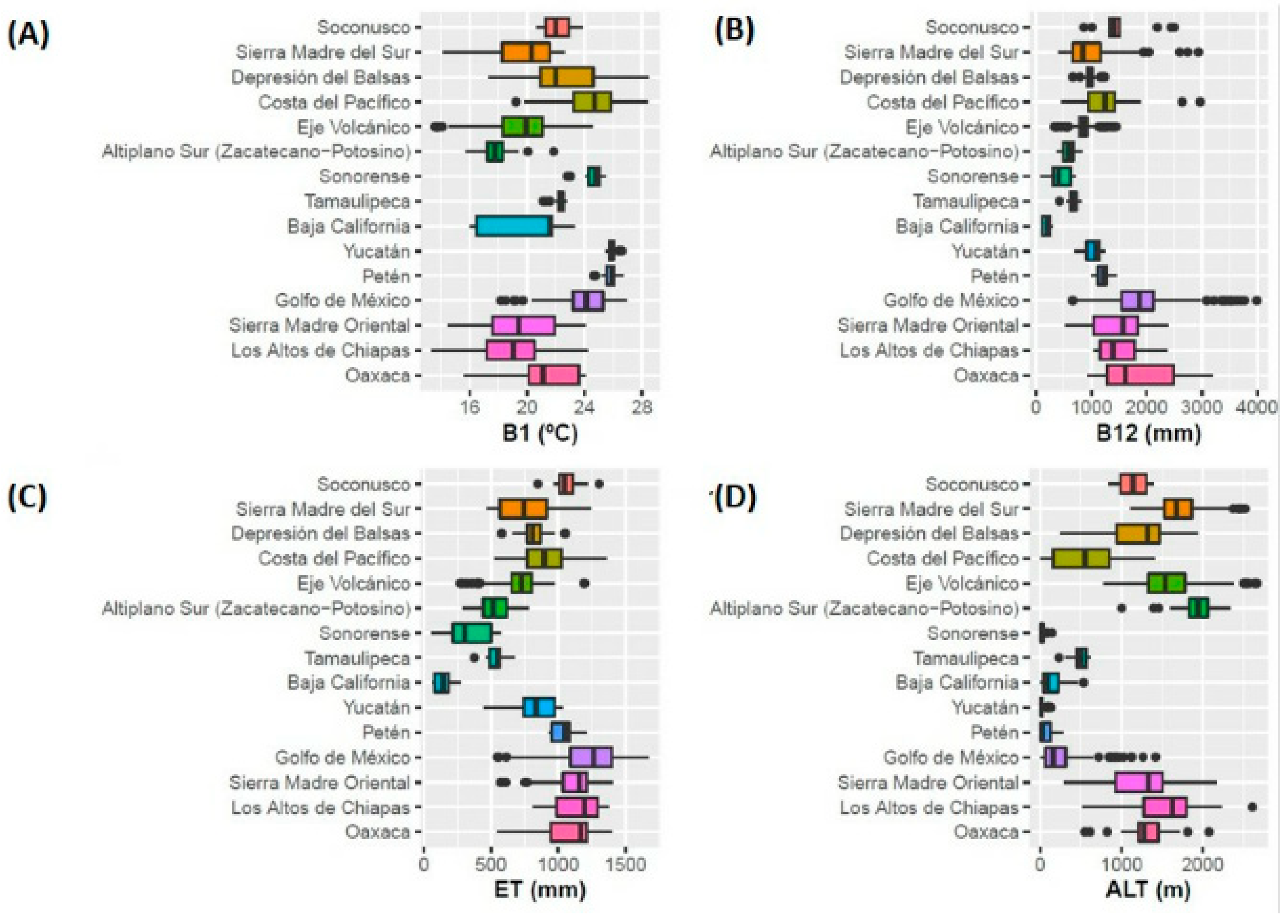

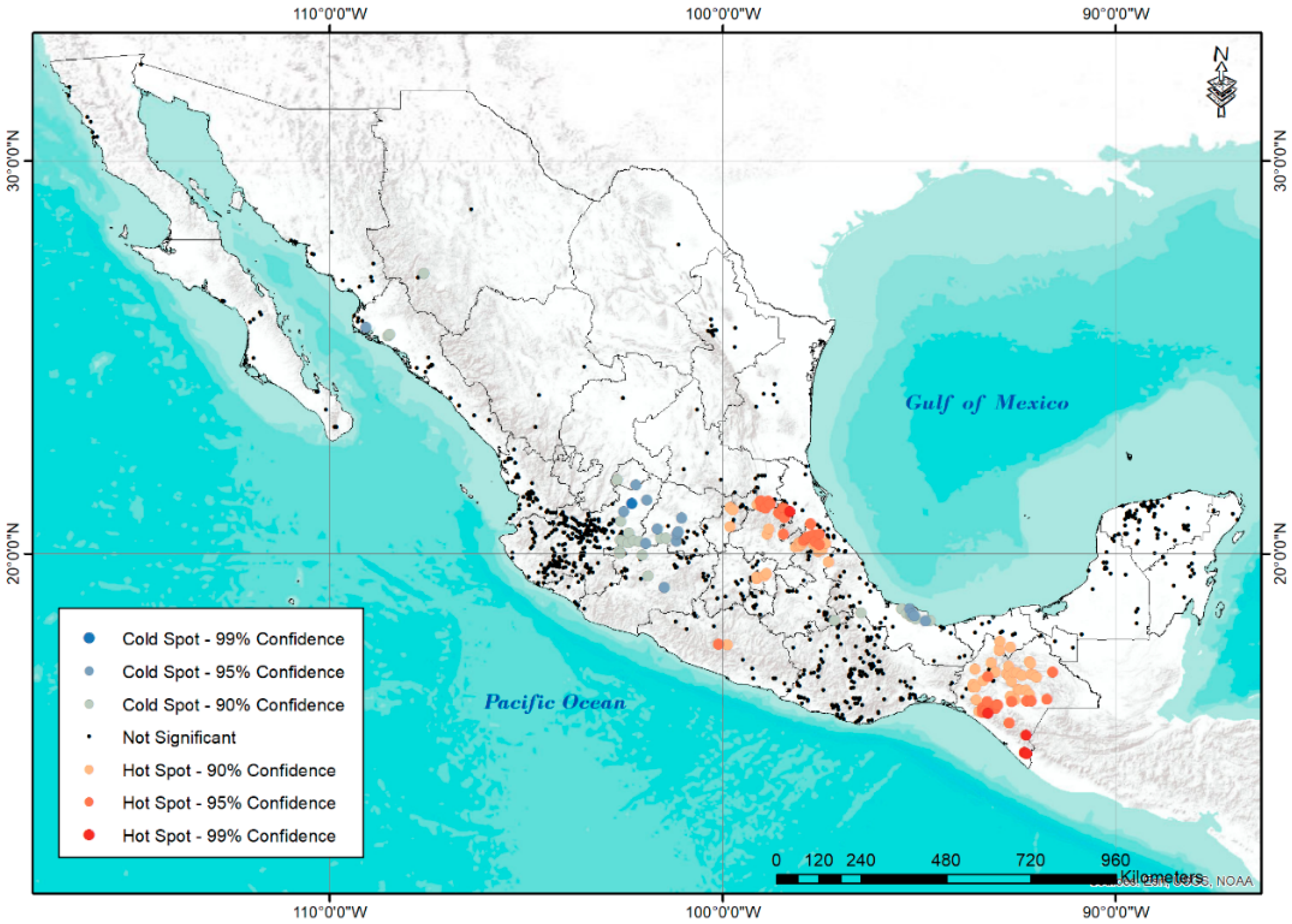
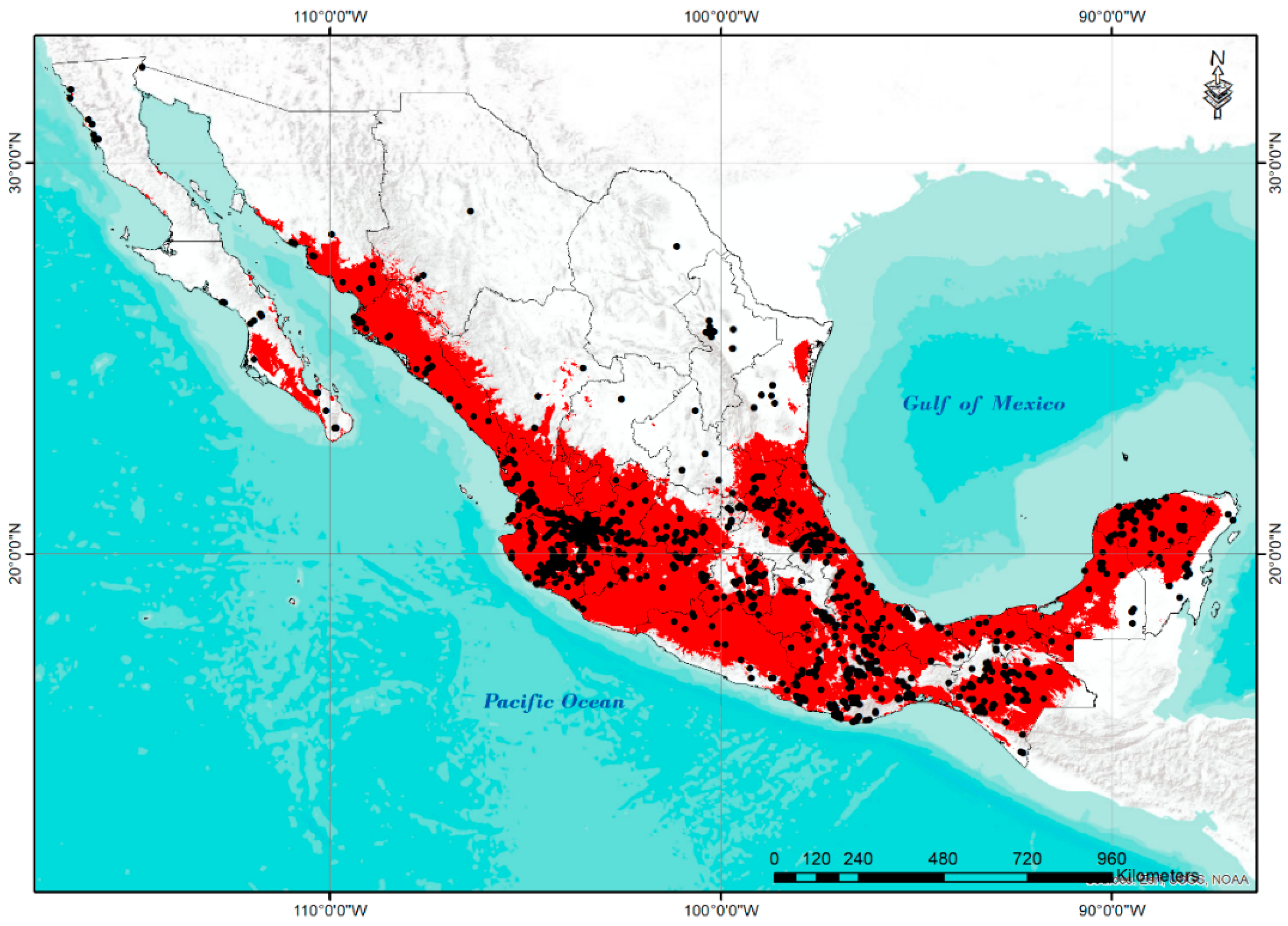

| CLUSTER | PHYSIOGRAPHIC PROVINCE | PC1 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B2 | B7 | B12 | B14 | B15 | ||||||||||||
| Range | Med | CV | Range | Med | CV | Range | Med | CV | Range | Med | CV | Range | Med | CV | ||
| 1 | Soconusco | 11.5–13.3 | 12.3 | 3.14 | 16.0–20.1 | 17.1 | 3.22 | 865–2497 | 1407 | 6.04 | 0–17 | 2 | 250 | 65.5–108.8 | 105.3 | 5.2 |
| Sierra Madre del Sur | 9.4–17.8 | 13.6 | 14.8 | 14.4–24.6 | 19.6 | 13 | 414–2938 | 849 | 29.93 | 01–64.0 | 5 | 65 | 76.1–105.3 | 92.5 | 2.4 | |
| Depresión del Balsas | 13.8–17.3 | 15.4 | 4.41 | 19.8–25.9 | 23.2 | 4.71 | 665–1240 | 969 | 3.99 | 1.0–9.0 | 3 | 33.33 | 93.8–119.5 | 108.4 | 1.07 | |
| Costa del Pacífico | 9.6–19.1 | 13.8 | 10.66 | 12.8–31.9 | 20.1 | 14.17 | 466–2968 | 1248 | 18.96 | 0–15.0 | 2 | 50 | 82.4–133.9 | 112.7 | 5.81 | |
| 2 | Eje Volcánico | 11.9–19.5 | 16.9 | 5.19 | 18.6–31.0 | 26.1 | 5.94 | 327–1449 | 844 | 8.12 | 0–36.0 | 4 | 37.5 | 73.6–124.7 | 110.4 | 5.87 |
| Altiplano Sur | 13.4–18.7 | 16.5 | 6.12 | 21.2–29.4 | 25.8 | 8.43 | 367–835 | 592 | 15.41 | 2.0–11.0 | 5 | 20 | 67.8–118.3 | 94.4 | 15.08 | |
| 3 | Sonorense | 12.0–17.9 | 14.8 | 4.16 | 23.7–35.4 | 25.7 | 6.6 | 78–722 | 402 | 39.86 | 0–2.0 | 1 | 37.5 | 49.7–122.4 | 115.2 | 4.4 |
| Tamaulipeca | 13.5–15.1 | 14.3 | 0.76 | 27.1–31.8 | 27.8 | 2.43 | 424–815 | 696 | 7.51 | 11.0–21.0 | 18 | 5.56 | 58.6–85.0 | 80.7 | 5.47 | |
| Baja California | 11.6–18.5 | 15.9 | 8.74 | 19.3–28.6 | 26.8 | 9.51 | 102–294 | 205 | 31.52 | 0–1.0 | 0 | 0 | 70.5–124.8 | 89.9 | 7.53 | |
| 4 | Yucatán | 10.7–14.9 | 13.4 | 4.34 | 16.1–22.0 | 19.8 | 3.47 | 694–1253 | 1075 | 10.57 | 16.0–39.0 | 23 | 8.15 | 59.1–87.6 | 67.6 | 4.78 |
| Petén | 9.7–14.5 | 12.4 | 6.49 | 14.5–20.6 | 17.5 | 7.43 | 1005–1449 | 1230 | 6.12 | 18–43 | 37 | 19.57 | 47.6–84.5 | 56.1 | 5.4 | |
| Golfo de México | 7.9–14.1 | 10.5 | 6.98 | 15.2–27.7 | 18.3 | 5.74 | 657–3995 | 1867 | 15.63 | 8–141 | 49.5 | 26.01 | 42.4–111.8 | 66 | 15.76 | |
| 5 | Sierra Madre Oriental | 9.8–14.6 | 12 | 3.92 | 17.3–25.2 | 20.5 | 5.37 | 525–2404 | 1573 | 24.95 | 8.0–56.0 | 37 | 25.68 | 67.4–90.8 | 78.9 | 5.59 |
| Los Altos de Chiapas | 10.7–14.1 | 12.5 | 6.07 | 16.8–19.7 | 18.7 | 2.81 | 1045–2376 | 1390 | 22.72 | 3.0–73.0 | 22 | 55.68 | 55.0–93.7 | 82.4 | 9.82 | |
| Oaxaca | 11.0–16.7 | 13.1 | 9.5 | 17.6–24.8 | 20.5 | 7.32 | 923–3192 | 1616 | 36.88 | 11.0–72.0 | 34 | 57.35 | 70.1–92.4 | 79.3 | 9.12 | |
| CLUSTER | PHYSIOGRAPHIC PROVINCE | PC1 | PC2 | PC3 | ||||||||||||
| ET | B3 | B4 | B1 | ALT | ||||||||||||
| Range | Med | CV | Range | Med | CV | Range | Med | CV | Range | Med | CV | Range | Med | CV | ||
| 1 | Soconusco | 3–121 | 13 | 134.6 | 66.3–75.3 | 72.1 | 2.38 | 93.6–173.8 | 134 | 6.63 | 20.7–23.9 | 21.9 | 3.62 | 839–1396 | 1141 | 14.02 |
| Sierra Madre del Sur | 12–310 | 26 | 52.4 | 61.2–79.4 | 71.5 | 2.98 | 83.9–197.8 | 143 | 12.6 | 14.2–22.6 | 20.3 | 8.09 | 1108–2523 | 1676 | 10.02 | |
| Depresión del Balsas | 18–21 | 29 | 12.07 | 64.4–69.9 | 67 | 1.76 | 135.9–213.8 | 183 | 5.01 | 17.3–28.4 | 21.9 | 8.28 | 243–1940 | 1320 | 19.95 | |
| Costa del Pacífico | 1–99 | 37 | 52.7 | 55.1–80.0 | 68.5 | 3.22 | 61.4–486.2 | 175 | 34.08 | 19.2–28.4 | 24.6 | 5.24 | 5–1406 | 550 | 63.51 | |
| 2 | Eje Volcánico | 9–122 | 34 | 20.59 | 59.8–73.0 | 64.4 | 2.57 | 124.3–309.0 | 240 | 9.62 | 13.6–24.5 | 19.9 | 6.99 | 773–2667 | 1539 | 14.62 |
| Altiplano Sur | 21–46 | 27 | 10.65 | 59.6–70.1 | 63.5 | 1.75 | 173.6–349.7 | 272 | 8.32 | 15.7–21.9 | 17.7 | 3 | 1002–2344 | 1939 | 5.75 | |
| 3 | Sonorense | 28–64 | 52 | 6.73 | 46.6–61.9 | 55.2 | 6.37 | 368.6–742.4 | 487 | 14.79 | 22.8–25.5 | 24.7 | 1.49 | 1–139 | 15 | 86.67 |
| Tamaulipeca | 43–73 | 60 | 4.17 | 46.3–53.2 | 50.6 | 1.44 | 485.3–644.1 | 504 | 3.72 | 21.1–22.7 | 22.3 | 0.76 | 229–613 | 504 | 12.05 | |
| Baja California | 32–155 | 54 | 63.43 | 56.8–65.9 | 60.8 | 2.72 | 279.2–499.4 | 368 | 13.44 | 16.0–23.3 | 21.5 | 12.17 | 4–532 | 86 | 108.2 | |
| 4 | Yucatán | 78–150 | 100 | 7.71 | 63.9–71.5 | 67.5 | 1.58 | 181.8–218.1 | 196 | 2.85 | 25.4–26.6 | 25.8 | 0.52 | 0–124 | 15 | 64.6 |
| Petén | 89–203 | 147 | 10.63 | 63.8–74.1 | 70.3 | 1.91 | 154.3–215.4 | 185 | 7.01 | 24.6–26.7 | 25.9 | 0.96 | 6–279 | 22 | 243.2 | |
| Golfo de México | 31–836 | 209 | 23.33 | 49.5–68.0 | 56.1 | 4.47 | 176.9–509.0 | 286 | 16.05 | 18.2–26.9 | 24.1 | 4.34 | 0–1419 | 156 | 79.63 | |
| 5 | Sierra Madre Oriental | 34–185 | 122 | 24.18 | 53.2–63.0 | 59.4 | 3.21 | 183.3–368.4 | 237 | 20.54 | 14.5–24.1 | 19.4 | 11.09 | 299–2166 | 1330 | 21.71 |
| Los Altos de Chiapas | 20–328 | 87 | 67.82 | 62.5–71.9 | 67.3 | 3.84 | 133.6–203.7 | 169 | 7.79 | 13.4–24.2 | 19.1 | 8.83 | 518–2613 | 1629 | 16.04 | |
| Oaxaca | 45–290 | 128 | 60.16 | 60.3–68.0 | 64.8 | 3.26 | 191.0–238.9 | 219 | 2.82 | 15.6–24.1 | 21.1 | 8.38 | 549–2078 | 1275 | 9.33 | |
| Climate Type |
|---|
| Af (tropical and rainforest), Am (tropical and monsoon), Aw (tropical and savannah), BWh (arid, desert, and hot), BWk (arid, desert, and cold), BSh (arid, steppe, and hot), BSk (arid, steppe, and cold), Csa (temperate, dry summer, and hot summer), Csb (temperate, dry summer, and warm summer), Csc (temperate and dry and cold summer), Cwa (temperate, dry winter, and hot summer), Cwb (temperate, dry winter, and warm summer), Cwc (temperate, dry winter, and cold summer), Cfa (temperate, no dry season, and hot summer), Cfb (temperate, no dry season, and warm summer), Cfc (temperate, no dry season, and cold summer), Dsa (cold, dry summer, and hot summer), Dsb (cold, dry summer, and warm summer), Dsc (cold, dry summer, and cold summer), Dsd (cold, dry summer, and very cold winter), Dwa (cold, dry winter, and hot summer), Dwb (cold, dry winter, and warm summer), Dwc (cold, dry winter, and cold summer), Dwd (cold, dry winter, and very cold winter), Dfa (cold, no dry season, and hot summer), Dfb (cold, no dry season, and warm summer), Dfc (cold, no dry season, and cold summer), Dfd (cold, no dry season, and very cold winter), ET (polar and tundra), and EF (polar and frost). |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramírez-Ojeda, G.; Rodríguez-Pérez, J.E.; Rodríguez-Guzmán, E.; Sahagún-Castellanos, J.; Chávez-Servia, J.L.; Peralta, I.E.; Barrera-Guzmán, L.Á. Distribution and Climatic Adaptation of Wild Tomato (Solanum lycopersicum L.) Populations in Mexico. Plants 2022, 11, 2007. https://doi.org/10.3390/plants11152007
Ramírez-Ojeda G, Rodríguez-Pérez JE, Rodríguez-Guzmán E, Sahagún-Castellanos J, Chávez-Servia JL, Peralta IE, Barrera-Guzmán LÁ. Distribution and Climatic Adaptation of Wild Tomato (Solanum lycopersicum L.) Populations in Mexico. Plants. 2022; 11(15):2007. https://doi.org/10.3390/plants11152007
Chicago/Turabian StyleRamírez-Ojeda, Gabriela, Juan Enrique Rodríguez-Pérez, Eduardo Rodríguez-Guzmán, Jaime Sahagún-Castellanos, José Luis Chávez-Servia, Iris E. Peralta, and Luis Ángel Barrera-Guzmán. 2022. "Distribution and Climatic Adaptation of Wild Tomato (Solanum lycopersicum L.) Populations in Mexico" Plants 11, no. 15: 2007. https://doi.org/10.3390/plants11152007
APA StyleRamírez-Ojeda, G., Rodríguez-Pérez, J. E., Rodríguez-Guzmán, E., Sahagún-Castellanos, J., Chávez-Servia, J. L., Peralta, I. E., & Barrera-Guzmán, L. Á. (2022). Distribution and Climatic Adaptation of Wild Tomato (Solanum lycopersicum L.) Populations in Mexico. Plants, 11(15), 2007. https://doi.org/10.3390/plants11152007







