Cytokinin Deficiency Alters Leaf Proteome and Metabolome during Effector-Triggered Immunity in Arabidopsis thaliana Plants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material, Growth Conditions, and flg22 Treatment
2.2. Histochemical Assays
2.3. Chlorophyll Fluorescence Imaging
2.4. Plant Hormone Analysis
2.5. LC-MS-Based Proteomics Assays and Bioinformatic Analysis
2.6. GC-MS-Based Metabolomics Analyses and Multivariate Statistical Analysis
2.7. Transcriptomic Data Mining
2.8. Statistical Analysis
3. Results
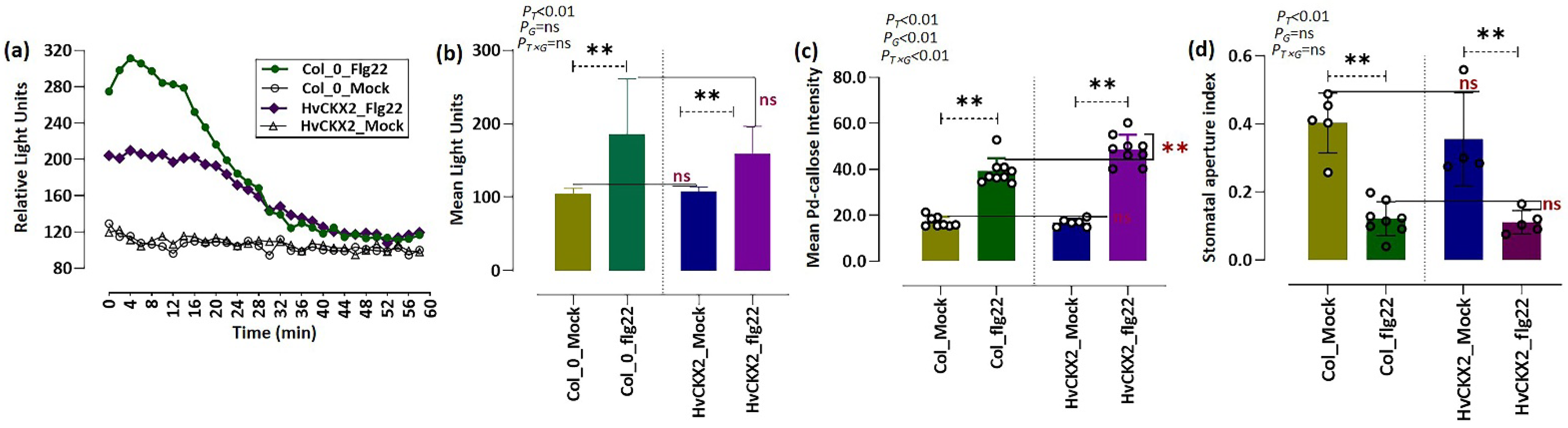
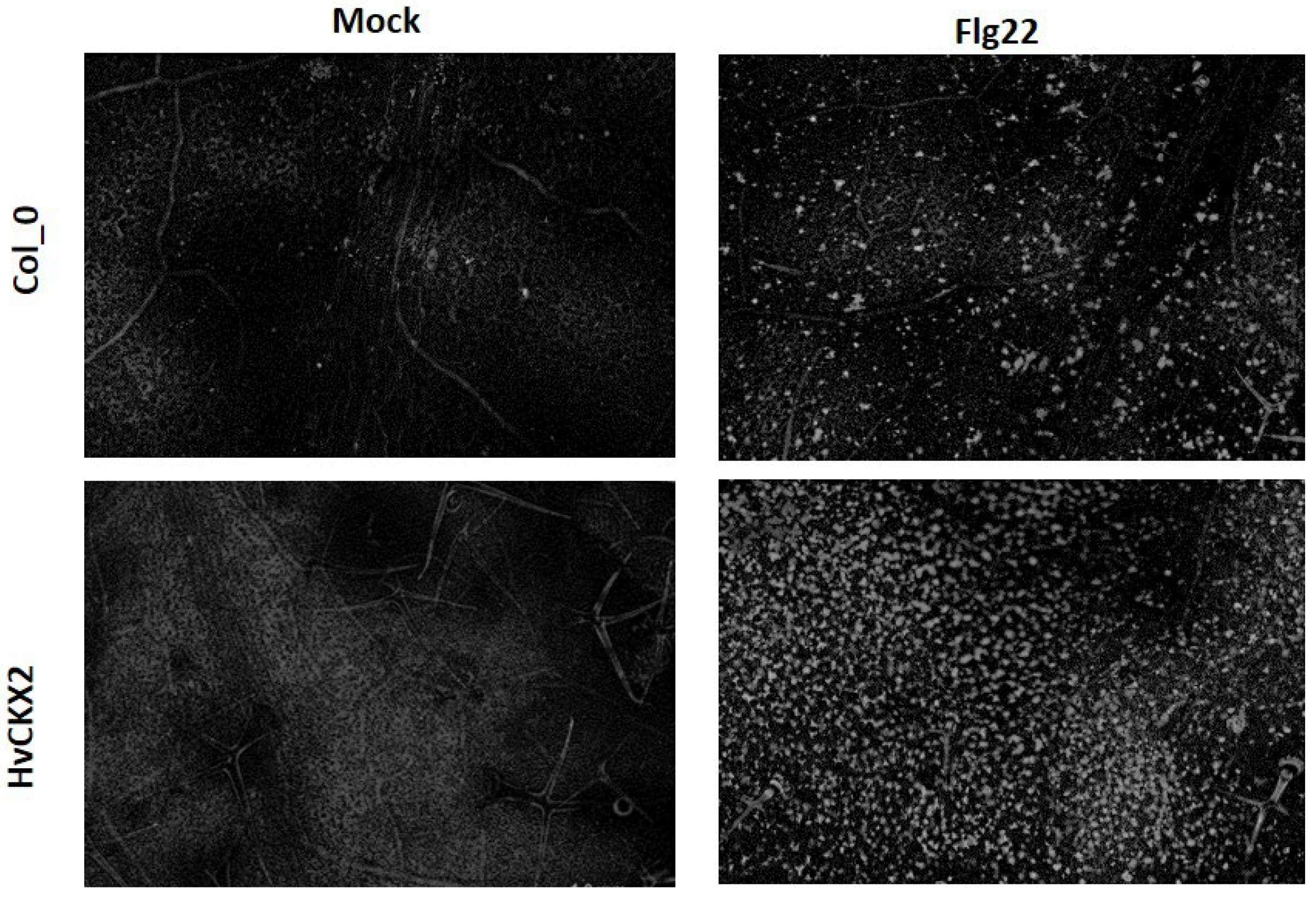
3.1. CK Deficiency Altered flg22-Triggered Early Defence Events
3.2. CK Deficiency Is Involved in the Regulation of Plant Photosynthesis upon Flg22 Peptide Application
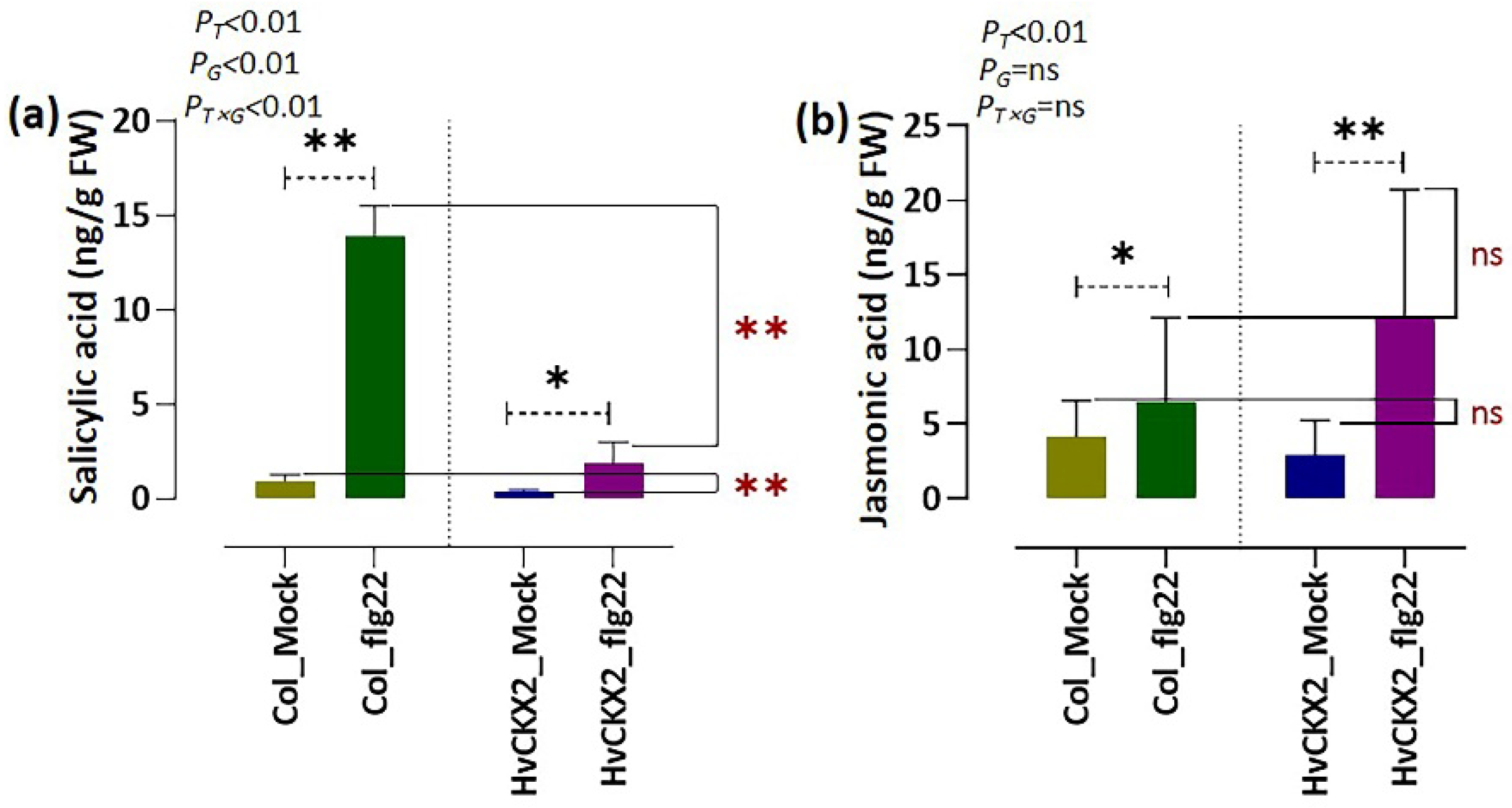
3.3. Flg22-Triggered SA Synthesis Was Inhibited by CK Deficiency, While JA Synthesis Seems to Be CK-Independent
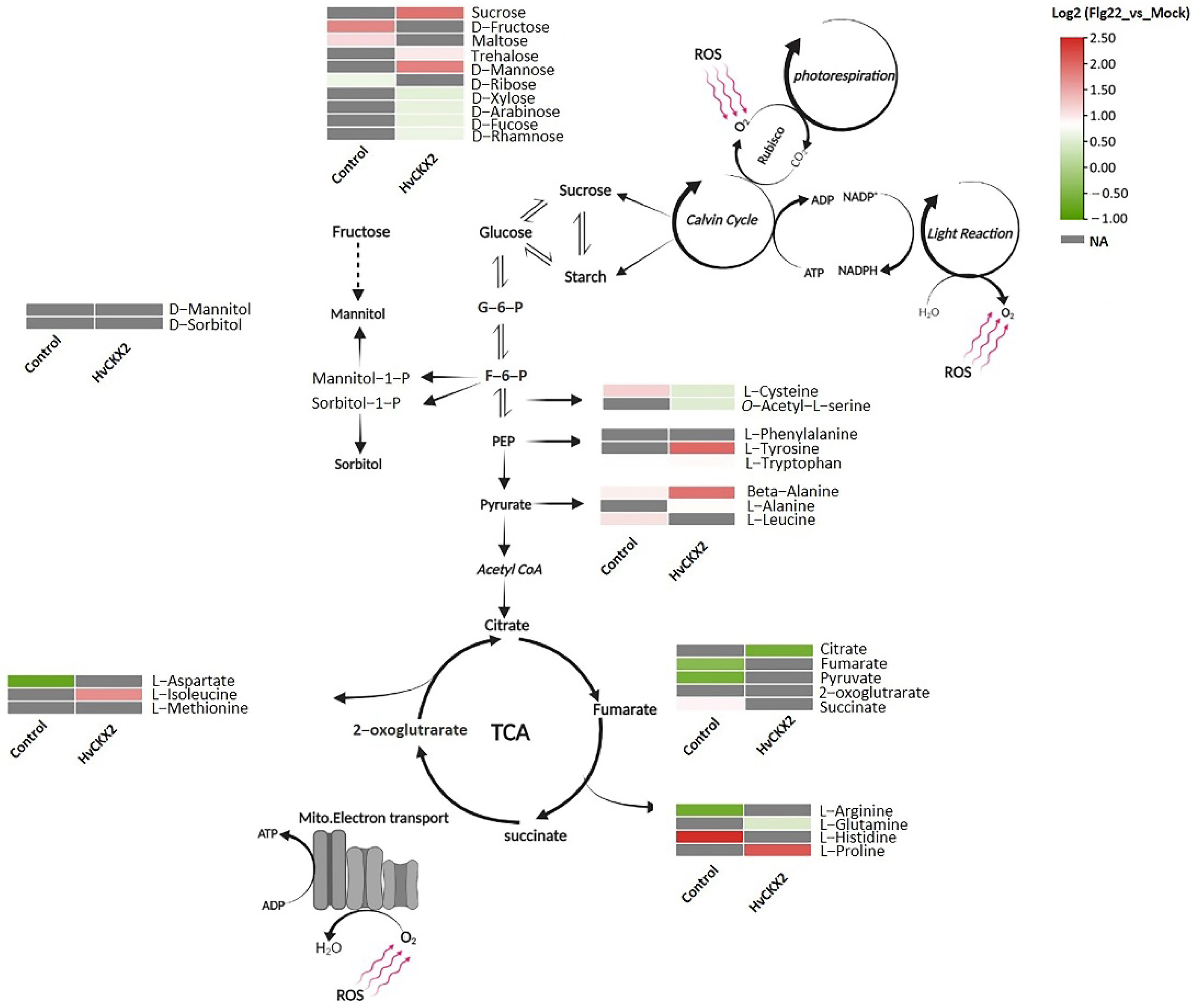
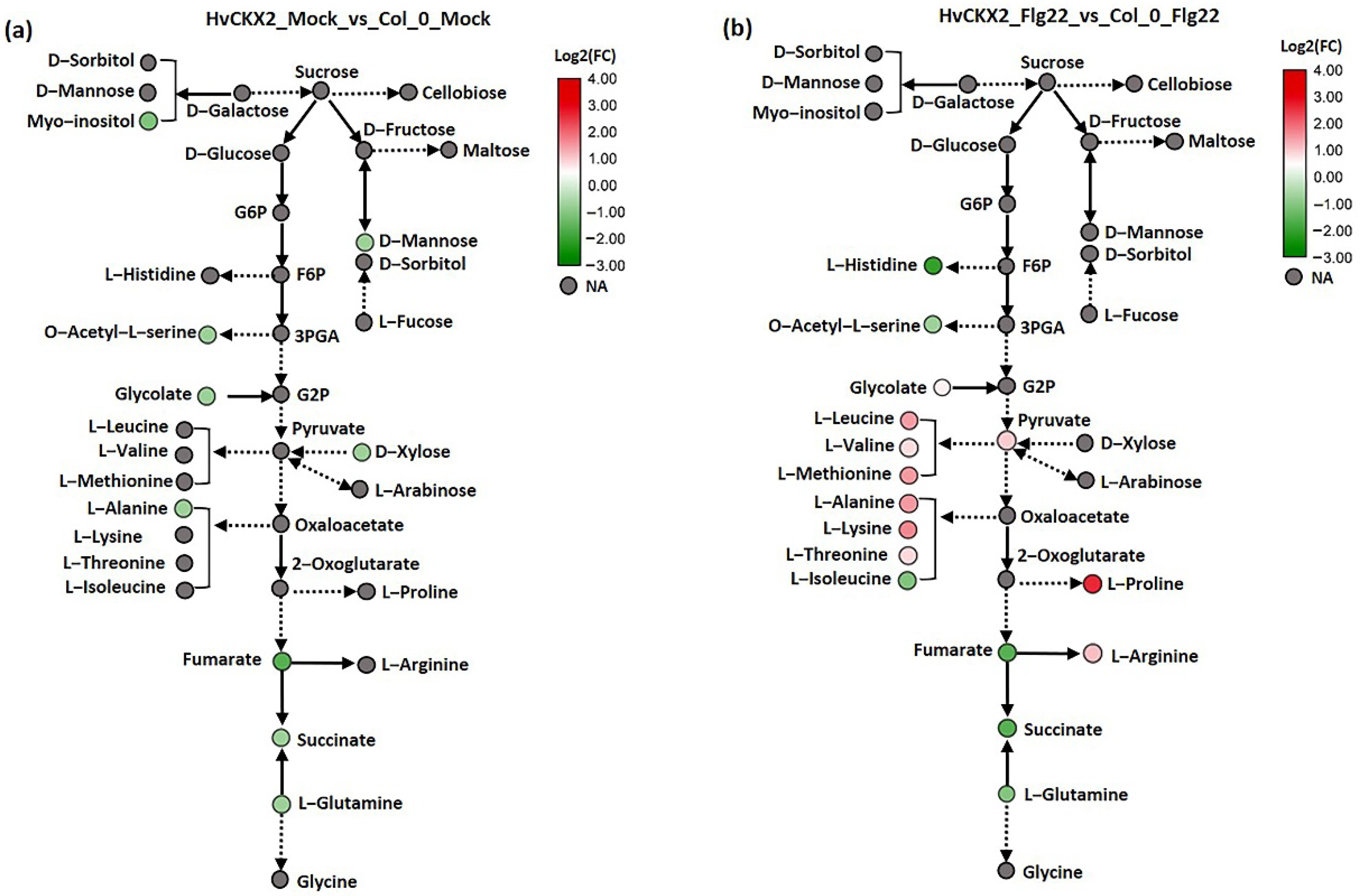
3.4. CK Deficiency Specifically Enhanced Amino Acid Biosynthesis and Increased the Sucrose and D-Mannose Concentration in Arabidopsis Leaves upon Flg22 Peptide Application
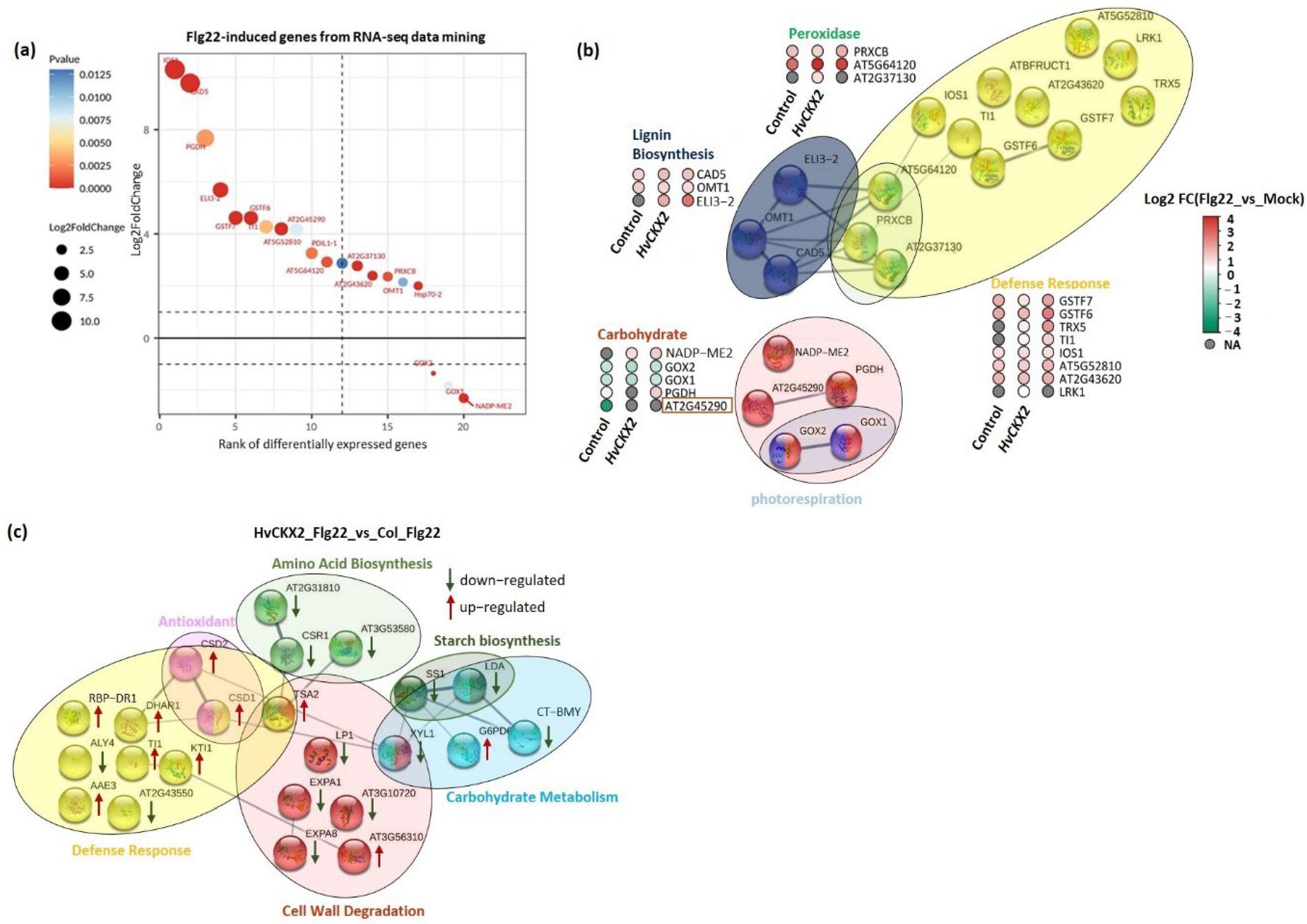
3.5. CK Deficiency Altered the Abundance of Proteins Related to Defence Response, Amino Acid Biosynthesis, and Cell Wall Integrity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Choi, J.; Choi, D.; Lee, S.; Ryu, C.-M.; Hwang, I. Cytokinins and plant immunity: Old foes or new friends? Trends Plant Sci. 2011, 16, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Li, S.M.; Zheng, H.X.; Zhang, X.S.; Sui, N. Cytokinins as central regulators during plant growth and stress response. Plant Cell Rep. 2021, 40, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Pizarro, L.; Leibman-Markus, M.; Marash, I.; Bar, M. Cytokinin response induces immunity and fungal pathogen resistance, and modulates trafficking of the PRR LeEIX2 in tomato. Mol. Plant Pathol. 2020, 21, 1287–1306. [Google Scholar] [CrossRef] [PubMed]
- Amasino, R.J.P.P. 1955: Kinetin arrives. The 50th anniversary of a new plant hormone. Plant Physiol. 2005, 138, 1177–1184. [Google Scholar] [CrossRef] [PubMed]
- Schmülling, T.; Werner, T.; Riefler, M.; Krupková, E.; Manns, I.B.Y. Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J. Plant Res. 2003, 116, 241–252. [Google Scholar] [CrossRef]
- Trifunović, M.; Cingel, A.; Simonović, A.; Jevremović, S.; Petrić, M.; Dragićević, I.Č.; Motyka, V.; Dobrev, P.I.; Zahajská, L.; Subotić, A. Overexpression of Arabidopsis cytokinin oxidase/dehydrogenase genes AtCKX1 and AtCKX2 in transgenic Centaurium erythraea Rafn. Plant Cell Tissue Organ Cult. 2013, 115, 139–150. [Google Scholar] [CrossRef]
- Černý, M.; Kuklová, A.; Hoehenwarter, W.; Fragner, L.; Novák, O.; Rotková, G.; Jedelský, P.L.; Žáková, K.; Šmehilová, M.; Strnad, M.; et al. Proteome and metabolome profiling of cytokinin action in Arabidopsis identifying both distinct and similar responses to cytokinin down- and up-regulation. J. Exp. Bot. 2013, 64, 4193–4206. [Google Scholar] [CrossRef]
- Galuszka, P.; Frébortová, J.; Werner, T.; Yamada, M.; Strnad, M.; Schmülling, T.; Frébort, I. Cytokinin oxidase/dehydrogenase genes in barley and wheat. Eur. J. Biochem. 2004, 271, 3990–4002. [Google Scholar] [CrossRef]
- Skalák, J.; Černý, M.; Jedelský, P.; Dobrá, J.; Ge, E.; Novák, J.; Hronková, M.; Dobrev, P.; Vanková, R.; Brzobohatý, B. Stimulation of ipt overexpression as a tool to elucidate the role of cytokinins in high temperature responses of Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 2861–2873. [Google Scholar] [CrossRef]
- Prerostova, S.; Dobrev, P.I.; Gaudinova, A.; Knirsch, V.; Körber, N.; Pieruschka, R.; Fiorani, F.; Brzobohatý, B.; černý, M.; Spichal, L.; et al. Cytokinins: Their Impact on Molecular and Growth Responses to Drought Stress and Recovery in Arabidopsis. Front. Plant Sci. 2018, 9, 655. [Google Scholar] [CrossRef]
- Skalák, J.; Vercruyssen, L.; Claeys, H.; Hradilová, J.; Černý, M.; Novák, O.; Plačková, L.; Saiz-Fernández, I.; Skaláková, P.; Coppens, F.; et al. Multifaceted activity of cytokinin in leaf development shapes its size and structure in Arabidopsis. Plant J. 2019, 97, 805–824. [Google Scholar] [CrossRef] [PubMed]
- Cortleven, A.; Leuendorf, J.E.; Frank, M.; Pezzetta, D.; Bolt, S.; Schmülling, T. Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ. 2019, 42, 998–1018. [Google Scholar] [CrossRef] [PubMed]
- Argueso, C.T.; Ferreira, F.J.; Epple, P.; To, J.P.C.; Hutchison, C.E.; Schaller, G.E.; Dangl, J.L.; Kieber, J.J. Two-Component Elements Mediate Interactions between Cytokinin and Salicylic Acid in Plant Immunity. PLoS Genet. 2012, 8, e1002448. [Google Scholar] [CrossRef] [PubMed]
- Baillie, A.L.; Fleming, A.J. The developmental relationship between stomata and mesophyll airspace. New Phytol. 2020, 225, 1120–1126. [Google Scholar] [CrossRef]
- Gupta, R.; Leibman-Markus, M.; Pizarro, L.; Bar, M. Cytokinin induces bacterial pathogen resistance in tomato. Plant Pathol. 2021, 70, 318–325. [Google Scholar] [CrossRef]
- Großkinsky, D.K.; Naseem, M.; Abdelmohsen, U.R.; Plickert, N.; Engelke, T.; Griebel, T.; Zeier, J.; Novák, O.; Strnad, M.; Pfeifhofer, H.; et al. Cytokinins Mediate Resistance against Pseudomonas syringae in Tobacco through Increased Antimicrobial Phytoalexin Synthesis Independent of Salicylic Acid Signaling. Plant Physiol. 2011, 157, 815–830. [Google Scholar] [CrossRef]
- Novák, J.; Pavlů, J.; Novák, O.; Nožková-Hlaváčková, V.; Špundová, M.; Hlavinka, J.; Koukalová, Š.; Skalák, J.; Černý, M.; Brzobohatý, B. High cytokinin levels induce a hypersensitive-like response in tobacco. Ann. Bot. 2013, 112, 41–55. [Google Scholar] [CrossRef]
- Barna, B.; Smigocki, A.C.; Baker, J.C. Transgenic Production of Cytokinin Suppresses Bacterially Induced Hypersensitive Response Symptoms and Increases Antioxidative Enzyme Levels in Nicotiana spp. Phytopathology 2008, 98, 1242–1247. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, D.; Gao, D.; Zhao, W.; Du, H.; Qiu, Z.; Huang, J.; Wen, P.; Wang, Y.; Li, Q.; et al. Cytokinin Confers Brown Planthopper Resistance by Elevating Jasmonic Acid Pathway in Rice. Int. J. Mol. Sci. 2022, 23, 5946. [Google Scholar] [CrossRef]
- Choi, J.; Huh, S.U.; Kojima, M.; Sakakibara, H.; Paek, K.-H.; Hwang, I. The Cytokinin-Activated Transcription Factor ARR2 Promotes Plant Immunity via TGA3/NPR1-Dependent Salicylic Acid Signaling in Arabidopsis. Dev. Cell 2010, 19, 284–295. [Google Scholar] [CrossRef]
- Arnaud, D.; Lee, S.; Takebayashi, Y.; Choi, D.; Choi, J.; Sakakibara, H.; Hwang, I. Cytokinin-Mediated Regulation of Reactive Oxygen Species Homeostasis Modulates Stomatal Immunity in Arabidopsis. Plant Cell 2017, 29, 543. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Elkabetz, D.; Leibman-Markus, M.; Sayas, T.; Schneider, A.; Jami, E.; Kleiman, M.; Bar, M. Cytokinin drives assembly of the phyllosphere microbiome and promotes disease resistance through structural and chemical cues. ISME J. 2022, 16, 122–137. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Gómez, L.; Boller, T. FLS2: An LRR Receptor–like Kinase Involved in the Perception of the Bacterial Elicitor Flagellin in Arabidopsis. Mol. Cell 2000, 5, 1003–1011. [Google Scholar] [CrossRef]
- Yi, S.Y.; Shirasu, K.; Moon, J.S.; Lee, S.-G.; Kwon, S.-Y. The Activated SA and JA Signaling Pathways Have an Influence on flg22-Triggered Oxidative Burst and Callose Deposition. PLoS ONE 2014, 9, e88951. [Google Scholar] [CrossRef] [PubMed]
- Pizarro, L.; Munoz, D.; Marash, I.; Gupta, R.; Anand, G.; Leibman-Markus, M.; Bar, M. Cytokinin Modulates Cellular Trafficking and the Cytoskeleton, Enhancing Defense Responses. Cells 2021, 10, 1634. [Google Scholar] [CrossRef]
- Gigli-Bisceglia, N.; Engelsdorf, T.; Strnad, M.; Vaahtera, L.; Khan, G.A.; Yamoune, A.; Alipanah, L.; Novák, O.; Persson, S.; Hejatko, J.; et al. Cell wall integrity modulates Arabidopsis thaliana cell cycle gene expression in a cytokinin- and nitrate reductase-dependent manner. Development 2018, 145, dev166678. [Google Scholar] [CrossRef]
- Ha, S.; Vankova, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.-S.P. Cytokinins: Metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci. 2012, 17, 172–179. [Google Scholar] [CrossRef]
- Schippers, J.H.; Jing, H.-C.; Hille, J.; Dijkwel, P.P. Developmental and hormonal control of leaf senescence. In Senescence Process in Plants; Wiley-Blackwell: Hoboken, NJ, USA, 2007; Volume 26, pp. 145–170. [Google Scholar]
- Eisele, J.F.; Fäßler, F.; Bürgel, P.F.; Chaban, C. A Rapid and Simple Method for Microscopy-Based Stomata Analyses. PLoS ONE 2016, 11, e0164576. [Google Scholar] [CrossRef]
- Berka, M.; Luklová, M.; Dufková, H.; Berková, V.; Novák, J.; Saiz-Fernández, I.; Rashotte, A.M.; Brzobohatý, B.; Černý, M. Barley Root Proteome and Metabolome in Response to Cytokinin and Abiotic Stimuli. Front. Plant Sci. 2020, 11, 590337. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Mach, J.M.; Castillo, A.R.; Hoogstraten, R.; Greenberg, J.T. The Arabidopsis-accelerated cell death gene ACD encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc. Natl. Acad. Sci. USA 2001, 98, 771. [Google Scholar] [CrossRef] [PubMed]
- Zurbriggen, M.D.; Carrillo, N.; Hajirezaei, M.-R. ROS signaling in the hypersensitive response. Plant Signal. Behav. 2010, 5, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Espinoza, V.A.; López-Climent, M.F.; Casaretto, J.A.; Gómez-Cadenas, A. Water Stress Responses of Tomato Mutants Impaired in Hormone Biosynthesis Reveal Abscisic Acid, Jasmonic Acid and Salicylic Acid Interactions. Front. Plant Sci. 2015, 6, 997. [Google Scholar] [CrossRef]
- Brouwer, S.M.; Odilbekov, F.; Burra, D.D.; Lenman, M.; Hedley, P.E.; Grenville-Briggs, L.; Alexandersson, E.; Liljeroth, E.; Andreasson, E. Intact salicylic acid signalling is required for potato defence against the necrotrophic fungus Alternaria solani. Plant Mol. Biol. 2020, 104, 1–19. [Google Scholar] [CrossRef]
- Abreu, M.E.; Munné-Bosch, S. Salicylic acid deficiency in NahG transgenic lines and sid2 mutants increases seed yield in the annual plant Arabidopsis thaliana. J. Exp. Bot. 2009, 60, 1261–1271. [Google Scholar] [CrossRef] [PubMed]
- Howlader, P.; Bose, S.K.; Jia, X.; Zhang, C.; Wang, W.; Yin, H. Oligogalacturonides induce resistance in Arabidopsis thaliana by triggering salicylic acid and jasmonic acid pathways against Pst DC3000. Int. J. Biol. Macromol. 2020, 164, 4054–4064. [Google Scholar] [CrossRef] [PubMed]
- Dewdney, J.; Reuber, T.L.; Wildermuth, M.C.; Devoto, A.; Cui, J.; Stutius, L.M.; Drummond, E.P.; Ausubel, F.M. Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J. 2000, 24, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Redox Homeostasis and Antioxidant Signaling: A Metabolic Interface between Stress Perception and Physiological Responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar] [CrossRef]
- Bela, K.; Horváth, E.; Gallé, Á.; Szabados, L.; Tari, I.; Csiszár, J. Plant glutathione peroxidases: Emerging role of the antioxidant enzymes in plant development and stress responses. J. Plant Physiol. 2015, 176, 192–201. [Google Scholar] [CrossRef]
- Xu, N.; Chen, G.; Liu, H. Antioxidative Categorization of Twenty Amino Acids Based on Experimental Evaluation. Molecules 2017, 22, 2066. [Google Scholar] [CrossRef]
- Filiz, E.; Ozyigit, I.I.; Saracoglu, I.A.; Uras, M.E.; Sen, U.; Yalcin, B. Abiotic stress-induced regulation of antioxidant genes in different Arabidopsis ecotypes: Microarray data evaluation. Biotechnol. Biotechnol. Equip. 2019, 33, 128–143. [Google Scholar] [CrossRef]
- Hématy, K.; Cherk, C.; Somerville, S. Host–pathogen warfare at the plant cell wall. Curr. Opin. Plant Biol. 2009, 12, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Vojta, P.; Kokáš, F.; Husičková, A.; Grúz, J.; Bergougnoux, V.; Marchetti, C.F.; Jiskrová, E.; Ježilová, E.; Mik, V.; Ikeda, Y.; et al. Whole transcriptome analysis of transgenic barley with altered cytokinin homeostasis and increased tolerance to drought stress. New Biotechnol. 2016, 33, 676–691. [Google Scholar] [CrossRef]
- Sun, L.; Ren, H.; Liu, R.; Li, B.; Wu, T.; Sun, F.; Liu, H.; Wang, X.; Dong, H. An h-type thioredoxin functions in tobacco defense responses to two species of viruses and an abiotic oxidative stress. Mol. Plant Microbe Interact. 2010, 23, 1470–1485. [Google Scholar] [CrossRef]
- Luna, E.; Pastor, V.; Robert, J.; Flors, V.; Mauch-Mani, B.; Ton, J. Callose deposition: A multifaceted plant defense response. Mol. Plant Microbe Interact. 2011, 24, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Abou-Saleh, R.H.; Hernandez-Gomez, M.C.; Amsbury, S.; Paniagua, C.; Bourdon, M.; Miyashima, S.; Helariutta, Y.; Fuller, M.; Budtova, T.; Connell, S.D.; et al. Interactions between callose and cellulose revealed through the analysis of biopolymer mixtures. Nat. Commun. 2018, 9, 4538. [Google Scholar] [CrossRef]
- Schenke, D.; Böttcher, C.; Scheel, D. Crosstalk between abiotic ultraviolet-B stress and biotic (flg22) stress signalling in Arabidopsis prevents flavonol accumulation in favor of pathogen defence compound production. Plant Cell Environ. 2011, 34, 1849–1864. [Google Scholar] [CrossRef] [PubMed]
- Samalova, M.; Elsayad, K.; Melnikava, A.; Peaucelle, A.; Gahurova, E.; Gumulec, J.; Spyroglou, I.; Zemlyanskaya, E.V.; Ubogoeva, E.V.; Hejatko, J. Expansin-controlled cell wall stiffness regulates root growth in Arabidopsis. BioRxiv 2020, 170969. [Google Scholar] [CrossRef]
- Dumez, S.; Wattebled, F.; Dauvillee, D.; Delvalle, D.; Planchot, V.r.; Ball, S.G.; D’Hulst, C. Mutants of Arabidopsis Lacking Starch Branching Enzyme II Substitute Plastidial Starch Synthesis by Cytoplasmic Maltose Accumulation. Plant Cell 2006, 18, 2694–2709. [Google Scholar] [CrossRef]
- Tang, X.-J.; Peng, C.; Zhang, J.; Cai, Y.; You, X.-M.; Kong, F.; Yan, H.-G.; Wang, G.-X.; Wang, L.; Jin, J.; et al. ADP-glucose pyrophosphorylase large subunit 2 is essential for storage substance accumulation and subunit interactions in rice endosperm. Plant Sci. 2016, 249, 70–83. [Google Scholar] [CrossRef]
- Miyazawa, Y.; Sakai, A.; Miyagishima, S.-y.; Takano, H.; Kawano, S.; Kuroiwa, T.J.P.P. Auxin and cytokinin have opposite effects on amyloplast development and the expression of starch synthesis genes in cultured bright yellow-2 tobacco cells. Plant Physiol. 1999, 121, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Peterhansel, C.; Horst, I.; Niessen, M.; Blume, C.; Kebeish, R.; Kürkcüoglu, S.; Kreuzaler, F. Photorespiration. Arab. Book 2010, 8, e0130. [Google Scholar] [CrossRef] [PubMed]
- Dellero, Y.; Jossier, M.; Glab, N.; Oury, C.; Tcherkez, G.; Hodges, M. Decreased glycolate oxidase activity leads to altered carbon allocation and leaf senescence after a transfer from high CO2 to ambient air in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 3149–3163. [Google Scholar] [CrossRef]
- Ritchie, G.A.J.P.R. Chlorophyll Fluorescence: What Is It and. Proceedings RMRS 1998, 2, 34. [Google Scholar]
- Ruban, A.V. Nonphotochemical Chlorophyll Fluorescence Quenching: Mechanism and Effectiveness in Protecting Plants from Photodamage. Plant Physiol. 2016, 170, 1903–1916. [Google Scholar] [CrossRef] [PubMed]
- Göhre, V.; Jones, A.M.E.; Sklenář, J.; Robatzek, S.; Weber, A.P.M. Molecular Crosstalk Between PAMP-Triggered Immunity and Photosynthesis. Mol. Plant Microbe Interact. 2012, 25, 1083–1092. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, L.; Berka, M.; Černý, M.; Novák, J.; Luklová, M.; Brzobohatý, B.; Saiz-Fernández, I. Cytokinin Deficiency Alters Leaf Proteome and Metabolome during Effector-Triggered Immunity in Arabidopsis thaliana Plants. Plants 2022, 11, 2123. https://doi.org/10.3390/plants11162123
Pan L, Berka M, Černý M, Novák J, Luklová M, Brzobohatý B, Saiz-Fernández I. Cytokinin Deficiency Alters Leaf Proteome and Metabolome during Effector-Triggered Immunity in Arabidopsis thaliana Plants. Plants. 2022; 11(16):2123. https://doi.org/10.3390/plants11162123
Chicago/Turabian StylePan, Ling, Miroslav Berka, Martin Černý, Jan Novák, Markéta Luklová, Břetislav Brzobohatý, and Iñigo Saiz-Fernández. 2022. "Cytokinin Deficiency Alters Leaf Proteome and Metabolome during Effector-Triggered Immunity in Arabidopsis thaliana Plants" Plants 11, no. 16: 2123. https://doi.org/10.3390/plants11162123
APA StylePan, L., Berka, M., Černý, M., Novák, J., Luklová, M., Brzobohatý, B., & Saiz-Fernández, I. (2022). Cytokinin Deficiency Alters Leaf Proteome and Metabolome during Effector-Triggered Immunity in Arabidopsis thaliana Plants. Plants, 11(16), 2123. https://doi.org/10.3390/plants11162123





